INTRODUCTION
Transcription is the process of synthesizing an RNA strand using one template DNA strand of the genomic DNA using RNA polymerase II (RNA pol II) in eukaryotes[1,2]. It is one main rate-limiting processes in gene expression, and understanding transcriptional regulation is critical to elucidating the diversity in biological processes. Eukaryotic mRNA synthesis requires the assembly of the general transcription factor complex[3,4] prior to the recruitment of RNA pol II. Such mRNA transcriptional processes are the centerpiece of central dogma to produce functional protein molecules to orchestrate cellular signaling, including stem cell (SC) self-renewal, and a variety of transcription factors assist in fine-tuning of mRNA synthesis.
The complete human genome contains 19969 protein-coding genes[5]. Although the majority of genes are autosomal, there is sex divergence based on differential gene expression between males and females, as recently analyzed systematically[6]. Besides sex hormones, there are many genes involved in transcription on the X and Y chromosomes. One human X chromosome carries 829 protein-coding[7] (835 genes as of 2024; Supplementary Table 1) and the human Y chromosome contains 196 protein-coding genes[8] yet 107 out of 196 proteins have not been characterized. Although this number was published in 2009, still only 66 protein-coding genes on the Y chromosomes are listed with their loci as of 2024 (Supplementary Table 2). The middle region of the X and Y chromosomes are non-recombined regions (these are called non-pseudoautosomal regions, NPX and NPY, respectively). Shortening of the Y chromosome over 100 million years resulted in the loss of genes, thus, the Y chromosome only carries approximately 1/8 of the known protein-coding genes contained in the X chromosome. Even if all 196 protein-coding genes on the Y chromosome were characterized, it still would be 25% of those found on the X chromosome. Despite this divergent evolution of the X and Y chromosomes, many paralogs exist between the X and Y chromosomes. And more importantly, certain genes on these sex chromosomes are transcriptional regulators. A study published last year revealed that 10 X chromosomal genes have been discovered as candidates driving sex differences in common diseases and sex chromosome aneuploidies[9] The following study by the same group showed the autosomal gene regulation by X and Y chromosomes[10], implying that some of X- or Y-linked transcription factors may govern (SC self-renewal signals. Zinc-finger transcription factors have been reported in their role in the self-renewal of SCs; for example, KLF4[11], zinc-finger protein X-linked (ZFX; in glioblastoma SCs)[12], and Sall4B[13]. Some of the listed X and Y chromosomal transcriptional regulators are likely to play a pivotal role in the maintenance of self-renewal. The outstanding hypothesis from the line of studies is that some of the transcription factors on the X or Y chromosome (i.e., ZFX) may activate the transcription of the gene(s) necessary for the self-renewal of SCs.
In this review, we would like to open the topic with a very recent paper describing autosomal gene regulation by X and Y chromosomes, then develop the discussion to be centered around zinc finger protein X-linked (ZFX) and its role in the self-renewal of SCs and possible subsequent pathways. We would like to conclude the discussion with newly emerging, biological questions.
REGULATION OF AUTOSOMAL GENE EXPRESSION BY SEX CHROMOSOME GENES (QUESTION: ZFX EXPRESSION ON THE ACTIVE X CHROMOSOME)
Both X and Y chromosomes are responsible for the expression of approximately half of autosomal genes in lymphoblastoid and dermal fibroblast cell lines. A very recent study by the Page lab highlighted the role of several genes in the NPX and NPY genes on the X and Y chromosomes as potential candidates; ZFX/ZFY, DDX3X/DDX3Y, KDM5C/KDM5D, and KDM6A/UTY)[10]. Using extensive analyses including X-isochromosomes and X-Y translocated chromosomes, the study confirmed that the dose-dependent autosomal gene activation is not caused by the pseudoautosomal region of the sex chromosomes – but rather, a missing part of the NPX/NPY negatively impacted autosomal gene regulation. Although both KDM6A and ZFX escape X chromosome inactivation, the role of ZFX as a transcription factor certainly makes sense to explain global autosomal transcriptional regulation. Intriguing to find out if there were differences between ZFX and zinc finger gene on the Y chromosome (ZFY) in autosomal gene regulation.
THE STRUCTURAL DIFFERENCE BETWEEN ZFX AND ZFY
Both ZFX and ZFY are encoded on the sex-linked part of the mammalian X- and Y-chromosomes, respectively. Both gene products are Zinc-finger transcriptional factors[14-16]. As uncovered from studies in the 1990s, indeed both ZFX and ZFY function as transcriptional activators. ZFX’s target sequence is AGGCCTAG[17] and ZFY's target sequence is AGGCCY[18]. So, essentially both transcription factors share a similar consensus sequence as the target.
Structurally, ZFX and ZFY are fairly similar, paralog, and show 64% homology in their DNA alignment[19]. When ZFX and ZFY sequences are aligned at the protein level, 92% of the amino acids are identical (Figure 1). Human ZFX is composed of 805 amino acids (NP_003401.2) and ZFY is 801 amino acids (NP_003402.2), respectively. Thus, both proteins are structurally almost identical. However, as described later, the majority of research exclusively revealed the cellular function of ZFX. The role of ZFY in cellular functions might be similar to what ZFX does, although it remains elusive.
ZFX AND ZFY IN SELF-MAINTENANCE OF STEM CELLS
The first study describing the role of ZFX in the self-renewal of stem cells (SCs) was published by the Reizis lab in 2007[20]. One significance of this study is the revelation of the role of ZFX as a self-renewal factor in both embryonic stem cells (ESCs) as well as hematopoietic stem cells (HSCs) using mouse model systems. This study was soon followed by a report that ZFX was one of the 13 sequence-specific transcription factors[21]. Interestingly, ZFX deficiency only affected adult HSCs but not fetal HSCs and erythromyeloid progenitors[20]. Although it is not difficult to imagine the global role of ZFX in SC self-renewal, there is only one more study reporting the confirmation of ZFX in SC self-renewal several years later using human ESCs[22]. As induced pluripotent SCs can be established with three minimum transcription factors, Klf4, Oct4, and Sox2[23], it is logical to predict the induction of these three factors by ZFX. Nevertheless, to date, the direct link of ZFX to Klf4, Oct4, and Sox2 has been poorly understood. Therefore, such a pathway may be unlikely to explain the contribution of ZFX in the self-renewal of SCs. As the initial reports of induced pluripotent SCs used c-Myc in addition to Klf4, Oct4, and Sox2[24,25], the other possibility is that ZFX may induce c-Myc expression to enhance the capability of SC self-renewal. ZFX can regulate the expression of c-Myc as well as a few other embryonic stem cell-specific, self-renewal regulators such as Tbx1 and Tcl1 directly[20], ZFX regulation of c-Myc may make much more sense. Although this study focused on glioblastoma, it was shown that ZFX specifically binds to the GGGCCCCG sequence on the human c-Myc promoter region[12]. Around the same time, ZFX was shown to act as a preventive factor for the differentiation of acute T-lymphoblastic and myeloid leukemia SCs[26].
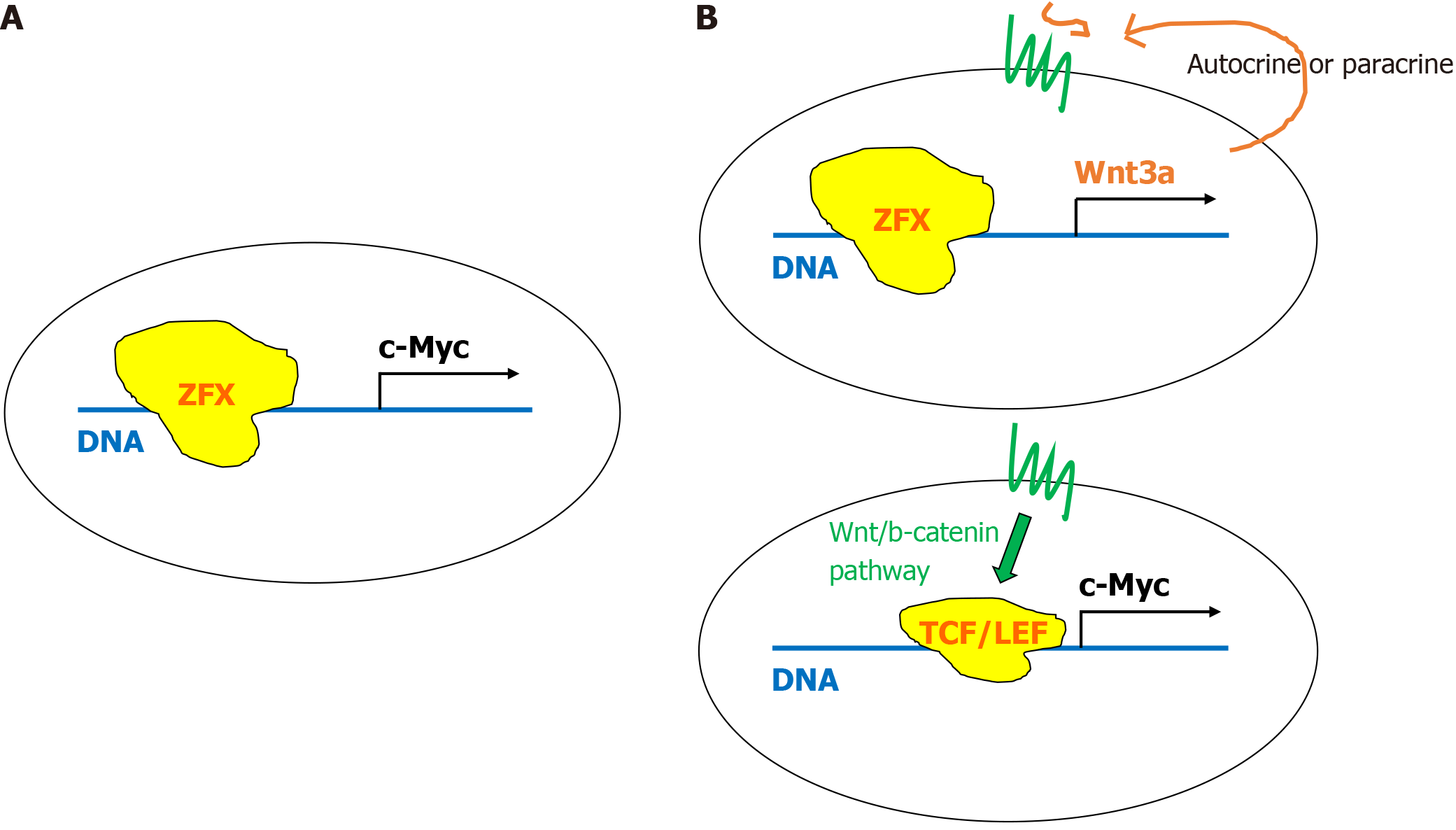
Thus, one possible downstream pathway of ZFX regulation in SC self-renewal may be the direct induction of c-Myc expression (Figure 2A). Nevertheless, there are still limited numbers of studies that have been done to dissect the transcriptional mechanism of c-Myc mRNA expression by ZFX. We should note that the putative ZFX binding sequence, GGCCCCG[12] is apparently quite different from the ZFX sequence discussed in the previous section[17], and still slightly different from the other sequence discussed in the next section. Despite such controversy, it is possible that ZFX contributes to the SC self-renewal, probably via c-Myc expression. In addition, the genome-wide nucleosome occupancy study employing chromatin immunoprecipitation and DNA sequencing demonstrated that the nucleosome occupancies at c-Myc and ZFX sites do not show similar trends when compared between mouse embryonic fibroblasts, ESCs, and neural progenitor cells[27]. Besides the target sequence found in the c-Myc promoter, one remaining unanswered question in humans may be whether or not ZFX is a specific self-renewal factor for adult HSCs. If the case in mice is the same in humans, we can assume that the same mechanism governs the self-renewal of adult HSCs in humans, although there is no experimental proof showing it so far. Regarding this point, the commentary published in 2007 is very insightful[28]. Although ZFX is illustrated as an anti-apoptotic factor, the other factor(s) cooperating with ZFX is/are different between HSCs and ESCs. It would be interesting to investigate if such different factors can be found between fetal and adult HSCs.
Figure 2 The schematic figure summarizes the potential self-renewal signaling driving c-Myc expression.
A: Direct control of c-Myc expression by ZFX; B: Indirect control of c-Myc expression through canonical Wnt/-catenin signaling. ZFX first binds the consensus sequence upstream of the Wnt3a gene to induce the expression of the Wnt3a protein. Secreted Wnt3a binds to its receptor by either autocrine or paracrine. TCF/LEF: T-cell factor/lymphoid enhancer factor.
The role of ZFY in the self-renewal of SCs has not been described as an independent study yet, although the most recent study by the Page lab[10] demonstrated similar autosomal gene regulation by ZFX on the inactive chromosome(s) and ZFY on the Y chromosome. Thus, additional studies may eventually reveal the similar (or differential) functions of ZFY. Because many fewer individual studies are dissecting the role of ZFY in self-renewal or other roles, we will focus on ZFX in the following sections. In short conclusion, it may be more reasonable to hypothesize indirect control of c-Myc expression by ZFX.
ZFX AND WNT SIGNALING
A very recent study reported that the overexpression of ZFX can promote Wnt3 expression and thus the growth of chronic myeloid leukemia stem/progenitor cells[29]. Although this study used cancer SCs, it clearly showed the link between ZFX and Wnt3 signaling in self-renewal. It should also be noted that this study conducted a comparable biological analysis and found that all tested vertebrates (human, mouse, rat, cattle, and dog) share the 100% conserved consensus ZFX-binding sequence (GGGCCGGGCGG) in the promoter region of the Wnt3a gene. There is no other study showing the link between ZFX and other Wnt genes to date, however, the outcome of this study really supports the pioneering discovery showing the role of Wnt (Wnt3a) as an SC growth factor[30,31]. In fact, the Wnt3a knockout was an embryonic lethal due to a reduction of HSCs and progenitor cells in the fetal liver[32].
Wnt signaling has been well-documented as one of the key-signaling pathways in regulating the fate of SCs through asymmetric cell division of SCs. The study by the Weissman lab[30] would be the first that demonstrated the role of Wnt signaling in SC (in this case, HSCs) self-renewal. The role of Wnt in asymmetric cell division and self-renewal is well-explained in a review by Clevers et al[33]. The binding of Wnt to the cell surface receptor, Frizzled, prevents b-catenin from degradation and thus promotes transcription controlled by TCF/LEF. Interestingly, a cell on the side of Wnt binding keeps the progenitor, losing the capability of SCs to divide
In humans, there are a total of 19 Wnt genes[34]. Among them, the role of Wnt in self-renewal seems to be specific to Wnt3a, at least in the self-renewal of ESCs. One study showed that Wnt3a, but not Wnt11, specifically supports ESC renewal[35]. Note that this study used mouse ESCs and feeder cells expressing Wnt3a. In the same year, a different study also showed the role of Wnt3a in mouse ESC renewal[36]. This study confirmed that recombinant Wnt3a is not effective in maintaining the pluripotency of mouse ESCs. However, one key point in this study was that the synergistic action of LIF can maintain the pluripotency of mouse ESCs, if both ZFX and ZFY are added as supplements in the media.
It is not surprising to imagine the correlation between Wnt3a and cancer SCs―In fact, there are several studies reporting the role of Wnt3a in colorectal and breast cancer[37,38].
In summary, the ZFX-Wnt3a axis appears to be a key pathway in maintaining the stemness of certain SCs as well as some cancer cells. Because it is relatively well-acknowledged in the field of the link of Wnt/b-catenin signaling with c-Myc expression[39-41] and the role of Wnt3a[42], it may be more reasonable to presume that ZFX-induced expression of Wnt3a causes either autocrine or paracrine activation of canonical Wnt/b-catenin signaling, leading to the induction of c-Myc (Figure 2B). Additional studies would still be informative to confirm the ZFX-Wnt3a axis in SCs, especially in HSC self-renewal and potential roles in cancer. As mentioned in the introduction, we would like to focus on SC self-renewal in this review.
CONCLUSION
ZFX has emerged as a global transcriptional regulator[10], and it has already been shown as a key transcriptional factor for the self-renewal of SCs[20,22]. Based on published studies, c-Myc and Wnt3a have emerged as molecules linking ZFX with SC self-renewal. Although ZFX appears to be a key factor in assisting SC self-renewal, additional time and studies will be necessary to see if ZFY also shares the same role as ZFX. This is currently one unanswered question. Besides the difference between ZFX and ZFY, there would be a few potentially novel biological questions associated with these studies: (1) Is the transcriptional activity of ZFX maintained in the same way between males and females? (2) If there is a chromosome dosage-dependent difference in ZFX activity; does it affect the self-renewal capability between females and males? and (3) What may be the difference between ZFX on active and inactive X chromosomes?
There are a few points that might command our attention. Although the ZFX target sequence(s) may need to be further investigated, the ZFY target sequence is AGGCCY, which is different from reported ZFX target sequences[12,29], maybe except the closest one[17]. Thus, ZFY may still not have the same capacity, such as SC self-renewal, that ZFX has. Supporting this notion, ZFY was not listed as one of the 13 sequence-specific transcriptional factors in genome-wide, chromatin immunoprecipitation-coupled DNA sequencing[21]. Although the structure of ZFY in solution was reported using nuclear magnetic resonance three decades ago[43], there is no experimentally confirmed structural information on ZFX protein. Therefore, despite very similar primary structures between the two proteins (Figure 1), it might be still worth revisiting and comparing both protein structures with more advanced technologies.
Females probably could express (slightly) more proteins that are regulated by ZFX, as ZFX can escape X chromosome inactivation. Does this mean that females may have higher regeneration potential, especially in the reconstruction of the hematopoietic system?
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author's Membership in Professional Societies: American Society for Cell Biology, 43778.
Specialty type: Cell biology
Country of origin: United States
Peer-review report’s classification
Scientific Quality: Grade B, Grade C
Novelty: Grade B, Grade C
Creativity or Innovation: Grade C, Grade C
Scientific Significance: Grade B, Grade B
P-Reviewer: Wang X; Zhou XC S-Editor: Liu JH L-Editor: A P-Editor: Zhang L