Published online Jun 20, 2024. doi: 10.5662/wjm.v14.i2.92982
Revised: April 15, 2024
Accepted: April 28, 2024
Published online: June 20, 2024
Processing time: 120 Days and 11.5 Hours
In accordance with the World Health Organization data, cancer remains at the forefront of fatal diseases. An upward trend in cancer incidence and mortality has been observed globally, emphasizing that efforts in developing detection and treatment methods should continue. The diagnostic path typically begins with learning the medical history of a patient; this is followed by basic blood tests and imaging tests to indicate where cancer may be located to schedule a needle biopsy. Prompt initiation of diagnosis is crucial since delayed cancer detection entails higher costs of treatment and hospitalization. Thus, there is a need for novel cancer detection methods such as liquid biopsy, elastography, synthetic biosensors, fluorescence imaging, and reflectance confocal microscopy. Conventional therapeutic methods, although still common in clinical practice, pose many limitations and are unsatisfactory. Nowadays, there is a dynamic advancement of clinical research and the development of more precise and effective methods such as oncolytic virotherapy, exosome-based therapy, nanotechnology, dendritic cells, chimeric antigen receptors, immune checkpoint inhibitors, natural product-based therapy, tumor-treating fields, and photodynamic therapy. The present paper compares available data on conventional and modern methods of cancer detection and therapy to facilitate an understanding of this rapidly advancing field and its future directions. As evidenced, modern methods are not without drawbacks; there is still a need to develop new detection strategies and therapeutic approaches to improve sensitivity, specificity, safety, and efficacy. Nevertheless, an appropriate route has been taken, as confirmed by the approval of some modern methods by the Food and Drug Administration.
Core Tip: Cancer remains at the forefront of fatal diseases, with an upward trend in incidence and mortality. Conventional methods have many limitations, necessitating the development of novel diagnostic and therapeutic approaches. The present paper reviews conventional and modern methods of cancer detection and therapy to facilitate an understanding of the rapidly advancing field and its future directions. Modern methods are not without drawbacks; there is still a need for new strategies to improve sensitivity, specificity, safety, and efficacy. Nevertheless, some novel techniques have been approved for use in clinical settings, certifying that an appropriate route was taken.
- Citation: Gromek P, Senkowska Z, Płuciennik E, Pasieka Z, Zhao LY, Gielecińska A, Kciuk M, Kłosiński K, Kałuzińska-Kołat Ż, Kołat D. Revisiting the standards of cancer detection and therapy alongside their comparison to modern methods. World J Methodol 2024; 14(2): 92982
- URL: https://www.wjgnet.com/2222-0682/full/v14/i2/92982.htm
- DOI: https://dx.doi.org/10.5662/wjm.v14.i2.92982
According to the World Health Organization data from 2019, cancer ranks as the first or second leading cause of death in the majority of countries[1]. In populations with higher levels of economic development, mortality associated with cancer is increasing, but those of stroke and coronary heart disease are on the decline[2]. Despite the large volume of research on innovative and more effective therapeutic methods, new cancer cases and related mortality index continue to increase[1,3]. Many causes have been postulated, including the extension of human lifespan and exposure to carcinogens. In addition, while socioeconomic development has improved the quality of life (better living conditions, infrastructure, and medical care), it may also facilitate cancer detection[4,5]. According to worldwide data from the Global Cancer Observatory (GLOBOCAN), there were an estimated 19.3 million cancer cases and nearly 10 million deaths in 2020[6]. Interestingly, males are generally 19% more likely to suffer from cancer. The most common tumors among them include prostate, lung, colorectal, and liver cancer. Females are most commonly diagnosed with breast cancer but also with colorectal, lung, and cervical tumors[1,7,8]. Nevertheless, lung cancer causes the highest mortality among both genders. Comparably important are neoplasms that occur less frequently but have high mortality rates. Examples include brain tumors, with over 300000 new cases and over 250000 related deaths in 2020. The most common and most aggressive primary intracranial tumor is glioblastoma; 95%-97% of patients survive less than three years[9-11]. According to the newest GLOBOCAN estimates, the global cancer burden independent of tumor type is expected to be 35.3 million cases and 18.5 million deaths in 2050.
Detecting tumors in an advanced state makes therapy less effective and more expensive. Despite their widespread use, conventional therapeutic approaches remain unsatisfactory and may cause many adverse effects[12]. There is arguably a need to review existing standards of cancer detection or therapy in light of modern methods due to the rapid advancement in cancer research, the clinical relevance of data generated from novel methods, unmet needs related to treatment resistance and toxicity, as well as an emphasis on tailoring therapy based on individual patient characteristics and tumor profiles. Therefore, the aim of this paper is to provide a clearer understanding of this rapidly advancing field and its future directions; to this end, it reviews available data on conventional and modern methods of cancer detection and therapy.
General practitioners play an important role in cancer diagnosis, as they are usually the first to be consulted by patients with worrisome symptoms. Physicians must correctly interpret the information obtained during an interview and schedule further tests[13]. The current diagnostic path begins with examining a patient and learning about disturbing symptoms or family history of cancer[14]. This is followed by basic blood tests (complete blood count, creatinine level, electrolyte level, liver function test, tumor markers) and imaging tests (X-ray scan, computer tomography, ultrasound, magnetic resonance spectroscopy) to indicate where cancer may be located[15]. Afterwards, the patient is scheduled for a needle biopsy; however, this can be subject to error due to interobserver variability and sample preparation. The results do not take into account heterogeneity because only a fragment of the tumor mass is sampled. Another limitation of this traditional approach is tissue availability[16]. The following step is an investigation of the cancer stage, carried out by an oncologist shortly after receiving all results to provide appropriate treatment on time[13,15,17]. Chemotherapy, radiotherapy, and surgery are standard therapeutic approaches that can be used in combination[18]. They are aimed at neutralizing the cancer but without an entirely individual approach to each patient[19]. These therapeutic approaches are outlined in consecutive paragraphs.
Chemotherapeutic drugs can be divided into four main classes depending on how they affect the cancer cell. These include alkylating agents (form byproducts with the DNA strand), antimetabolites (inactivate components necessary for DNA synthesis), topoisomerase inhibitors (cause DNA strand breaks), and spindle inhibitors (target microtubules to prevent cell division)[19-21]. Chemotherapeutics are harmful to both cancer cells and normal cells, which negatively affects the quality of life due to myelosuppression, inflammation, nausea, vomiting, diarrhea, fatigue, and infertility. For such therapy to be effective, a considerable amount of time is required, which might not be available[22]. Another limitation is the development of drug resistance, during which cancer cells are no longer sensitive to the administered drug(s), and the treatment becomes ineffective. Tumors become resistant due to the reduction in drug availability, alteration of drug targets, cell death inhibition, epithelial-mesenchymal transition, and oncogenic signaling pathways[23,24].
The second pillar of standard cancer treatment is radiotherapy which is administered to approximately 50% of patients[25,26]. It acts by delivering high-energy radiation to cells[25]. Currently, efforts are being made to minimize the exposure of normal tissues to radiation by using, e.g., four-dimensional computed tomography that allows for accurate mapping of the tumor but requires correct positioning and immobilization of the patient during the procedure[27]. Radiotherapy is often used in combination with surgery to reduce the cancerous lesion before surgical intervention and to eliminate any microscopic changes that may have remained after resection[25]. Typically, radiation is systematically administered via external beam radiotherapy; however, it can also be provided by brachytherapy – an invasive procedure delivering a source of radiation close to the cancerous lesion[28]. These methods can be used in combination, which usually improves the results of therapy. Despite continuous improvements, radiotherapy is bothersome for the patient. Side effects can develop during radiotherapy (such as acute mucositis, pneumonitis, bronchospasm, proctitis, radiation cystitis) or occur later (e.g., psychological effects after treatment, radiotherapy-induced hypothyroidism, dysphagia, sexual dysfunction, fertility concerns, and toxicity in different body regions)[28-30].
Finally, the tumor can be physically removed with an appropriate margin. Despite the development of surgical techniques, this strategy does not guarantee improvement in the patient’s health and carries several risks[31,32]. Surgical interventions can lead to bleeding and damage to surrounding organs and tissues[33,34]. The trauma caused by surgery triggers both local and systemic inflammatory responses, which can potentially cause wound-healing problems and the development of residual and micrometastatic diseases. Another complication is deep vein thrombosis caused by, e.g., prolonged periods of immobilization, blood vessel compression, and hypercoagulability[32,35,36]. Nevertheless, surgical techniques are being improved to minimize side effects. For example, laparoscopy is much less invasive than traditional open surgery. It is characterized by decreased intraoperative blood loss, expedited postoperative recovery, shorter hospitalization, and improved immune response[37]. Another technique is robot-assisted surgery, in which the surgeon manipulates instruments with enhanced precision using robotic arms controlled by a console; however, this method is more expensive and does not achieve better outcomes than laparoscopic techniques[38,39].
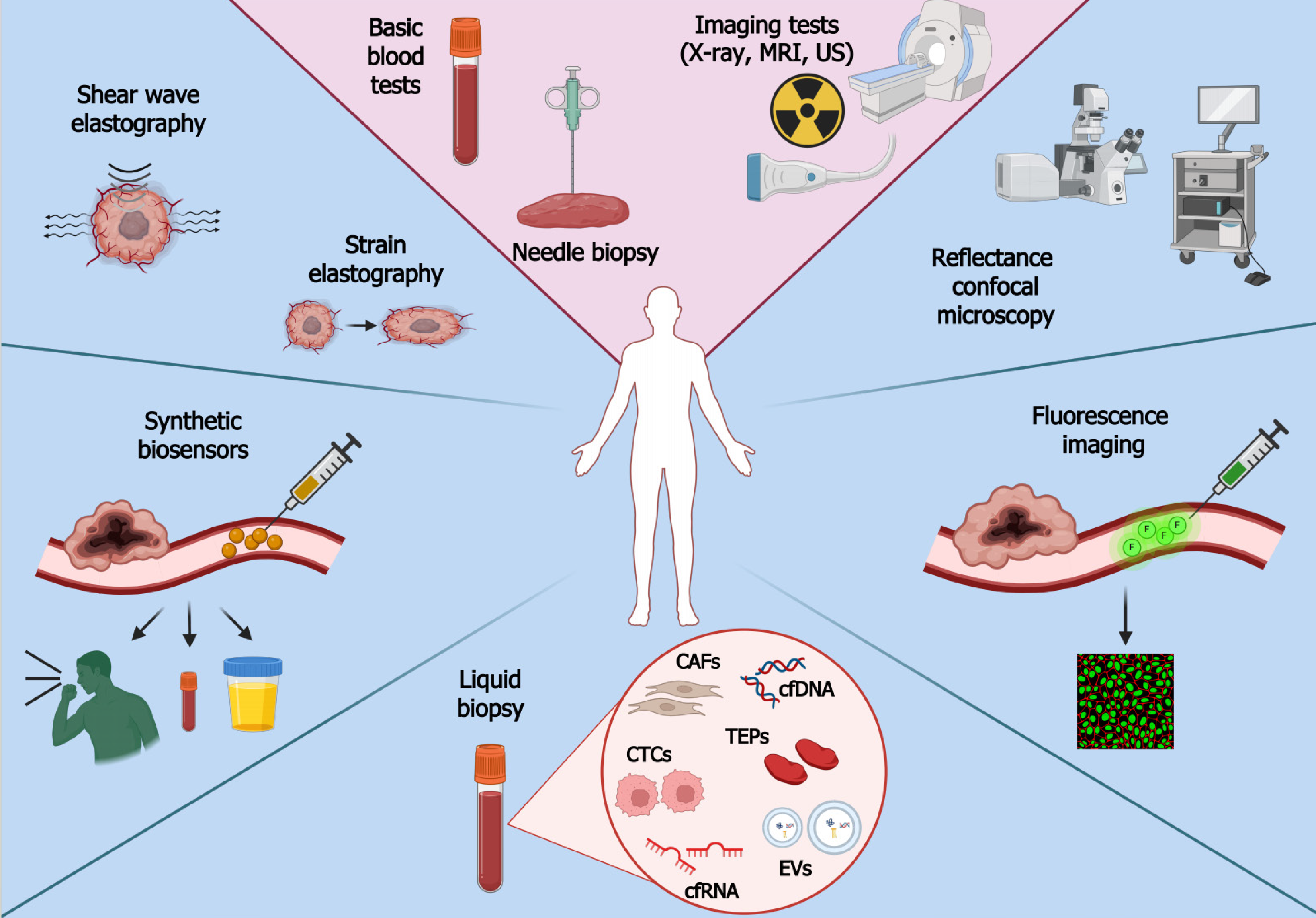
Regardless of the method, it is crucial to initiate diagnosis promptly. Patients diagnosed at an early stage of the disease have a much better prognosis and lower risk of death[17]. Survival rate data from 2012-2018 prepared by the SEER program (Surveillance, Epidemiology, and End Results) show that patients diagnosed with stage I cancer have the highest chance of survival (71%), whereas those diagnosed with stage IV have the lowest (14%). Early cancer detection is hindered by the poor sensitivity of screening tests: Only a small number of tumors are detected in non-invasive and rapid procedures[40,41]. Furthermore, the coronavirus disease 2019 outbreak had a considerable effect on early diagnosis as it limited access to health care and dissuaded potential patients from visiting a doctor[42]. Delayed cancer detection entails higher costs of treatment and hospitalization[43,44]. Therefore, novel and more accurate methods of detecting cancer are needed. Consecutive subsections are dedicated to various approaches for diagnosing and monitoring patients. In addition to relatively new methods such as elastography or synthetic biosensors, we will describe tumor-related molecules that can be evaluated using liquid biopsy, a popular solution to some problems of traditional biopsy. Modern and conventional methods of cancer detection are depicted in Figure 1.
Liquid biopsy holds promise in early cancer detection, prognosis, verifying therapeutic response, and monitoring relapses[45,46]. The method involves collecting body fluids such as blood, urine, saliva, cerebrospinal fluid, and bone marrow for analysis[47]. Liquid biopsy is superior to the traditional approach in various ways[48,49]. The procedure is non-invasive or minimally invasive, which reduces the risks and costs. It offers systemic and homogenous profiles of tumor lesions throughout the body, overcoming challenges related to intra- or inter-tumoral heterogeneity and thus treatment failure. It is also possible to monitor changes in cancer development in real-time. However, the technique also has limitations. One of them is an inability to provide histological evaluation, a crucial aspect for understanding the tissue architecture and cellular characteristics[50]. Secondly, it does not provide comprehensive information about the proteome of a tumor[48]. There is also a need for improvement in the precision and accuracy of detecting biomarkers in bodily fluids, which may be present at very low concentrations. Moreover, the technique is not entirely validated in the clinical spectrum, and reproducibility issues are present due to the use of multiple assays[51,52]. Two main groups of components can be tested. The first includes nucleic acids (DNA and RNA), lipids, proteins, carbohydrates, and various other metabolites or elements. The second one comprises cellular or vesicular components that are released from tumor masses, e.g., circulating tumor cells (CTCs), circulating cancer-related fibroblasts (CAFs), tumor-educated platelets (TEPs), and extracellular vesicles (EVs)[47]. Selected components are described in consecutive subsections.
CTCs: CTCs are components that have detached from the primary neoplasm and entered the bloodstream, which allows them to circulate throughout the body[53]. These cells can be analyzed to determine the ability of the tumor to metastasize[54] or to understand its heterogeneity[55] and allow for identifying prognostic markers[56] or evaluating treatment response[57]. A major limitation of CTC isolation is their limited number in the blood, necessitating the use of sensitive detection methods[47,58]. One such technique is immunomagnetic separation, which uses antibody-labeled magnetic beads to pull down CTCs with appropriate antigens. A widely used antigen is EpCAM which characterizes epithelial tumors, but others are also used, such as ERBB2 and ALK[47,59]. Another approach is to use microfluidic devices with microchannels that allow the size-based separation of CTCs from other blood components. Some of these approaches use antibody-coated surfaces to capture CTCs based on specific markers[54]. The isolation can also be performed using the Ficoll reagent, which separates blood components based on density gradient centrifugation[60]. Since biological cells have diverse electric properties, the components can be separated by dielectrophoresis; however, this is relatively slow and has low throughput[61]. Furthermore, whole genome sequencing can be used to detect mutational variants of CTCs, thus indicating therapeutic outcomes and patient survival. Sensitivity up to one CTC per milliliter of blood is possible through the use of droplet digital PCR combined with size-based enrichment[62,63]. As technology advances, the capabilities of liquid biopsy for cancer research and clinical applications extend further through the use of new and improved methods for CTC isolation.
CAFs: CAFs influence the tumor and its environment. They regulate proliferation and inflammation via the secretion of various growth factors, cytokines, and chemokines[64,65]. The release of proteases such as matrix metalloproteinase by CAFs entails remodeling of the extracellular matrix, leading to invasion and metastasis[66]. Several biomarkers are highly expressed in CAFs and not in other cells; for instance, FAP, FSP, alfa-SMA, CCL11, PDLIM3, AMIGO2, CD70, LOXL2, and UCH-L1[64,65]. Furthermore, cytokeratin and CD31 are considered negative markers since CAFs do not possess endothelial or epithelial characteristics[67]. CAFs may derive from myeloid precursor cells, normal fibroblasts that have acquired genetic mutations, epithelial cells that have undergone epithelial-to-mesenchymal transition, mesenchymal stem cells, or less commonly pericytes and smooth muscle cells[68]. The lack of a specific CAFs-related marker[69] and the diverse origin of these cells can be considered as limitations. In addition, no sufficient exclusion criteria exist among various mesenchymal lineages (such as pericytes or adipocytes), the correlation with specific cancer types/locations is unclear, and few experimental studies have been performed. Consequently, forthcoming studies will concentrate on new biomarkers and methods for selecting CAFs[68]. Nevertheless, the usage of CAFs may be advantageous due to their abundance in the microenvironment, ability to release an ample secretome, and the possibility for modulation instead of depletion, especially since they possess both oncogenic and tumor-restraining functions[70,71].
TEPs: Other components present in the bloodstream of cancer patients are TEPs, in which mRNA expression is influenced by interactions with cancer cells. Such interaction is classified as direct (face-to-face) or indirect (virtual) education. The former is based on a contact between platelets and cancer cells via surface adhesion molecules such as mucins, selectins, and integrins, whereas the latter is more distant and occurs via extracellular metabolites and enzymes. These interactions alter the transcriptome of platelets, which contribute to cancer progression and metastasis[72]. TEPs regulate various steps of tumor invasion, for example by secreting various growth factors and proteases, which promote the adhesion of the tumor cell to the endothelium and allow disruption of the extracellular matrix. They also help CTCs travel in the blood by protecting them from shear stress and allowing metastatic cells to exit from the blood vessel and form a micrometastatic niche[73]. TEPs can be isolated by gradient centrifugation, flow cytometry, gel filtration, and magnetic bead separation. The predominant approach is gradient centrifugation, enabling quick sample processing and achieving relatively high platelet recovery. Nevertheless, this method introduces notable leukocyte contamination, which can be diminished through gel filtration and magnetic bead separation. However, these methods entail extended duration, increased cost, and reduced yield[74,75]. In cancer research, the use of TEPs offers enhanced sensitivity in detecting cancer cells at an early stage of the disease. Moreover, there is no genomic DNA interference since platelets lack nuclei. Platelets are also continuously exposed to the tumor and its surroundings, which facilitates the exchange of biomolecules with cancer cells[76]. On the other hand, the tumor-derived mutant RNAs in TEPs are only present at relatively low levels, which are often below the detection limits of conventional PCR and RNA sequencing methods, but they can be improved by droplet digital PCR[77].
Cell-free DNA: Cell-free DNAs (cfDNA) are deoxyribonucleic acid fragments with a length of 160-180 bp that circulate freely in the bloodstream[78,79]. They are released into the blood by normal and abnormal cells mainly through apoptosis, necrosis, and active secretion[80]. Within the pool of cfDNA, a subset known as circulating tumor DNA (ctDNA) originates explicitly from tumor cells. Detecting cancer-associated genetic alterations using cfDNA enables early detection of multiple cancers and offers an insight into disease stage and prognosis[81]. ctDNA harbors tissue-specific methylation patterns, facilitating diagnosis and establishment of tumor localization[82]. This method provides a more comprehensive representation of the tumor’s genetic landscape than needle biopsy[83]. Furthermore, the ability to serially sample cfDNA allows for longitudinal assessment of dynamic changes in the concentration of cfDNA or tracking clonal evolution, which enables the monitoring of therapy response, acquired resistance, minimal residual disease and recurrence[84]. Technical improvements in detection techniques are needed, particularly given the scant presence of cfDNA and the prevalence of background signals. Moreover, ctDNA assays may exhibit reduced sensitivity in detecting fusion events and copy number changes[85]. It has been proposed that the effectiveness of ctDNA relies on its capacity to reflect the genetic changes identified in the tumor[86]. Because the concentration of ctDNA is the highest in blood[80], it is usually collected in tubes containing the K3 potassium salt of ethylenediaminetetraacetic acid to prevent blood clotting. Special kits have been developed for cfDNA isolation to minimize material loss[87]. Alterations in ctDNA such as translocations, inversions, copy number variations, deletions, and insertions are detected by methods such as digital PCR, cancer personalized profiling by deep sequencing (CAPP-Seq), or tagged amplicon deep sequencing (TAm-Seq)[47,88,89]. CAPP-Seq is ultrasensitive and adopts a hybrid-capture methodology that examines selected genomic regions; it has been applied for diagnosis and molecular profiling, therapeutic response, and detection of postsurgical residual disease[90]. TAm-Seq is an amplicon-based method that utilizes a target enrichment array with barcoded primers to prepare the amplicon library. The two-step amplification design enables the identification of mutations with a sensitivity of over 97%[89]. The enhanced TAm-Seq, denoted as eTAm-Seq, can detect the mutant allele fraction as low as 0.25% with a sensitivity of 94%[91].
Cell-free RNA: Another nucleic acid that can be analyzed by liquid biopsy is cell-free RNA (cfRNA). Among the wide variety of cfRNA, microRNAs (miRNAs) and long non-coding RNAs (lncRNAs) are of particular interest in cancer detection and research[92]. Similar to cfDNA, cfRNA originates from cell death, but it can provide more detailed information about the expression signature of cancer cells and their intercellular communication in the tumor microenvironment[93]. Despite this, detection and analysis are complicated by the low abundance and susceptibility of cfRNA to degradation. There is also a lack of standardization of procedures in cfRNA analysis, resulting in inconsistent results across different studies and laboratories[94,95]. cfRNA can be isolated from various body fluids such as serum, plasma, tears, or urine[96]. One method is based on the traditional phenol-chloroform extraction method followed by precipitation. However, this technique is rather prone to contamination, which can be avoided using column-based RNA adsorption[97]. Regarding cfRNA subtypes, the miRNA ranges in length between 20-25 nucleotides and regulates gene expression post-transcriptionally, thus affecting various cellular events. In cancer, they are responsible for sustaining angiogenesis, providing limitless replication potential, and evading apoptosis, but they also inhibit tumor growth or prevent invasion and metastasis[98]. In general, changes in miRNA levels are prevalent among tumors, and their expression is specific to the tissue[43,99]. Some cancer-specific miRNAs have shown promising results in disease detection, such as miR-185-5p and miR-362-5p in breast cancer[100] or miR-451a in pancreatic cancer[101]. Another type of cfRNA is lncRNA, a heterogeneous group of transcripts exceeding 200 nucleotides in length. They are involved in gene expression and various physiological processes but are also known to influence cancer initiation, progression, and metastasis[102]. Accumulating evidence underscores the pivotal roles of lncRNAs in mediating the reprogramming of energy metabolism in cancer[103]. Circulating lncRNAs proved to be useful in detecting tumors of lung (MALAT1, SPRY4-IT1, ANRIL, NEAT1), breast (RP11-445H22.4, H19), gastric (HULC, CUDR, LSINCT-5, PTENP1), liver (AF085935), and prostate (PCA3, LincRNA-p21)[104].
EVs: EVs are lipid bilayer-enclosed particles discharged from a wide range of cell types and present in biological fluids such as blood, urine, saliva, cerebrospinal fluid, breast milk, and seminal fluid. They contain a wide range of heterogeneous molecules that can be used to determine a patient’s health status[105]. A few general groups of EVs have been distinguished based on their size and origin, namely microvesicles (100-1000 nm), apoptotic bodies (50-4000 nm), and exosomes (50-200 nm)[106]. The latter have garnered significant interest due to their role in intercellular communication and their potential applications in cancer detection. They carry various biomolecules, including proteins, lipids, and nucleic acids such as miRNAs and mRNAs, reflecting the molecular content of their parent cells. Those deriving from cancer cells often contain a unique cargo of molecules that can serve as potential biomarkers for cancer detection[107]. Exosomes have some advantages over CTCs and cell-free nucleic acids; they are actively released by living cells, and their abundance in biofluids facilitates collection. Their stability, attributed to their lipid bilayers, enables them to circulate even within the challenging tumor microenvironment. This high biological stability allows for the long-term storage of specimens for further isolation and detection[108]. Furthermore, they can be used to identify the present disease by transporting specific molecules derived from parent cells[106]. Unfortunately, isolating these molecules is complicated due to their heterogeneity and small size[108]. Diagnostic accuracy can also be influenced by the physical characteristics of the studied body fluid, such as density and viscosity[105]. Isolation can depend on physical or biological properties since there are centrifugation and size-based techniques[109], or capture-based, polymer-based, and microfluidics-based methods[110].
The use of endogenous biomarkers detecting cancer at an early stage, e.g., cell-free nucleic acids, proteins, or lipids, is limited due to their relatively quick degradation and low concentrations. However, this can be improved by the use of synthetic biosensors, i.e., exogenous molecules introduced into the body to detect phenotypic changes and enhance cancer-related signals to a conveniently measurable level[111,112]. These synthetics are divided into two main classes, namely activity-based and genetically encoded biosensors.
The first group encompasses protease-activated and small-molecule biosensors. In the protease-activated approach, a biocompatible carrier (e.g., iron oxide nanoparticle or polyethylene glycol) is linked to peptide substrate and cleavable reporter (e.g., mass-barcoded and fluorescent peptide); the latter is detached and detected in body fluid by mass spectrometry or enzyme-linked immunosorbent assay. In contrast, small-molecule probes consist of an enzyme recognition site linked to the substrate molecule and a cleavable reporter such as volatile organic compounds (VOCs) or deuterated metabolites[111,113]. VOC analysis is particularly useful in the early detection of lung cancer, but fewer than 15 compounds (e.g., ethanol, acetone, isoprene) have been found useful. Based on the D5-ethyl-β-d-glucuronide example, it is enzymatically converted into D5-ethanol by β-glucuronidase, an extracellular enzyme secreted by solid tumors. The product is detected from the breath by gas chromatography coupled with high-resolution mass spectrometry[111,112]. Both types of activity-based biosensors described above are administered intravenously.
Genetically encoded biosensors use reporters detectable in biofluids, whose release is influenced by the tumor microenvironment[114]. These synthetic biomarkers are subdivided into vector-based, mammalian cell-based, and bacterial cell-based systems. Each category varies depending on a specific tumor-related characteristic, the “input signal” that is measured, and the method by which this signal is amplified for detection. As the synthetic biomarker is produced in cells of a specific phenotype, this approach reduces the number of false positives caused by background production in healthy tissues. The vector-based system exploits tumor-specific promoters and secretory reporters, which leads to the specific gene expression pattern associated with a tumor. The mammalian cell-based system takes advantage of the metabolic changes in tumor-infiltrating immune cells [typically macrophages but also T-cells, B-cells, and natural killers (NKs)] to induce the generation of a secreted biomarker[115-118]. Bacteria can colonize tumors due to the diminished immunosurveillance and elevated nutrient availability in the necrotic tumor core; the bacterial cell-based system makes it possible to discharge reporters at the tumor site[119]. Vector-based strategies raise concerns about immunogenicity and insertion mutagenesis, which is particularly challenging for early detection applications requiring longitudinal assessment and repeated administrations. Mammalian-based sensors might fail to detect necrotic tumors in high tumor burden scenarios, likely due to poor infiltration. Additionally, the high cost and complex pipeline make this approach unsuitable for routine screening. Using bacteria for early cancer detection poses safety concerns about the inherent toxicity of bacterial components and the potential for reversion to virulence. It is also unclear if systemically delivered bacteria can colonize all tumor types and nascent lesions lacking a necrotic core[111]. In conclusion, synthetic biosensors are sophisticated and promising but require thorough investigation and standardization.
Cancer detection through fluorescence imaging (FI) employs diverse optical imaging technologies to enhance the identification of early neoplasia by focusing on unique molecular signatures. This technique is characterized by low invasiveness and considerable specificity and sensitivity. Detecting lesions at an earlier stage leads to enhanced treatment outcomes and lowers treatment expenses by averting the necessity for multimodal care at an advanced disease stage[120]. FI is usually used for the identification of easily accessible tumors, such as those of the oral cavity[121]. Three main types of tumor-targeted fluorescent dyes (TTFDs) are distinguished based on their activation mechanism[122-125]. The first comprises passively-targeted TTFDs that accumulate in some tumors due to their enhanced permeability and retention properties. The second group interacts with an overexpressed tumor-specific ligand, which leads to the endocytosis of the complex and allows for the detection of cancer cells. The last group are activated in the presence of molecules such as enzymes that are predominant in the tumor and its surroundings; the specificity of this TTFD class varies depending on the target, the magnitude of enzyme upregulation, tumor-background ratios and quencher release rates. Limitations of TTFDs include the inability to detect malignant lesions > 2 cm beneath the tissue surface and the tendency to accumulate in some noncancerous tissues[126].
Another method used to detect cancer is reflectance confocal microscopy (RCM). It facilitates high-resolution imaging of tissues at considerable depths, enabling optical sectioning for the three-dimensional reconstruction of the samples of interest[127]. Produced images are presented in grayscale, with bright (white) structures indicating a higher refractive index compared to their surroundings. Examples of such structures include melanin, keratin, and collagen[128]. RCM is employed to detect skin lesions, which makes it a valuable tool for detecting skin cancers[129]. However, it is not limited to these tumors since it can also determine precancerous and cancerous conditions in the oral cavity or cervix[130,131]. In addition to non-invasiveness, this method has the ability to monitor dynamic processes like blood flow[132]. On the other hand, RCM can be outperformed by biopsy when diagnosing and subtyping primary basal cell carcinoma[133]. Other disadvantages are decreasing image resolution below a depth of 100-150 μm and less robustness in evaluating nuclear details relative to hematoxylin and eosin histopathology[132].
Elastography has emerged as a promising and innovative technique for cancer detection, providing valuable insights into tissue biomechanics to overcome the limitations of traditional imaging methods. This approach focuses on assessing tissue stiffness, a characteristic that often varies between normal and pathological tissues[134]. Two main types of this method are strain elastography and shear wave elastography. The first one determines the tissue stiffness by exerting external pressure, which alters tissue dimensions and results in deformation known as strain. Lesions with higher stiffness exhibit less deformation, which manifests in lower strain values and a higher Young's modulus, the latter being a property of materials that measures the tensile or compressive stiffness when the force is applied[135]. Inducing stress depends on the tissue; it can be elicited, e.g., through arterial and respiratory motion in the liver or via manual compression in the thyroid[136]. The second type of elastography uses a pushing beam of acoustic radiation force to displace tissue and generate shear waves, which are monitored at multiple off-axis locations. Shear wave speed provides a quantitative estimate of tissue elasticity, with faster propagation observed through stiffer and more contracted tissue[137-139]. Providing real-time visualizations of tissue deformation or measuring the shear wave speed enhances the accuracy of cancer detection. Moreover, elastography can differentiate between benign and malignant lesions, as investigated in breast cancer[140]. Shear waves have been extensively employed in liver imaging, where they assess liver fibrosis and cirrhosis, which are symptoms of liver cancer[136]. Like other techniques, elastography has some limitations. In particularly obese patients with thick subcutaneous fat and ascites, it might be impossible to measure tissue stiffness[141]. The visualization may also return artifacts instead of actual lesions. Nevertheless, integrating elastography into routine clinical practice holds promise for enhancing the precision and efficiency of cancer detection across various organs and tissues[142].
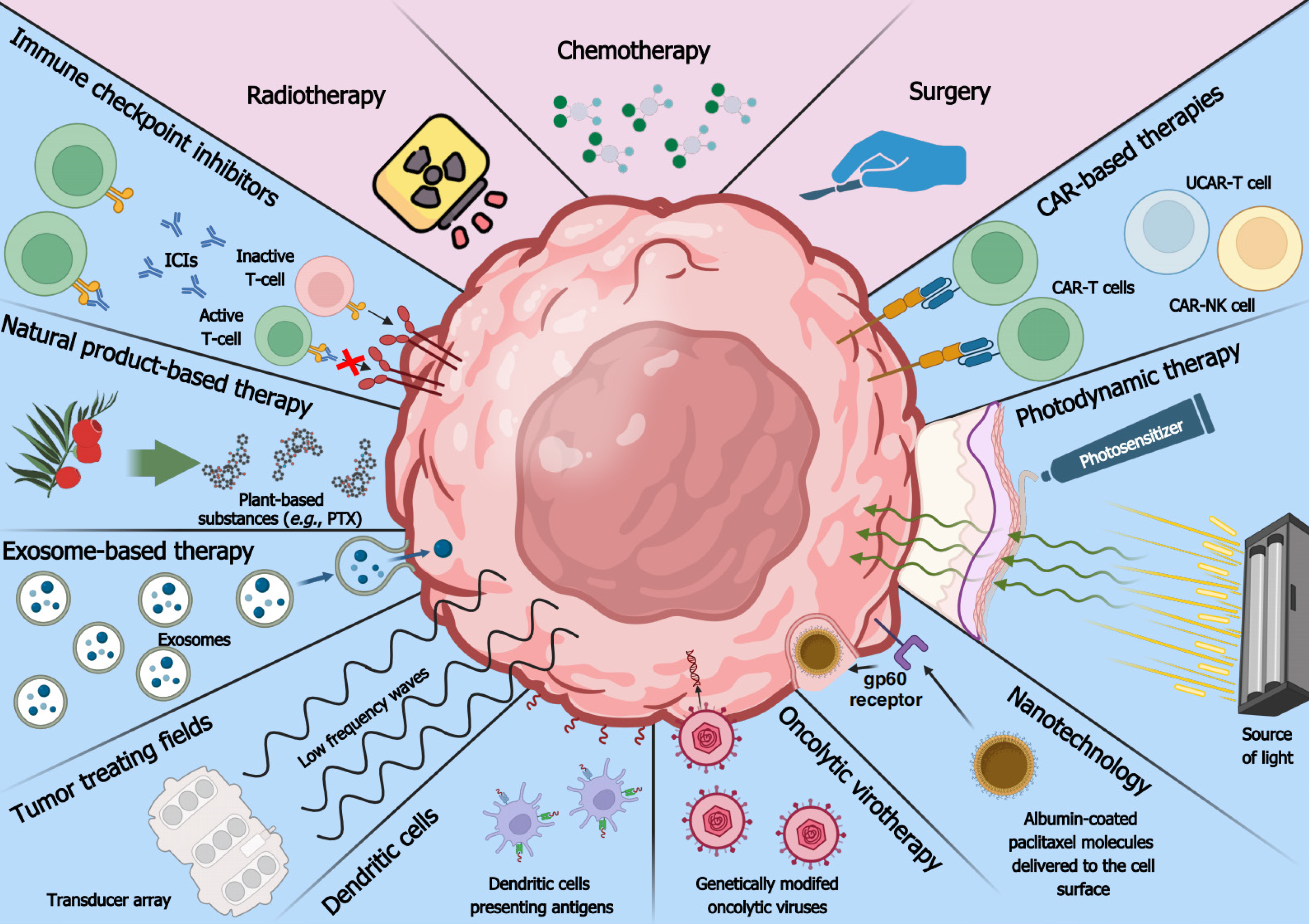
Tumor malignancy intensifies during extended periods without intervention, posing therapeutic challenges[143]. The current standard of care is unsatisfactory in most cancer types, necessitating the development of novel strategies[144]. Since therapies are more efficient against low-stage tumors, a combination of modern approaches with early detection should significantly improve the quality of life while causing the least side effects[27,145]. Consecutive sections outline novel methods targeting various cancer types and compare their effectiveness and safety to commonly-used therapies. Modern and conventional methods of cancer therapy are depicted in Figure 2.
Oncolytic viruses (OVs) represent an encouraging way of treating cancer since they selectively replicate in tumor cells and lyse them without targeting noncancerous cells. The excessive pool of nucleotides needed to supply rapidly dividing cancer cells creates a favorable environment for the replication of viruses, which are also able to pass the microenvironment[146,147]. OVs manifest their function via direct infection and anti-tumor immunity[148]. The former acts through viral replication and host cell death, causing a cascade reaction in which dying cells amplify the presence of OVs within the tumor microenvironment. The second mechanism involves immunogenic cell death, which alarms immune cells by ejecting various particles, such as tumor antigens and damage-associated molecular patterns. Although this process needs further research, it holds promise in counteracting cancer and metastasis[149].
One oncolytic virotherapy, Talimogene laherparepvec, also known as T-Vec or Imlygic, has been approved by the Food and Drug Administration (FDA) and European Medicines Agency for treating melanoma[150]. T-Vec is a modified herpes simplex virus 1, which has been generated from the JS1 viral strand. Modifications include the deletion of the ICP34.5 and ICP47 proteins encoded by γ34.5 and α47 genes, as well as the insertion of the human GM-CSF encoded by the CSF2 gene. Deletion of γ34.5 limits neurotoxicity and enhances cancer cell-specific viral replication, whereas α47 deletion enables immune recognition of the injected virus, causing the elimination of the virus outside the tumor microenvironment[151]. The insertion of the human CSF2 gene promotes dendritic cell (DC) infiltration and maturation, enhancing tumor antigens’ presentation to T-cells and improving anti-tumor immunity efficiency[152]. Imlygic is administered as a direct injection into melanoma lesions. A phase III trial showed an overall survival (OS) of 23.3 months, 50% or greater tumor regression in 64% of injected lesions, and mild treatment-associated symptoms in stage IIIB-IV melanoma patients[147]. In comparison, chemotherapy resulted in 5.1-month OS and a partial response rate of 15%-28%[153]. Another limiting factor of chemotherapy was high toxicity associated with severe or life-threatening adverse events, whereas T-Vec caused mild symptoms such as fever or swelling in the injection area[147,153]. Studies are underway into the effects of combining T-Vec with other therapies or its use in other cancer types[150,154].
Another example of an OV-based approach is G47∆ therapy against glioma. G47∆ is a triple mutant of herpes simplex virus 1, which was developed based on the G207 virus. The mutations present in G207 include the deletion of the γ34.5 and α47 genes, as well as the insertion of the lacZ gene into UL39, i.e., the gene encoding ribonucleotide reductase subunit ICP6, resulting in its inhibition[155,156]. Insertion of the lacZ gene limits growth, proliferation, and virulence[157,158]. Oncolytic virus G47∆ can be administered at several sites within the tumor to increase efficiency[156]. The average OS was 20.2 months after G47∆ initiation. Survival at 24 months was 41% (8 out of 19 patients), and at 30 months was 31.58%[156]. Five patients survived more than three years after the onset of therapy, and three patients survived more than eight years. The combination of surgery followed by chemoradiotherapy and adjuvant temozolomide (TMZ) achieved a median OS of 10 months and only 6.7% survival at 24 months[10]. Each patient receiving G47∆ demonstrated only minor side effects (mostly fever in 17 out of 19 patients), and only one patient needed prolonged hospitalization. There were no adverse events that discontinued the therapy. With regard to the standard of care consisting of an invasive operation and chemotherapy, 44% of patients showed systemic adverse events including dizziness, muscle weakness, trouble with memory, and hair loss[159].
Some cancer therapies end up ineffective due to the inability to infiltrate tumor microenvironment or cancer cells. The value of exosomes in an early diagnosis of oncological patients was described in the section about cancer detection, but their drug delivery characteristics are comparably promising[160]. Compared to other drug delivery methods, exosomes offer greater stability in body fluids, low cytotoxicity or immunogenicity, superior drug protection owing to lipid bilayer structure, the ability to pass the blood-brain barrier, convenient modification for specific tumor types, and the possibility to transport both hydrophobic and hydrophilic molecules[161]. The small size of exosomes alongside their ability to transport molecules of varying sizes and charges make them exquisite candidates for delivering drugs against different types of cancer.
Clinical trials on exosomes in anti-cancer drug delivery are ongoing but a few have yielded preliminary data. The combination of DC-derived exosomes with metronomic cyclophosphamide was tested in a phase II clinical trial on lung cancer. The primary goal of the study was to assess progression-free survival (PFS) in a four-month timeframe[162]. Twenty-two patients were evaluated and their median PFS was 2.2 months. A four-month PFS was achieved in 32% of patients, while OS reached a median value of 15 months. No objective tumor response or cancer-specific T-cell immune response was detected. Although the efficiency results were not exceptional, the treatment was found to be feasible and well tolerated[163]. The safety and tolerability of exosomes were also evaluated in pancreatic cancer and advanced hepatocellular carcinoma. In the case of pancreatic cancer, an ongoing study of KRAS G12D siRNA loaded into exosomes aims to identify maximum tolerated dose and dose-limiting toxicities[164]. The KRAS G12D mutation is the major driver for most pancreatic cancer cases, and its silencing might serve as a potential therapeutic strategy[165]. Although the planned completion date of the study is April 2025, it was already feasible to generate clinical-grade exosomes in mice implanted with patient-derived xenograft harboring KRAS G12D with no severe adverse events[166]. In advanced hepatocellular carcinoma, exoASO-STAT6 is becoming a promising therapy against STAT6, based on targeting with specific anti-sense oligonucleotides[167]. In preclinical in vitro trials, exoASO-STAT6 presented selective targeting towards tumor-associated macrophages, which inhibited M2 macrophage polarization and induced an anti-tumor immune response[168]. This could hinder the STAT6 signaling pathway and cancer proliferation and enhance the effectiveness of other methods such as immune checkpoint inhibitors (ICIs) and radiotherapy[169]. Overall, the use of exosomes as drug carriers is encouraging in cancer therapy, but more research is needed to assess their safety and efficiency comprehensively.
Antigen presentation is enabled by immune components such as macrophages, Langerhans cells, and B-lymphocytes. In addition, a superior approach involves the use of DCs, as they induce adaptive immunity and support the innate immune response against tumors[170,171]. DCs-based therapy utilizes the first and second classes of major histocompatibility complex (MHC) to stimulate the patient’s immune system and counteract tumor[172,173]. DCs are primarily involved in determining the compatibility of organs undergoing transplantation but can also be utilized to detect cancer neoantigens. DCs enforce the anti-tumor response, present tumor antigens, and activate T-lymphocytes. The vaccine can be available in an off-the-shelf formulation for repeated long-term administration to a patient[174].
The innovative DC-utilizing therapy termed DCVax-L ensures that the immunosuppressive effects of the tumor are avoided by injecting DCs with cancer-specific antigens extracted from the patient’s tumor. This personalized approach allows for a presentation of the tumor antigens and improves the response of the patient’s immune system[172,174]. Additionally, the drug can be used in combination with surgical resection of the tumor, chemotherapy, and radiation. Research on DCVax-L is already in the third phase of clinical trials conducted in several countries[174]. The drug was administered intradermally into the patient’s arm (with a change of arm for every dose), rendering a less invasive approach than intratumoral injection[174]. The average survival rate among patients with primary glioma was 19.3 months after randomization of the study group, in comparison to 16.5 months for the external control group (ECP) treated with TMZ. Forty-eight months after completion of DCVax-L therapy, the survival rate equaled 15.7% (9.9% in the external control group), whereas 60 months after treatment it was 13% (5.7% in the ECP)[174]. Thus, a more than twofold reduction in the risk of death after completing DC therapy was noted. Among 64 patients with recurrent cancer, a 42% reduction in risk of death was indicated following completion of DCVax-L therapy (13.2 vs 7.8 months in ECP). After administering a total of more than 2000 doses of the drug to 64 patients, only five cases of adverse events were detected, with three concerning intracranial edema, one related to severe nausea, and the last regarding lymph node infection. No signs of cytokine storm or autoimmune reaction were detected, and the drug has been proven safe for use[174].
Another example of DC-based therapy is Neo-DCVac, a novel take on counteracting non-small-cell lung cancer (NSCLC). A phase I clinical study on the feasibility and safety was performed among 12 patients who took five doses of the treatment[175]. All patients were in an advanced stage of disease. The treatment showed similar results to chemotherapy, with 7.9 months of OS after Neo-DCVac compared to a median of eight months of OS after chemotherapy[175,176]. The therapy showed synergistic effects with ICIs, only mild-to-moderate adverse events, and no dose-limiting factors. This certifies that DC therapy is a safe treatment option, although the effectiveness of some approaches needs improvement. Further research is required to enhance the Neo-DCVac therapy and determine the optimal dosage.
Chimeric antigen receptor T-cell (CAR-T) therapy involves the development of a chimeric antigen receptor that is inserted via genetic engineering techniques into T-cells collected from a patient or differentiated from mononuclear blood cells[177,178]. Genetic engineering is typically ensured via transfection of the lymphocytes with a lentiviral vector that carries the CAR sequence[178,179]. Following transfection, the chimeric antigen receptor is expressed on the T-lymphocyte surface, which enables precise recognition and elimination of cancer cells. The chimeric antigen receptor is made out of three parts: A signaling domain inside the T-cell, a transmembrane domain, and an antigen recognition domain on the surface of the T-cell[177]. Nowadays, novel CAR-T cells have a modified signaling domain to enhance efficacy or are enriched with an effector memory phenotype[180,181]. Despite showing efficiency in some neoplasms, CAR-T application is challenging in solid tumors and is often correlated with expensive and complicated manufacturing processes, as well as various adverse events such as cytopenia, cytokine release syndrome, and neurotoxicity[181,182].
The first two CAR-T therapies (Kymriah and Yescarta) were approved by the FDA in 2017. Both products were designed to treat B-cell malignancies, namely leukemia and lymphoma, and are targeted to recognize CD19. This antigen is expressed on the surface of B-lymphocytes and their precursors, which guarantees high specificity and limits off-target toxicity[179]. Yescarta showed reliable effectiveness in treating relapsed/refractory large B-cell lymphoma. Survival without the incidence of adverse events, cytokine release syndrome, and neurologic events reached 21.5 months for CAR-T therapy, compared to only 2.5 months for chemotherapy with autologous stem-cell transplantation. In addition, the OS reached 23.9 months for CAR-T vs 11.2 months for chemotherapy, with respective response rates of 88% compared to 52% and a complete response rate of 75% compared to only 33%[183]. Unfortunately, 94% of the treated patients experienced severe and life-threatening adverse events, but the toxicity was manageable[183]. Since 2017, various new CAR-T therapies have emerged, and some have already been approved by the FDA, such as Cervykti targeting CD38 in multiple myeloma[184].
To reduce the time and expense involved in classical CAR-T therapies, a new solution has emerged: Universal CAR-T (UCAR-T). Each currently approved CAR-T therapy is made of patient-derived T-lymphocytes to avoid immune rejection. UCAR-T breaks this trend by using allogenic CAR-T cells with T-lymphocytes harvested from healthy donors. This enables the mass production of CAR-T, which decreases the costs and speeds up the time between diagnosis and onset of treatment from two-to-five weeks to only eight days[185,186]. The main issue with UCAR-T is graft vs host disease (GvHD), which can manifest when foreign cells are injected into the patient’s body, triggering immune rejection[185,187]. Fortunately, this issue can be handled by gene editing, with the most commonly used technique, CRISPR/Cas9, utilized to insert the CAR sequence into a T-cell locus and knockout GvHD-causing genes such as TRAC[187]. This increases the safety of UCAR-T and opens avenues for further research on this type of approach. In the future, UCAR-T could substitute current autologous CAR-T therapies, as evidenced by a dual-targeting CD19/CD22 UCAR-T therapy denoted CTA101 that was developed to counteract relapsed/refractory large B-cell lymphoblastic leukemia[186]. CTA101 was tested on six patients in a phase I study; it yielded no GvHD while achieving an 83.3% complete remission rate and a negative result for minimal residual disease after 28 days of treatment[186]. This is higher than treatment via Yescarta, which targets CD19 and only obtained a complete response rate of about 75%[183]. Another alternative to CAR-T is CAR-NK, which substitutes the T-lymphocytes with NK cells. Utilizing NK cells reduces toxicity, inflammation, and cytokine release syndrome, as well as improves production efficiency by harvesting cells from different sources or differentiating them from umbilical cord blood[188,189]. Similar to regular CAR-T, NK cells can be transduced with viral vectors and edited by CRISPR/Cas9. A clinical trial featuring CAR-NK cells targeted at CD19-positive lymphoid tumors showed a response rate of 73% and complete remission in 64% of treated patients. No GvHD or cytokine increase has been detected[190]. Novel platforms such as UCAR-T and CAR-NK warrant further investigation in solid tumors.
ICIs are biologicals, usually monoclonal antibodies, that prevent the immune-evasive effect of tumors. ICIs enhance the activation, proliferation, and differentiation of T lymphocytes[191,192], improving the patient’s immune system as monotherapy or enhancing the activity of another type of treatment. ICIs can bind to various proteins on the cellular surface, but the most common target is the programmed death 1 (PD-1) receptor or its ligand[191]. ICIs attach to the checkpoint protein and block its function. For example, the first ICI approved by the FDA is a human IgG4 monoclonal antibody called nivolumab. It binds to the programmed death 1 receptor on T-cells and prevents its interaction with programmed cell death ligand 1 on tumor cells[191]. This ensures proper lymphocyte T activation, proliferation, and anti-cancer cytokine secretion[193]. In addition to conventional ICIs, it is possible to target phagocytosis checkpoints such as LILRB1/MHC-1 and LILRB2/MHC-1 or NK-cell checkpoints such as KIRs/MHC-1 and PVRIG/CD112[194]; this limits the immunosuppressive effect of tumors and stimulates the elimination of cancer cells via phagocytosis or NK-mediated cell death[194].
Not every cancer type is suitable for ICI therapy, e.g., glioma creates a microenvironment unfavorable for binding antigens to the cancer cell surface. However, ICIs hold promise in lung cancer. It was found that a frequently used ICI known as durvalumab increased OS to 12.9 months when combined with chemotherapy in comparison to chemotherapy alone which reached 10.5 months[195]. Although ICIs can be effective, new inhibitors should be developed to improve clinical endpoints further. One of the newer antibodies targeting PD-1 is serplulimab. This humanized IgG4 monoclonal antibody was tested in a phase III randomized clinical trial on extensive-stage small-cell lung cancer, where each patient received the treatment intravenously every three weeks[196]. Serplulimab improved OS to a greater extent than durvalumab: The median OS of patients receiving serplulimab combined with chemotherapy reached 15.4 months compared to 10.9 months for individuals receiving chemotherapy. However, ICI also caused severe or life-threatening adverse events in 33.2% of patients[197]. Although ICIs show promising results, further development is needed to enhance their efficacy, limit adverse effects, and apply them to a broader range of cancer types. A deeper understanding of immune checkpoints on cancer cells and discovering novel targets or inhibitors will facilitate progress in ICI therapy.
The search for anti-cancer compounds has expanded to include substances found in natural sources and the everyday diet. Plant-based substances are of particular interest, despite sometimes providing only small amount of compound. Nowadays, it is possible to synthesize them chemically in a laboratory. A prime example is the natural substance paclitaxel (PTX), a strong anti-cancer drug that laid the foundations for new therapies against many malignancies such as ovarian, colorectal, bladder, pancreatic, lung, and squamous cell carcinoma[198-201]. PTX was originally produced from yew tree bark but is now manufactured from precursors such as 10-deacetylbaccatin III[202]. PTX belongs to the group of anti-cancer drugs so-called “mitotic poisons” that bind tubulin. The compound prevents microtubule depolymerization, impairs the function of the mitotic spindle, stops cell cycle progression, and ultimately inhibits cancer cell proliferation[199,200]. While mitotic arrest is the primary mechanism, PTX also induces p53-mediated apoptosis. In some cancer cells, PTX has been connected to elevated levels of reactive oxygen species (ROS) and upregulated genes associated with endoplasmic reticulum stress. It remains to be determined whether the endoplasmic reticulum stress derives from gene dysregulation caused by p53 activation[203].
A phase III clinical trial examined the use of PTX in combination with carboplatin for the treatment of ovarian cancer. This combination was compared to the conventional PTX-doxorubicin-cisplatin regimen. Although PTX-carboplatin treatment resulted in lower OS (37 vs 41 months), it was a preferred choice due to the improved quality of life and reduced toxicity for patients. This therapy was also recommended as a first-line approach against advanced endometrial cancer[204,205]. Another trial with 861 metastatic pancreatic cancer patients found that gemcitabine plus albumin-bound PTX was superior to gemcitabine alone. Improvements in response rates (23% vs 7%), PFS (5.5 vs 3.7 months), and OS (8.5 vs 6.7 months) were noted in the combination therapy. The nab-PTX-gemcitabine group showed higher one-year survival (35% vs 22%) and two-year survival (9% vs 4%) compared to the gemcitabine group[206]. The distinct mechanism of action of PTX highlights its potential to enhance patient outcomes.
Nanotechnology is a wide scientific branch utilizing submicrometer-sized materials. It can be used for detecting and marking tumors via different fluorescent or light-emitting probes[207]. Nanotechnological vesicles with a size of < 100 nm are being used for drug and probe delivery to tumor cells. The process includes the formation of the nanostructure, surface modification of the nanocarrier (to target cancer cells), transportation of the nanocarrier, and release of the drug in the target site or binding with the attached probe[208]. For safe and effective therapy, it is necessary to target cancer cells specifically. Fortunately, tumors are characterized by elevated vascular permeability and decreased lymphatic drainage, allowing passive targeting[209]. In contrast, active targeting involves the interaction between targets and ligands; the latter are located on nanoparticles and specifically recognize targets overexpressed in cancer cells (peptides, amino acids, carbohydrates, antibodies)[210]. This results in endocytosis of the nanoparticle together with its payload, e.g., proteins or siRNA[211]. Nanotechnological vesicles come in various forms, such as polymeric or nonpolymeric nanoparticles, quantum specks, nanotubules, dendrimers, and lipid- or micelle-based nanoparticles.
Nanoparticle albumin-bound paclitaxel (nab-PTX), a novel form of the well-known agent, has demonstrated good effectiveness and safety. Nanoparticles enable the delivery of higher PTX levels in shorter infusion times compared to the commonly used solvent-based formulation[212]. Regarding stomach, breast, and lung tumors, the effectiveness of nab-PTX was found to be comparably effective as a standard of care[213-215]; however, it turned out to be of greater efficacy against NSCLC than docetaxel[216]. A phase III multicenter, randomized clinical trial investigated the efficiency of nab-PTX against NSCLC, presenting longer survival of patients treated with nab-PTX than those treated with docetaxel: OS of 16.2 months compared to 13.6 months and PFS of 4.2 months vs 3.4 months. Another significant difference was the objective response rate, which reached 29.9% for nanoparticle therapy compared to 15.4% for docetaxel[212]. This suggests that nanotechnology can be superior to standard treatment and perhaps will facilitate the development of future anti-cancer approaches.
Another method employs tumor-treating fields (TTFs), in which alternating, low-intensity, intermediate-frequency electric fields are transmitted transdermally to the tumor site; they selectively affect dividing cancer cells with minimal effect on non-extensively proliferating normal cells. The frequency of the alternating current can range from 100 to 400 kHz but is typically 150 kHz for NSCLC and pleural mesothelioma or 200 kHz for glioblastoma[159,217,218]. TTFs are approved by the FDA for the last two mentioned tumors, and are expected to receive approval soon for NSCLC[219-221]. Other frequencies are being evaluated to suit other cancer types. TTFs are applied by using special transducer arrays with electrodes that are activated sequentially to cause a change in the direction of the current[217]. The direction of electric charge flow and electrode placement can be determined using special software such as NovoTAL[159]. At a certain frequency, such alternating fields affect cellular structures. TTFs can be delivered to various parts of the human body, such as the patient’s scalp for glioma treatment or the thoracic region for NSCLC treatment. Electric fields can be applied for several hours a day (most often over 18 h daily) without significant harm to healthy normal cells but causing various effects on tumor cells[159,218]. TTFs cause an aberrant mitotic exit, prevent mitotic spindle assembly, perturb mitotic tubulin, and adjust cell membrane potential to increase Ca2+ influx and reduce microtubule polymerization[159,222]. They can also increase blood-brain barrier permeability, decrease cell dispersion, reverse markers of epithelial-to-mesenchymal transition, as well as inhibit cancer cell proliferation and metastasis[217]. Moreover, TTFs can induce autophagy, replication stress, MHC class II expression, macrophage differentiation, and various immunological effects such as immunogenic cell death[223,224]. This multimodality leads to substantial efficiency that is combined with great safety.
TTFs have been proven effective in targeting glioma, pleural mesothelioma, and NSCLC. For the latter, standard therapy (docetaxel and nivolumab, pembrolizumab, or atezolizumab) combined with TTFs achieved a median OS of 13.2 months compared to 9.9 months for standard treatment alone. Related adverse events were reported in 95 out of 133 patients; 81 patients had mild-to-moderate disorders of the skin and subcutaneous tissue[218]. In glioma, one of the clinical trials reported an OS of 27.2 months in newly diagnosed glioma patients receiving TTF with TMZ compared to 15.2 months in those with TMZ only[225]. Another study showed an OS of 20.9 months in the TTFs + TMZ group compared to 16 months in the TMZ group with no serious adverse events: Only mild-to-moderate skin toxicity underneath transducer arrays[159]. A phase II study on pleural mesothelioma showed a median OS of 18.2 months compared to 14.2 months in the control group and predominantly mild-to-moderate adverse events[220]. These results hold promise for future research on the efficiency of TTFs and their use in various cancer types.
Photodynamic therapy (PDT) is an approach targeting lesions that can be pre-cancerous such as actinic keratosis, or cancerous such as mycosis fungoides and basal cell carcinoma[226,227]. Despite being predominantly investigated in skin lesions, there is ongoing research on PDT in glioma, leukemia, and tumors of the bladder, colon, breast, and cervix[228,229]. Effective therapy requires three essential components, namely a photosensitizer (PS), light, and oxygen[230]. The PS is often an ointment or cream accumulating in pathological tissues[231]. It is designed to absorb light of a given wavelength to minimize unwanted activation after sunlight exposure. The PS only manifests its toxicity in the region of application, improving precision and safety[230]. The most commonly tested PS is hypericin[227,228]. After light absorption, the PS may transition to an excited singlet state and emit fluorescence as it returns to the ground state. The singlet state can also convert to a triplet state that interacts with endogenous substances to produce free radicals such as H2O2 and O2–. Alternatively, the triplet state can react with molecular oxygen to form 1O2; this is the most common scenario for ROS generation by PS in PDT[232-234]. Singlet oxygen generation leads to significant toxicity, signaling pathway alterations, apoptosis, and necrosis in PS-absorbing areas[228,230].
Clinical trials have shown that hypericin-associated PDT can effectively treat mycosis fungoides, a type of cutaneous T-cell lymphoma[227,228]. A phase III, double-blind, randomized clinical trial featuring 169 patients utilized hypericin as a PS, a light panel providing a 500-650 nm spectrum, and oxygen to enable ROS generation[227]. The results indicated that hypericin offers better efficiency than a placebo comparator, which was chosen due to the lack of FDA-approved standard skin-directed therapy for mycosis fungoides. The primary outcome was index lesion response rate (ILRR) measured after each of the three treatment cycles, the third of which was optional. The hypericin group demonstrated better ILRR than the placebo after the first cycle (16% vs 4%) and similar after the second cycle (40% vs 22%). In the third cycle, ILRR increased to 49% for patients who participated in all three cycles, but no data from the comparative placebo group was provided. Both hypericin and placebo achieved similar safety profiles but slightly in favor of the placebo ointment. The most common adverse events were subcutaneous tissue-related disorders (13.5%-17.3% across treatment cycles vs 10.5% for placebo) and application-site reactions (3.2%-6.9% across treatment cycles vs 4% for placebo). Since there were no serious adverse events related to PDT, further research is advisable to develop new PSs and enhance their universal application and effectiveness.
The continuously increasing incidence of cancer cases and the related mortality index emphasize the need for more precise detection and therapeutic approaches. Conventional methods, although still common in clinical practice, pose many limitations which justify the development of novel strategies for earlier diagnosis or more reliable and precise targeting of cancer. The present paper integrates available data on conventional and modern methods of cancer detection and therapy to facilitate an understanding of the rapidly advancing field and its future directions. Modern techniques have their drawbacks, and there is still a need to develop new detection strategies and therapeutic approaches to improve sensitivity, specificity, safety, and efficacy. Strategies utilizing DCs, nanotechnology, and plant-based agents are promising, but their effectiveness depends on the specific therapy, with some being superior or comparable to the standard of care. Oncolytic virotherapy greatly improves survival and entails only minor side effects, but multiple administrations may raise safety concerns. PDT or an exosome-based approach requires more data to understand related outcomes. ICIs and classical chimeric antigen receptor-based therapies pose many adverse effects, emphasizing the need for improved solutions. TTFs predominantly entail mild adverse events, and their ability to improve survival is encouraging. All things considered, the approval of some modern methods by the FDA emphasizes that efforts should continue.
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64442] [Article Influence: 16110.5] [Reference Citation Analysis (176)] |
| 2. | Lin L, Li Z, Yan L, Liu Y, Yang H, Li H. Global, regional, and national cancer incidence and death for 29 cancer groups in 2019 and trends analysis of the global cancer burden, 1990-2019. J Hematol Oncol. 2021;14:197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 214] [Article Influence: 53.5] [Reference Citation Analysis (0)] |
| 3. | Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87-108. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18694] [Cited by in RCA: 21363] [Article Influence: 2136.3] [Reference Citation Analysis (3)] |
| 4. | Lortet-Tieulent J, Georges D, Bray F, Vaccarella S. Profiling global cancer incidence and mortality by socioeconomic development. Int J Cancer. 2020;147:3029-3036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 5. | Fidler MM, Soerjomataram I, Bray F. A global view on cancer incidence and national levels of the human development index. Int J Cancer. 2016;139:2436-2446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 213] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 6. | Ferlay J, Colombet M, Soerjomataram I, Parkin DM, Piñeros M, Znaor A, Bray F. Cancer statistics for the year 2020: An overview. Int J Cancer. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2411] [Cited by in RCA: 2944] [Article Influence: 736.0] [Reference Citation Analysis (7)] |
| 7. | Almasan I, Piciu D. Triple Primary Malignancies: Tumor Associations, Survival, and Clinicopathological Analysis: A 25-Year Single-Institution Experience. Healthcare (Basel). 2023;11. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Chen Z, Xu L, Shi W, Zeng F, Zhuo R, Hao X, Fan P. Trends of female and male breast cancer incidence at the global, regional, and national levels, 1990-2017. Breast Cancer Res Treat. 2020;180:481-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 86] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 9. | Zhu B, Wu X, Piao H, Xu S, Yao B. A Comparison of Epidemiological Characteristics of Central Nervous System Tumours in China and Globally from 1990 to 2019. Neuroepidemiology. 2021;55:460-472. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 10. | Mohammed S, Dinesan M, Ajayakumar T. Survival and quality of life analysis in glioblastoma multiforme with adjuvant chemoradiotherapy: a retrospective study. Rep Pract Oncol Radiother. 2022;27:1026-1036. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 94] [Reference Citation Analysis (0)] |
| 11. | Ilic I, Ilic M. International patterns and trends in the brain cancer incidence and mortality: An observational study based on the global burden of disease. Heliyon. 2023;9:e18222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 30] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 12. | Debela DT, Muzazu SG, Heraro KD, Ndalama MT, Mesele BW, Haile DC, Kitui SK, Manyazewal T. New approaches and procedures for cancer treatment: Current perspectives. SAGE Open Med. 2021;9:20503121211034366. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 692] [Cited by in RCA: 666] [Article Influence: 166.5] [Reference Citation Analysis (0)] |
| 13. | Emery J, Chiang P. The role of risk tools in diagnosing cancer in primary care. Aust Fam Physician. 2014;43:508-512. [PubMed] |
| 14. | Yazdanian A. Oncology Information System: A Qualitative Study to Identify Cancer Patient Care Workflows. J Med Life. 2020;13:469-474. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Wilkinson AN. Cancer diagnosis in primary care: Six steps to reducing the diagnostic interval. Can Fam Physician. 2021;67:265-268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 16. | Dabeer S, Khan MM, Islam S. Cancer diagnosis in histopathological image: CNN based approach. Informatics in Medicine Unlocked. 2019;16:100231. [RCA] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 17. | Zeng H, Ran X, An L, Zheng R, Zhang S, Ji JS, Zhang Y, Chen W, Wei W, He J; HBCR Working Group. Disparities in stage at diagnosis for five common cancers in China: a multicentre, hospital-based, observational study. Lancet Public Health. 2021;6:e877-e887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 115] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 18. | Liao CK, Kuo YT, Lin YC, Chern YJ, Hsu YJ, Yu YL, Chiang JM, Hsieh PS, Yeh CY, You JF. Neoadjuvant Short-Course Radiotherapy Followed by Consolidation Chemotherapy before Surgery for Treating Locally Advanced Rectal Cancer: A Systematic Review and Meta-Analysis. Curr Oncol. 2022;29:3708-3727. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Tilsed CM, Fisher SA, Nowak AK, Lake RA, Lesterhuis WJ. Cancer chemotherapy: insights into cellular and tumor microenvironmental mechanisms of action. Front Oncol. 2022;12:960317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 80] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 20. | Ali JH, Walter M. Combining old and new concepts in targeting telomerase for cancer therapy: transient, immediate, complete and combinatory attack (TICCA). Cancer Cell Int. 2023;23:197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 21. | Amjad MT, Chidharla A, Kasi A. Cancer Chemotherapy. 2023 Feb 27. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2024 Jan-. [PubMed] |
| 22. | Yousefi Rizi HA, Hoon Shin D, Yousefi Rizi S. Polymeric Nanoparticles in Cancer Chemotherapy: A Narrative Review. Iran J Public Health. 2022;51:226-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 23. | Yeldag G, Rice A, Del Río Hernández A. Chemoresistance and the Self-Maintaining Tumor Microenvironment. Cancers (Basel). 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 155] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 24. | Wei G, Wang Y, Yang G, Ju R. Recent progress in nanomedicine for enhanced cancer chemotherapy. Theranostics. 2021;11:6370-6392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 264] [Article Influence: 66.0] [Reference Citation Analysis (0)] |
| 25. | Baskar R, Lee KA, Yeo R, Yeoh KW. Cancer and radiation therapy: current advances and future directions. Int J Med Sci. 2012;9:193-199. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1193] [Cited by in RCA: 1570] [Article Influence: 120.8] [Reference Citation Analysis (0)] |
| 26. | Olivares-Urbano MA, Griñán-Lisón C, Marchal JA, Núñez MI. CSC Radioresistance: A Therapeutic Challenge to Improve Radiotherapy Effectiveness in Cancer. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 148] [Article Influence: 29.6] [Reference Citation Analysis (0)] |
| 27. | Vinod SK, Hau E. Radiotherapy treatment for lung cancer: Current status and future directions. Respirology. 2020;25 Suppl 2:61-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 218] [Article Influence: 43.6] [Reference Citation Analysis (0)] |
| 28. | Fendler W, Tomasik B, Atkins K, Stawiski K, Chałubińska-Fendler J, Kozono D. The clinician's guide to radiotherapy complications. Pol Arch Intern Med. 2022;132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 29. | Dilalla V, Chaput G, Williams T, Sultanem K. Radiotherapy side effects: integrating a survivorship clinical lens to better serve patients. Curr Oncol. 2020;27:107-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 82] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 30. | Kishan AU, Cook RR, Ciezki JP, Ross AE, Pomerantz MM, Nguyen PL, Shaikh T, Tran PT, Sandler KA, Stock RG, Merrick GS, Demanes DJ, Spratt DE, Abu-Isa EI, Wedde TB, Lilleby W, Krauss DJ, Shaw GK, Alam R, Reddy CA, Stephenson AJ, Klein EA, Song DY, Tosoian JJ, Hegde JV, Yoo SM, Fiano R, D'Amico AV, Nickols NG, Aronson WJ, Sadeghi A, Greco S, Deville C, McNutt T, DeWeese TL, Reiter RE, Said JW, Steinberg ML, Horwitz EM, Kupelian PA, King CR. Radical Prostatectomy, External Beam Radiotherapy, or External Beam Radiotherapy With Brachytherapy Boost and Disease Progression and Mortality in Patients With Gleason Score 9-10 Prostate Cancer. JAMA. 2018;319:896-905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 254] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 31. | Pędziwiatr M, Małczak P, Major P, Witowski J, Kuśnierz-Cabala B, Ceranowicz P, Budzyński A. Minimally invasive pancreatic cancer surgery: What is the current evidence? Med Oncol. 2017;34:125. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Chen Z, Zhang P, Xu Y, Yan J, Liu Z, Lau WB, Lau B, Li Y, Zhao X, Wei Y, Zhou S. Surgical stress and cancer progression: the twisted tango. Mol Cancer. 2019;18:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 143] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 33. | Johnstone C, Rich SE. Bleeding in cancer patients and its treatment: a review. Ann Palliat Med. 2018;7:265-273. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 118] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 34. | Chaput G, Ibrahim M, Towers A. Cancer-related lymphedema: clinical pearls for providers. Curr Oncol. 2020;27:336-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 35. | Tohme S, Simmons RL, Tsung A. Surgery for Cancer: A Trigger for Metastases. Cancer Res. 2017;77:1548-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 255] [Cited by in RCA: 481] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 36. | Coffey JC, Smith MJ, Wang JH, Bouchier-Hayes D, Cotter TG, Redmond HP. Cancer surgery: risks and opportunities. Bioessays. 2006;28:433-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 21] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 37. | Salibasic M, Pusina S, Bicakcic E, Pasic A, Gavric I, Kulovic E, Rovcanin A, Beslija S. Colorectal Cancer Surgical Treatment, our Experience. Med Arch. 2019;73:412-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 42] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 38. | Guerrini GP, Esposito G, Magistri P, Serra V, Guidetti C, Olivieri T, Catellani B, Assirati G, Ballarin R, Di Sandro S, Di Benedetto F. Robotic versus laparoscopic gastrectomy for gastric cancer: The largest meta-analysis. Int J Surg. 2020;82:210-228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 117] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 39. | Liu G, Zhang S, Zhang Y, Fu X, Liu X. Robotic Surgery in Rectal Cancer: Potential, Challenges, and Opportunities. Curr Treat Options Oncol. 2022;23:961-979. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 34] [Reference Citation Analysis (0)] |
| 40. | Schiffman JD, Fisher PG, Gibbs P. Early detection of cancer: past, present, and future. Am Soc Clin Oncol Educ Book. 2015;57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 166] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 41. | Smith RA, Andrews KS, Brooks D, Fedewa SA, Manassaram-Baptiste D, Saslow D, Wender RC. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA Cancer J Clin. 2019;69:184-210. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 422] [Article Influence: 70.3] [Reference Citation Analysis (0)] |
| 42. | Rucinska M, Nawrocki S. COVID-19 Pandemic: Impact on Cancer Patients. Int J Environ Res Public Health. 2022;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 43. | Connal S, Cameron JM, Sala A, Brennan PM, Palmer DS, Palmer JD, Perlow H, Baker MJ. Liquid biopsies: the future of cancer early detection. J Transl Med. 2023;21:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 138] [Article Influence: 69.0] [Reference Citation Analysis (0)] |
| 44. | Mariotto AB, Enewold L, Zhao J, Zeruto CA, Yabroff KR. Medical Care Costs Associated with Cancer Survivorship in the United States. Cancer Epidemiol Biomarkers Prev. 2020;29:1304-1312. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 393] [Article Influence: 78.6] [Reference Citation Analysis (0)] |
| 45. | Alix-Panabières C, Pantel K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021;11:858-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 581] [Article Influence: 145.3] [Reference Citation Analysis (0)] |
| 46. | Rossi G, Ignatiadis M. Promises and Pitfalls of Using Liquid Biopsy for Precision Medicine. Cancer Res. 2019;79:2798-2804. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 98] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 47. | Armakolas A, Kotsari M, Koskinas J. Liquid Biopsies, Novel Approaches and Future Directions. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 57] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 48. | Ding Z, Wang N, Ji N, Chen ZS. Proteomics technologies for cancer liquid biopsies. Mol Cancer. 2022;21:53. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 97] [Cited by in RCA: 121] [Article Influence: 40.3] [Reference Citation Analysis (0)] |
| 49. | Hemenway G, Tierno MB, Nejati R, Sosa R, Zibelman M. Clinical Utility of Liquid Biopsy to Identify Genomic Heterogeneity and Secondary Cancer Diagnoses: A Case Report. Case Rep Oncol. 2022;15:78-85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 50. | Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, El-Rifai W, Bedognetti D, Batra SK, Haris M, Bhat AA, Macha MA. Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer. 2022;21:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 416] [Article Influence: 138.7] [Reference Citation Analysis (0)] |
| 51. | Karimi F, Azadbakht O, Veisi A, Sabaghan M, Owjfard M, Kharazinejad E, Dinarvand N. Liquid biopsy in ovarian cancer: advantages and limitations for prognosis and diagnosis. Med Oncol. 2023;40:265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 52. | Nikanjam M, Kato S, Kurzrock R. Liquid biopsy: current technology and clinical applications. J Hematol Oncol. 2022;15:131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 423] [Reference Citation Analysis (0)] |
| 53. | Deng Z, Wu S, Wang Y, Shi D. Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine. 2022;83:104237. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 147] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 54. | Gogoi P, Sepehri S, Zhou Y, Gorin MA, Paolillo C, Capoluongo E, Gleason K, Payne A, Boniface B, Cristofanilli M, Morgan TM, Fortina P, Pienta KJ, Handique K, Wang Y. Development of an Automated and Sensitive Microfluidic Device for Capturing and Characterizing Circulating Tumor Cells (CTCs) from Clinical Blood Samples. PLoS One. 2016;11:e0147400. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 72] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 55. | Tellez-Gabriel M, Heymann MF, Heymann D. Circulating Tumor Cells as a Tool for Assessing Tumor Heterogeneity. Theranostics. 2019;9:4580-4594. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 67] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 56. | Scher HI, Jia X, de Bono JS, Fleisher M, Pienta KJ, Raghavan D, Heller G. Circulating tumour cells as prognostic markers in progressive, castration-resistant prostate cancer: a reanalysis of IMMC38 trial data. Lancet Oncol. 2009;10:233-239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 514] [Cited by in RCA: 490] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 57. | Maas M, Hegemann M, Rausch S, Bedke J, Stenzl A, Todenhöfer T. Circulating tumor cells and their role in prostate cancer. Asian J Androl. 2017;21:24-31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 58. | Oshi M, Murthy V, Takahashi H, Huyser M, Okano M, Tokumaru Y, Rashid OM, Matsuyama R, Endo I, Takabe K. Urine as a Source of Liquid Biopsy for Cancer. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 59. | Eslami-S Z, Cortés-Hernández LE, Alix-Panabières C. Epithelial Cell Adhesion Molecule: An Anchor to Isolate Clinically Relevant Circulating Tumor Cells. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 80] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 60. | Theil G, Bialek J, Weiß C, Lindner F, Fornara P. Strategies for Isolating and Propagating Circulating Tumor Cells in Men with Metastatic Prostate Cancer. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 61. | Gascoyne PR, Shim S. Isolation of circulating tumor cells by dielectrophoresis. Cancers (Basel). 2014;6:545-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 183] [Cited by in RCA: 190] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 62. | Denis JA, Patroni A, Guillerm E, Pépin D, Benali-Furet N, Wechsler J, Manceau G, Bernard M, Coulet F, Larsen AK, Karoui M, Lacorte JM. Droplet digital PCR of circulating tumor cells from colorectal cancer patients can predict KRAS mutations before surgery. Mol Oncol. 2016;10:1221-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 76] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 63. | Auwal A, Hossain MM, Pronoy TUH, Rashel K, Nurujjaman M, Lam AK, Islam F. Clinical significance of genomic sequencing of circulating tumour cells (CTCs) in cancer. The Journal of Liquid Biopsy. 2024;3:100135. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 64. | Mirza S, Bhadresha K, Mughal MJ, McCabe M, Shahbazi R, Ruff P, Penny C. Liquid biopsy approaches and immunotherapy in colorectal cancer for precision medicine: Are we there yet? Front Oncol. 2022;12:1023565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 65. | Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci (Landmark Ed). 2010;15:166-179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 409] [Cited by in RCA: 557] [Article Influence: 37.1] [Reference Citation Analysis (0)] |
| 66. | Akolkar D, Patil D, Crook T, Limaye S, Page R, Datta V, Patil R, Sims C, Ranade A, Fulmali P, Srivastava N, Devhare P, Apurwa S, Patel S, Patil S, Adhav A, Pawar S, Ainwale A, Chougule R, Apastamb M, Srinivasan A, Datar R. Circulating ensembles of tumor-associated cells: A redoubtable new systemic hallmark of cancer. Int J Cancer. 2020;146:3485-3494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 67. | Sukowati CH, Anfuso B, Crocé LS, Tiribelli C. The role of multipotent cancer associated fibroblasts in hepatocarcinogenesis. BMC Cancer. 2015;15:188. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 68. | Fotsitzoudis C, Koulouridi A, Messaritakis I, Konstantinidis T, Gouvas N, Tsiaoussis J, Souglakos J. Cancer-Associated Fibroblasts: The Origin, Biological Characteristics and Role in Cancer-A Glance on Colorectal Cancer. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 69. | Shiga K, Hara M, Nagasaki T, Sato T, Takahashi H, Takeyama H. Cancer-Associated Fibroblasts: Their Characteristics and Their Roles in Tumor Growth. Cancers (Basel). 2015;7:2443-2458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 465] [Cited by in RCA: 593] [Article Influence: 59.3] [Reference Citation Analysis (0)] |
| 70. | Rizzolio S, Giordano S, Corso S. The importance of being CAFs (in cancer resistance to targeted therapies). J Exp Clin Cancer Res. 2022;41:319. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 33] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 71. | Wang W, Cheng B, Yu Q. Cancer-associated fibroblasts as accomplices to confer therapeutic resistance in cancer. Cancer Drug Resist. 2022;5:889-901. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 72. | Najafi S, Asemani Y, Majidpoor J, Mahmoudi R, Aghaei-Zarch SM, Mortezaee K. Tumor-educated platelets. Clin Chim Acta. 2024;552:117690. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 73. | Varkey J, Nicolaides T. Tumor-Educated Platelets: A Review of Current and Potential Applications in Solid Tumors. Cureus. 2021;13:e19189. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 74. | Chebbo M, Assou S, Pantesco V, Duez C, Alessi MC, Chanez P, Gras D. Platelets Purification Is a Crucial Step for Transcriptomic Analysis. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 75. | Jin J, Shao Y, Zhang J, Cao J, Tao Z, Hu X. High-purity isolation platelets by gradient centrifugation plus filtration. Int J Lab Hematol. 2023;45:187-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 76. | Yang L, Jiang Q, Li DZ, Zhou X, Yu DS, Zhong J. TIMP1 mRNA in tumor-educated platelets is diagnostic biomarker for colorectal cancer. Aging (Albany NY). 2019;11:8998-9012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 50] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 77. | Roweth HG, Battinelli EM. Lessons to learn from tumor-educated platelets. Blood. 2021;137:3174-3180. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 91] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 78. | Stejskal P, Goodarzi H, Srovnal J, Hajdúch M, van 't Veer LJ, Magbanua MJM. Circulating tumor nucleic acids: biology, release mechanisms, and clinical relevance. Mol Cancer. 2023;22:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 158] [Cited by in RCA: 126] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 79. | Vymetalkova V, Cervena K, Bartu L, Vodicka P. Circulating Cell-Free DNA and Colorectal Cancer: A Systematic Review. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 79] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 80. | Hu Z, Chen H, Long Y, Li P, Gu Y. The main sources of circulating cell-free DNA: Apoptosis, necrosis and active secretion. Crit Rev Oncol Hematol. 2021;157:103166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 79] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 81. | Gao Q, Zeng Q, Wang Z, Li C, Xu Y, Cui P, Zhu X, Lu H, Wang G, Cai S, Wang J, Fan J. Circulating cell-free DNA for cancer early detection. Innovation (Camb). 2022;3:100259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 61] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 82. | Spector BL, Harrell L, Sante D, Wyckoff GJ, Willig L. The methylome and cell-free DNA: current applications in medicine and pediatric disease. Pediatr Res. 2023;94:89-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 83. | Zhou Q, Moser T, Perakis S, Heitzer E. Untargeted profiling of cell-free circulating DNA. Transl Cancer Res. 2018;7:S140-S152. [RCA] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 84. | Bronkhorst AJ, Ungerer V, Holdenrieder S. The emerging role of cell-free DNA as a molecular marker for cancer management. Biomol Detect Quantif. 2019;17:100087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 376] [Cited by in RCA: 384] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 85. | Pascual J, Attard G, Bidard FC, Curigliano G, De Mattos-Arruda L, Diehn M, Italiano A, Lindberg J, Merker JD, Montagut C, Normanno N, Pantel K, Pentheroudakis G, Popat S, Reis-Filho JS, Tie J, Seoane J, Tarazona N, Yoshino T, Turner NC. ESMO recommendations on the use of circulating tumour DNA assays for patients with cancer: a report from the ESMO Precision Medicine Working Group. Ann Oncol. 2022;33:750-768. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 351] [Article Influence: 117.0] [Reference Citation Analysis (0)] |
| 86. | Telekes A, Horváth A. The Role of Cell-Free DNA in Cancer Treatment Decision Making. Cancers (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 87. | Polatoglou E, Mayer Z, Ungerer V, Bronkhorst AJ, Holdenrieder S. Isolation and Quantification of Plasma Cell-Free DNA Using Different Manual and Automated Methods. Diagnostics (Basel). 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 88. | Fettke H, Kwan EM, Azad AA. Cell-free DNA in cancer: current insights. Cell Oncol (Dordr). 2019;42:13-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 89. | Chen M, Zhao H. Next-generation sequencing in liquid biopsy: cancer screening and early detection. Hum Genomics. 2019;13:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 162] [Cited by in RCA: 328] [Article Influence: 54.7] [Reference Citation Analysis (0)] |
| 90. | Kato R, Hayashi H, Sakai K, Suzuki S, Haratani K, Takahama T, Tanizaki J, Nonagase Y, Tanaka K, Yoshida T, Takeda M, Yonesaka K, Kaneda H, Nishio K, Nakagawa K. CAPP-seq analysis of circulating tumor DNA from patients with EGFR T790M-positive lung cancer after osimertinib. Int J Clin Oncol. 2021;26:1628-1639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 91. | Gale D, Lawson ARJ, Howarth K, Madi M, Durham B, Smalley S, Calaway J, Blais S, Jones G, Clark J, Dimitrov P, Pugh M, Woodhouse S, Epstein M, Fernandez-Gonzalez A, Whale AS, Huggett JF, Foy CA, Jones GM, Raveh-Amit H, Schmitt K, Devonshire A, Green E, Forshew T, Plagnol V, Rosenfeld N. Development of a highly sensitive liquid biopsy platform to detect clinically-relevant cancer mutations at low allele fractions in cell-free DNA. PLoS One. 2018;13:e0194630. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 125] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 92. | Wu J, Hu S, Zhang L, Xin J, Sun C, Wang L, Ding K, Wang B. Tumor circulome in the liquid biopsies for cancer diagnosis and prognosis. Theranostics. 2020;10:4544-4556. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 93] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 93. | Kan CM, Pei XM, Yeung MHY, Jin N, Ng SSM, Tsang HF, Cho WCS, Yim AK, Yu AC, Wong SCC. Exploring the Role of Circulating Cell-Free RNA in the Development of Colorectal Cancer. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 94. | Debattista J, Grech L, Scerri C, Grech G. Copy Number Variations as Determinants of Colorectal Tumor Progression in Liquid Biopsies. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 95. | Boussios S, Ozturk MA, Moschetta M, Karathanasi A, Zakynthinakis-Kyriakou N, Katsanos KH, Christodoulou DK, Pavlidis N. The Developing Story of Predictive Biomarkers in Colorectal Cancer. J Pers Med. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 138] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 96. | Ortiz-Quintero B. Cell-free microRNAs in blood and other body fluids, as cancer biomarkers. Cell Prolif. 2016;49:281-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 97. | Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141:1202-1207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 1814] [Article Influence: 226.8] [Reference Citation Analysis (0)] |
| 98. | Menon A, Abd-Aziz N, Khalid K, Poh CL, Naidu R. miRNA: A Promising Therapeutic Target in Cancer. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 189] [Reference Citation Analysis (0)] |
| 99. | Miyoshi J, Zhu Z, Luo A, Toden S, Zhou X, Izumi D, Kanda M, Takayama T, Parker IM, Wang M, Gao F, Zaidi AH, Baba H, Kodera Y, Cui Y, Wang X, Liu Z, Goel A. A microRNA-based liquid biopsy signature for the early detection of esophageal squamous cell carcinoma: a retrospective, prospective and multicenter study. Mol Cancer. 2022;21:44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 100. | Zhang K, Wang YY, Xu Y, Zhang L, Zhu J, Si PC, Wang YW, Ma R. A two-miRNA signature of upregulated miR-185-5p and miR-362-5p as a blood biomarker for breast cancer. Pathol Res Pract. 2021;222:153458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 101. | Chen J, Yao D, Chen W, Li Z, Guo Y, Zhu F, Hu X. Serum exosomal miR-451a acts as a candidate marker for pancreatic cancer. Int J Biol Markers. 2022;37:74-80. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 102. | Aprile M, Katopodi V, Leucci E, Costa V. LncRNAs in Cancer: From garbage to Junk. Cancers (Basel). 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 103. | Tan YT, Lin JF, Li T, Li JJ, Xu RH, Ju HQ. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun (Lond). 2021;41:109-120. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 451] [Article Influence: 90.2] [Reference Citation Analysis (0)] |
| 104. | Beylerli O, Gareev I, Sufianov A, Ilyasova T, Guang Y. Long noncoding RNAs as promising biomarkers in cancer. Noncoding RNA Res. 2022;7:66-70. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 103] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 105. | Liu J, Chen Y, Pei F, Zeng C, Yao Y, Liao W, Zhao Z. Extracellular Vesicles in Liquid Biopsies: Potential for Disease Diagnosis. Biomed Res Int. 2021;2021:6611244. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 106. | Urabe F, Kosaka N, Ito K, Kimura T, Egawa S, Ochiya T. Extracellular vesicles as biomarkers and therapeutic targets for cancer. Am J Physiol Cell Physiol. 2020;318:C29-C39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 222] [Article Influence: 37.0] [Reference Citation Analysis (0)] |
| 107. | Dai J, Su Y, Zhong S, Cong L, Liu B, Yang J, Tao Y, He Z, Chen C, Jiang Y. Exosomes: key players in cancer and potential therapeutic strategy. Signal Transduct Target Ther. 2020;5:145. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 256] [Cited by in RCA: 789] [Article Influence: 157.8] [Reference Citation Analysis (0)] |
| 108. | Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X. Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer. 2022;21:56. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 481] [Article Influence: 160.3] [Reference Citation Analysis (0)] |
| 109. | Muller L, Hong CS, Stolz DB, Watkins SC, Whiteside TL. Isolation of biologically-active exosomes from human plasma. J Immunol Methods. 2014;411:55-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 364] [Article Influence: 33.1] [Reference Citation Analysis (0)] |
| 110. | Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, Shen XT, Zhu WW, Geng Y, Lu L, Jia HL, Qin LX, Dong QZ. Isolation and characterization of exosomes for cancer research. J Hematol Oncol. 2020;13:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 323] [Cited by in RCA: 322] [Article Influence: 64.4] [Reference Citation Analysis (0)] |
| 111. | Kwong GA, Ghosh S, Gamboa L, Patriotis C, Srivastava S, Bhatia SN. Synthetic biomarkers: a twenty-first century path to early cancer detection. Nat Rev Cancer. 2021;21:655-668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |
| 112. | Pulumati A, Pulumati A, Dwarakanath BS, Verma A, Papineni RVL. Technological advancements in cancer diagnostics: Improvements and limitations. Cancer Rep (Hoboken). 2023;6:e1764. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 94] [Article Influence: 47.0] [Reference Citation Analysis (0)] |
| 113. | Hao L, Zhao RT, Welch NL, Tan EKW, Zhong Q, Harzallah NS, Ngambenjawong C, Ko H, Fleming HE, Sabeti PC, Bhatia SN. CRISPR-Cas-amplified urinary biomarkers for multiplexed and portable cancer diagnostics. Nat Nanotechnol. 2023;18:798-807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 58] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 114. | Ovechkina VS, Zakian SM, Medvedev SP, Valetdinova KR. Genetically Encoded Fluorescent Biosensors for Biomedical Applications. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 14] [Reference Citation Analysis (0)] |
| 115. | Banerjee P, Franz B, Bhunia AK. Mammalian cell-based sensor system. Adv Biochem Eng Biotechnol. 2010;117:21-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 116. | Kouidhi S, Noman MZ, Kieda C, Elgaaied AB, Chouaib S. Intrinsic and Tumor Microenvironment-Induced Metabolism Adaptations of T Cells and Impact on Their Differentiation and Function. Front Immunol. 2016;7:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 117. | Somasundaram R, Zhang G, Fukunaga-Kalabis M, Perego M, Krepler C, Xu X, Wagner C, Hristova D, Zhang J, Tian T, Wei Z, Liu Q, Garg K, Griss J, Hards R, Maurer M, Hafner C, Mayerhöfer M, Karanikas G, Jalili A, Bauer-Pohl V, Weihsengruber F, Rappersberger K, Koller J, Lang R, Hudgens C, Chen G, Tetzlaff M, Wu L, Frederick DT, Scolyer RA, Long GV, Damle M, Ellingsworth C, Grinman L, Choi H, Gavin BJ, Dunagin M, Raj A, Scholler N, Gross L, Beqiri M, Bennett K, Watson I, Schaider H, Davies MA, Wargo J, Czerniecki BJ, Schuchter L, Herlyn D, Flaherty K, Herlyn M, Wagner SN. Tumor-associated B-cells induce tumor heterogeneity and therapy resistance. Nat Commun. 2017;8:607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 109] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 118. | Vitale M, Cantoni C, Pietra G, Mingari MC, Moretta L. Effect of tumor cells and tumor microenvironment on NK-cell function. Eur J Immunol. 2014;44:1582-1592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 119. | Zhou S, Gravekamp C, Bermudes D, Liu K. Tumour-targeting bacteria engineered to fight cancer. Nat Rev Cancer. 2018;18:727-743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 529] [Article Influence: 75.6] [Reference Citation Analysis (0)] |
| 120. | Tipirneni KE, Rosenthal EL, Moore LS, Haskins AD, Udayakumar N, Jani AH, Carroll WR, Morlandt AB, Bogyo M, Rao J, Warram JM. Fluorescence Imaging for Cancer Screening and Surveillance. Mol Imaging Biol. 2017;19:645-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 46] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 121. | Vonk J, de Wit JG, Voskuil FJ, Witjes MJH. Improving oral cavity cancer diagnosis and treatment with fluorescence molecular imaging. Oral Dis. 2021;27:21-26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 122. | Nguyen QT, Olson ES, Aguilera TA, Jiang T, Scadeng M, Ellies LG, Tsien RY. Surgery with molecular fluorescence imaging using activatable cell-penetrating peptides decreases residual cancer and improves survival. Proc Natl Acad Sci U S A. 2010;107:4317-4322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 361] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 123. | Miampamba M, Liu J, Harootunian A, Gale AJ, Baird S, Chen SL, Nguyen QT, Tsien RY, González JE. Sensitive in vivo Visualization of Breast Cancer Using Ratiometric Protease-activatable Fluorescent Imaging Agent, AVB-620. Theranostics. 2017;7:3369-3386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 71] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 124. | Jiang T, Olson ES, Nguyen QT, Roy M, Jennings PA, Tsien RY. Tumor imaging by means of proteolytic activation of cell-penetrating peptides. Proc Natl Acad Sci U S A. 2004;101:17867-17872. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 641] [Cited by in RCA: 651] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 125. | Keating JJ, Okusanya OT, De Jesus E, Judy R, Jiang J, Deshpande C, Nie S, Low P, Singhal S. Intraoperative Molecular Imaging of Lung Adenocarcinoma Can Identify Residual Tumor Cells at the Surgical Margins. Mol Imaging Biol. 2016;18:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 32] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 126. | Low PS, Singhal S, Srinivasarao M. Fluorescence-guided surgery of cancer: applications, tools and perspectives. Curr Opin Chem Biol. 2018;45:64-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 57] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 127. | Levine A, Markowitz O. Introduction to reflectance confocal microscopy and its use in clinical practice. JAAD Case Rep. 2018;4:1014-1023. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 67] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 128. | Farabi B, Khan S, Jamgochian M, Atak MF, Jain M, Rao BK. The role of reflectance confocal microscopy in the diagnosis and management of pigmentary disorders: A review. J Cosmet Dermatol. 2023;22:3213-3222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 129. | Guida S, Pellacani G, Ciardo S, Longo C. Reflectance Confocal Microscopy of Aging Skin and Skin Cancer. Dermatol Pract Concept. 2021;11:e2021068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 130. | Menditti D, Santagata M, Imola G, Staglianò S, Vitagliano R, Boschetti CE, Inchingolo AM. Personalized Medicine in Oral Oncology: Imaging Methods and Biological Markers to Support Diagnosis of Oral Squamous Cell Carcinoma (OSCC): A Narrative Literature Review. J Pers Med. 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 131. | Collier T, Guillaud M, Follen M, Malpica A, Richards-Kortum R. Real-time reflectance confocal microscopy: comparison of two-dimensional images and three-dimensional image stacks for detection of cervical precancer. J Biomed Opt. 2007;12:024021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 132. | Ahlgrimm-Siess V, Laimer M, Rabinovitz HS, Oliviero M, Hofmann-Wellenhof R, Marghoob AA, Scope A. Confocal Microscopy in Skin Cancer. Curr Dermatol Rep. 2018;7:105-118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 133. | Woliner-van der Weg W, Peppelman M, Elshot YS, Visch MB, Crijns MB, Alkemade HAC, Bronkhorst EM, Adang E, Amir A, Gerritsen MJP, van Erp PEJ, Lubeek SFK. Biopsy outperforms reflectance confocal microscopy in diagnosing and subtyping basal cell carcinoma: results and experiences from a randomized controlled multicentre trial. Br J Dermatol. 2021;184:663-671. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 134. | Hoang P, Wallace A, Sugi M, Fleck A, Pershad Y, Dahiya N, Albadawi H, Knuttinen G, Naidu S, Oklu R. Elastography techniques in the evaluation of deep vein thrombosis. Cardiovasc Diagn Ther. 2017;7:S238-S245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 135. | Emig R, Zgierski-Johnston CM, Timmermann V, Taberner AJ, Nash MP, Kohl P, Peyronnet R. Passive myocardial mechanical properties: meaning, measurement, models. Biophys Rev. 2021;13:587-610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 34] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 136. | Ozturk A, Grajo JR, Dhyani M, Anthony BW, Samir AE. Principles of ultrasound elastography. Abdom Radiol (NY). 2018;43:773-785. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 159] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 137. | Sigrist RMS, Liau J, Kaffas AE, Chammas MC, Willmann JK. Ultrasound Elastography: Review of Techniques and Clinical Applications. Theranostics. 2017;7:1303-1329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 954] [Cited by in RCA: 1090] [Article Influence: 136.3] [Reference Citation Analysis (0)] |
| 138. | Doherty JR, Trahey GE, Nightingale KR, Palmeri ML. Acoustic radiation force elasticity imaging in diagnostic ultrasound. IEEE Trans Ultrason Ferroelectr Freq Control. 2013;60:685-701. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 191] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 139. | Taljanovic MS, Gimber LH, Becker GW, Latt LD, Klauser AS, Melville DM, Gao L, Witte RS. Shear-Wave Elastography: Basic Physics and Musculoskeletal Applications. Radiographics. 2017;37:855-870. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 451] [Article Influence: 56.4] [Reference Citation Analysis (0)] |
| 140. | Liu C, Zhou J, Chang C, Zhi W. Feasibility of Shear Wave Elastography Imaging for Evaluating the Biological Behavior of Breast Cancer. Front Oncol. 2021;11:820102. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 141. | Srinivasa Babu A, Wells ML, Teytelboym OM, Mackey JE, Miller FH, Yeh BM, Ehman RL, Venkatesh SK. Elastography in Chronic Liver Disease: Modalities, Techniques, Limitations, and Future Directions. Radiographics. 2016;36:1987-2006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 152] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 142. | Bruce M, Kolokythas O, Ferraioli G, Filice C, O'Donnell M. Limitations and artifacts in shear-wave elastography of the liver. Biomed Eng Lett. 2017;7:81-89. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 46] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 143. | Hanna TP, King WD, Thibodeau S, Jalink M, Paulin GA, Harvey-Jones E, O'Sullivan DE, Booth CM, Sullivan R, Aggarwal A. Mortality due to cancer treatment delay: systematic review and meta-analysis. BMJ. 2020;371:m4087. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 383] [Cited by in RCA: 768] [Article Influence: 153.6] [Reference Citation Analysis (0)] |
| 144. | Wait S, Han D, Muthu V, Oliver K, Chrostowski S, Florindi F, de Lorenzo F, Gandouet B, Spurrier G, Ryll B, Wierinck L, Szucs T, Hess R, Rosvall-puplett T, Roediger A, Arora J, Yared W, Hanna S, Steinmann K, Aapro M. Towards sustainable cancer care: Reducing inefficiencies, improving outcomes-A policy report from the All. Can initiative. J Cancer Policy. 2017;13:47-64. [RCA] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 37] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 145. | Heinhuis KM, Ros W, Kok M, Steeghs N, Beijnen JH, Schellens JHM. Enhancing antitumor response by combining immune checkpoint inhibitors with chemotherapy in solid tumors. Ann Oncol. 2019;30:219-235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 385] [Article Influence: 64.2] [Reference Citation Analysis (0)] |
| 146. | Peters C, Rabkin SD. Designing Herpes Viruses as Oncolytics. Mol Ther Oncolytics. 2015;2:15010-15010. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 80] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 147. | Bommareddy PK, Silk AW, Kaufman HL. Intratumoral Approaches for the Treatment of Melanoma. Cancer J. 2017;23:40-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 148. | Martinez-Quintanilla J, Seah I, Chua M, Shah K. Oncolytic viruses: overcoming translational challenges. J Clin Invest. 2019;129:1407-1418. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 149. | Jiang S, Chai H, Tang Q, Shi Z, Zhou L. Clinical advances in oncolytic virus therapy for malignant glioma: a systematic review. Discov Oncol. 2023;14:183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 150. | Salloum A, Koblinski J, Bazzi N, Zeitouni NC. Talimogene Laherparepvec in Non-Melanoma Cancers. J Clin Aesthet Dermatol. 2021;14:18-25. [PubMed] |
| 151. | Kohlhapp FJ, Kaufman HL. Molecular Pathways: Mechanism of Action for Talimogene Laherparepvec, a New Oncolytic Virus Immunotherapy. Clin Cancer Res. 2016;22:1048-1054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 225] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 152. | Macedo N, Miller DM, Haq R, Kaufman HL. Clinical landscape of oncolytic virus research in 2020. J Immunother Cancer. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 232] [Article Influence: 46.4] [Reference Citation Analysis (0)] |
| 153. | Ernst M, Giubellino A. The Current State of Treatment and Future Directions in Cutaneous Malignant Melanoma. Biomedicines. 2022;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 36] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 154. | Dummer R, Hoeller C, Gruter IP, Michielin O. Combining talimogene laherparepvec with immunotherapies in melanoma and other solid tumors. Cancer Immunol Immunother. 2017;66:683-695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 155. | Olivet MM, Brown MC, Reitman ZJ, Ashley DM, Grant GA, Yang Y, Markert JM. Clinical Applications of Immunotherapy for Recurrent Glioblastoma in Adults. Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 156. | Todo T, Ito H, Ino Y, Ohtsu H, Ota Y, Shibahara J, Tanaka M. Intratumoral oncolytic herpes virus G47∆ for residual or recurrent glioblastoma: a phase 2 trial. Nat Med. 2022;28:1630-1639. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 275] [Cited by in RCA: 279] [Article Influence: 93.0] [Reference Citation Analysis (0)] |
| 157. | Hu M, Liao X, Tao Y, Chen Y. Advances in oncolytic herpes simplex virus and adenovirus therapy for recurrent glioma. Front Immunol. 2023;14:1285113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 158. | Todo T, Ino Y, Ohtsu H, Shibahara J, Tanaka M. A phase I/II study of triple-mutated oncolytic herpes virus G47∆ in patients with progressive glioblastoma. Nat Commun. 2022;13:4119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 109] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 159. | Stupp R, Taillibert S, Kanner A, Read W, Steinberg D, Lhermitte B, Toms S, Idbaih A, Ahluwalia MS, Fink K, Di Meco F, Lieberman F, Zhu JJ, Stragliotto G, Tran D, Brem S, Hottinger A, Kirson ED, Lavy-Shahaf G, Weinberg U, Kim CY, Paek SH, Nicholas G, Bruna J, Hirte H, Weller M, Palti Y, Hegi ME, Ram Z. Effect of Tumor-Treating Fields Plus Maintenance Temozolomide vs Maintenance Temozolomide Alone on Survival in Patients With Glioblastoma: A Randomized Clinical Trial. JAMA. 2017;318:2306-2316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1175] [Cited by in RCA: 1750] [Article Influence: 218.8] [Reference Citation Analysis (0)] |
| 160. | Zhang Y, Liu Q, Zhang X, Huang H, Tang S, Chai Y, Xu Z, Li M, Chen X, Liu J, Yang C. Recent advances in exosome-mediated nucleic acid delivery for cancer therapy. J Nanobiotechnology. 2022;20:279. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 181] [Article Influence: 60.3] [Reference Citation Analysis (0)] |
| 161. | Kim H, Jang H, Cho H, Choi J, Hwang KY, Choi Y, Kim SH, Yang Y. Recent Advances in Exosome-Based Drug Delivery for Cancer Therapy. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 162. | Roussy G. Trial of a Vaccination With Tumor Antigen-loaded Dendritic Cell-derived Exosomes (CSET 1437). [accessed 2024 Apr 25]. In: ClinicalTrials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://clinicaltrials.gov/study/NCT01159288 ClinicalTrials.gov Identifier: NCT01159288. |
| 163. | Besse B, Charrier M, Lapierre V, Dansin E, Lantz O, Planchard D, Le Chevalier T, Livartoski A, Barlesi F, Laplanche A, Ploix S, Vimond N, Peguillet I, Théry C, Lacroix L, Zoernig I, Dhodapkar K, Dhodapkar M, Viaud S, Soria JC, Reiners KS, Pogge von Strandmann E, Vély F, Rusakiewicz S, Eggermont A, Pitt JM, Zitvogel L, Chaput N. Dendritic cell-derived exosomes as maintenance immunotherapy after first line chemotherapy in NSCLC. Oncoimmunology. 2016;5:e1071008. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 602] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 164. | MD Anderson Cancer Center. iExosomes in Treating Participants With Metastatic Pancreas Cancer With KrasG12D Mutation. [accessed 2024 Apr 25]. In: ClinicalTrials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://classic.clinicaltrials.gov/ct2/show/NCT03608631 ClinicalTrials.gov Identifier: NCT03608631. |
| 165. | Huang R, Du H, Cheng L, Zhang P, Meng F, Zhong Z. Targeted nanodelivery of siRNA against KRAS G12D inhibits pancreatic cancer. Acta Biomater. 2023;168:529-539. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 166. | Mendt M, Kamerkar S, Sugimoto H, McAndrews KM, Wu CC, Gagea M, Yang S, Blanko EVR, Peng Q, Ma X, Marszalek JR, Maitra A, Yee C, Rezvani K, Shpall E, LeBleu VS, Kalluri R. Generation and testing of clinical-grade exosomes for pancreatic cancer. JCI Insight. 2018;3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 576] [Article Influence: 82.3] [Reference Citation Analysis (0)] |
| 167. | Codiak BioSciences. A Study of exoASO-STAT6 (CDK-004) in Patients With Advanced Hepatocellular Carcinoma (HCC) and Patients With Liver Metastases From EIther Primary Gastric Cancer or Colorectal Cancer (CRC).[accessed 2024 Apr 25]. In: ClinicalTrials.gov [Internet]. Bethesda (MD): U.S. National Library of Medicine. Available from: https://classic.clinicaltrials.gov/ct2/show/NCT05375604 ClinicalTrials.gov Identifier: NCT05375604. |
| 168. | Liu M, Liu L, Song Y, Li W, Xu L. Targeting macrophages: a novel treatment strategy in solid tumors. J Transl Med. 2022;20:586. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 42] [Reference Citation Analysis (0)] |
| 169. | He K, Barsoumian HB, Puebla-Osorio N, Hu Y, Sezen D, Wasley MD, Bertolet G, Zhang J, Leuschner C, Yang L, Kettlun Leyton CS, Fowlkes NW, Green MM, Hettrick L, Chen D, Masrorpour F, Gu M, Maazi H, Revenko AS, Cortez MA, Welsh JW. Inhibition of STAT6 with Antisense Oligonucleotides Enhances the Systemic Antitumor Effects of Radiotherapy and Anti-PD-1 in Metastatic Non-Small Cell Lung Cancer. Cancer Immunol Res. 2023;11:486-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 170. | Wylie B, Macri C, Mintern JD, Waithman J. Dendritic Cells and Cancer: From Biology to Therapeutic Intervention. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 108] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 171. | Galati D, Zanotta S. Dendritic Cell and Cancer Therapy. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 172. | Polyzoidis S, Ashkan K. DCVax®-L--developed by Northwest Biotherapeutics. Hum Vaccin Immunother. 2014;10:3139-3145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 47] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 173. | Najafi S, Mortezaee K. Advances in dendritic cell vaccination therapy of cancer. Biomed Pharmacother. 2023;164:114954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 42] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 174. | Liau LM, Ashkan K, Brem S, Campian JL, Trusheim JE, Iwamoto FM, Tran DD, Ansstas G, Cobbs CS, Heth JA, Salacz ME, D'Andre S, Aiken RD, Moshel YA, Nam JY, Pillainayagam CP, Wagner SA, Walter KA, Chaudhary R, Goldlust SA, Lee IY, Bota DA, Elinzano H, Grewal J, Lillehei K, Mikkelsen T, Walbert T, Abram S, Brenner AJ, Ewend MG, Khagi S, Lovick DS, Portnow J, Kim L, Loudon WG, Martinez NL, Thompson RC, Avigan DE, Fink KL, Geoffroy FJ, Giglio P, Gligich O, Krex D, Lindhorst SM, Lutzky J, Meisel HJ, Nadji-Ohl M, Sanchin L, Sloan A, Taylor LP, Wu JK, Dunbar EM, Etame AB, Kesari S, Mathieu D, Piccioni DE, Baskin DS, Lacroix M, May SA, New PZ, Pluard TJ, Toms SA, Tse V, Peak S, Villano JL, Battiste JD, Mulholland PJ, Pearlman ML, Petrecca K, Schulder M, Prins RM, Boynton AL, Bosch ML. Association of Autologous Tumor Lysate-Loaded Dendritic Cell Vaccination With Extension of Survival Among Patients With Newly Diagnosed and Recurrent Glioblastoma: A Phase 3 Prospective Externally Controlled Cohort Trial. JAMA Oncol. 2023;9:112-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 224] [Article Influence: 112.0] [Reference Citation Analysis (0)] |
| 175. | Ding Z, Li Q, Zhang R, Xie L, Shu Y, Gao S, Wang P, Su X, Qin Y, Wang Y, Fang J, Zhu Z, Xia X, Wei G, Wang H, Qian H, Guo X, Gao Z, Wei Y, Xu Q, Xu H, Yang L. Personalized neoantigen pulsed dendritic cell vaccine for advanced lung cancer. Signal Transduct Target Ther. 2021;6:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 176] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 176. | Schad F, Thronicke A, Steele ML, Merkle A, Matthes B, Grah C, Matthes H. Overall survival of stage IV non-small cell lung cancer patients treated with Viscum album L. in addition to chemotherapy, a real-world observational multicenter analysis. PLoS One. 2018;13:e0203058. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 40] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 177. | Chen YJ, Abila B, Mostafa Kamel Y. CAR-T: What Is Next? Cancers (Basel). 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 129] [Reference Citation Analysis (0)] |
| 178. | Khan AN, Chowdhury A, Karulkar A, Jaiswal AK, Banik A, Asija S, Purwar R. Immunogenicity of CAR-T Cell Therapeutics: Evidence, Mechanism and Mitigation. Front Immunol. 2022;13:886546. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 65] [Article Influence: 21.7] [Reference Citation Analysis (0)] |
| 179. | Salmikangas P, Kinsella N, Chamberlain P. Chimeric Antigen Receptor T-Cells (CAR T-Cells) for Cancer Immunotherapy - Moving Target for Industry? Pharm Res. 2018;35:152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 180. | Csaplár M, Szöllősi J, Gottschalk S, Vereb G, Szöőr Á. Cytolytic Activity of CAR T Cells and Maintenance of Their CD4+ Subset Is Critical for Optimal Antitumor Activity in Preclinical Solid Tumor Models. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 181. | Sterner RC, Sterner RM. CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 2021;11:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 1524] [Article Influence: 381.0] [Reference Citation Analysis (0)] |
| 182. | Choi G, Shin G, Bae S. Price and Prejudice? The Value of Chimeric Antigen Receptor (CAR) T-Cell Therapy. Int J Environ Res Public Health. 2022;19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 47] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 183. | Westin JR, Locke FL, Dickinson M, Ghobadi A, Elsawy M, van Meerten T, Miklos DB, Ulrickson ML, Perales MA, Farooq U, Wannesson L, Leslie L, Kersten MJ, Jacobson CA, Pagel JM, Wulf G, Johnston P, Rapoport AP, Du L, Vardhanabhuti S, Filosto S, Shah J, Snider JT, Cheng P, To C, Oluwole OO, Sureda A. Safety and Efficacy of Axicabtagene Ciloleucel versus Standard of Care in Patients 65 Years of Age or Older with Relapsed/Refractory Large B-Cell Lymphoma. Clin Cancer Res. 2023;29:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 30] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 184. | Ciltacabtagene Autoleucel (Carvykti): CADTH Reimbursement Review: Therapeutic area: Relapsed or refractory multiple myeloma [Internet]. Ottawa (ON): Canadian Agency for Drugs and Technologies in Health; 2023 Aug- . [PubMed] |
| 185. | Young RM, Engel NW, Uslu U, Wellhausen N, June CH. Next-Generation CAR T-cell Therapies. Cancer Discov. 2022;12:1625-1633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 101] [Article Influence: 33.7] [Reference Citation Analysis (0)] |
| 186. | Hu Y, Zhou Y, Zhang M, Ge W, Li Y, Yang L, Wei G, Han L, Wang H, Yu S, Chen Y, Wang Y, He X, Zhang X, Gao M, Yang J, Li X, Ren J, Huang H. CRISPR/Cas9-Engineered Universal CD19/CD22 Dual-Targeted CAR-T Cell Therapy for Relapsed/Refractory B-cell Acute Lymphoblastic Leukemia. Clin Cancer Res. 2021;27:2764-2772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 170] [Article Influence: 42.5] [Reference Citation Analysis (0)] |
| 187. | Lin H, Cheng J, Mu W, Zhou J, Zhu L. Advances in Universal CAR-T Cell Therapy. Front Immunol. 2021;12:744823. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 113] [Article Influence: 28.3] [Reference Citation Analysis (0)] |
| 188. | Du N, Guo F, Wang Y, Cui J. NK Cell Therapy: A Rising Star in Cancer Treatment. Cancers (Basel). 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 53] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 189. | Myers JA, Miller JS. Exploring the NK cell platform for cancer immunotherapy. Nat Rev Clin Oncol. 2021;18:85-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 822] [Article Influence: 164.4] [Reference Citation Analysis (0)] |
| 190. | Liu E, Marin D, Banerjee P, Macapinlac HA, Thompson P, Basar R, Nassif Kerbauy L, Overman B, Thall P, Kaplan M, Nandivada V, Kaur I, Nunez Cortes A, Cao K, Daher M, Hosing C, Cohen EN, Kebriaei P, Mehta R, Neelapu S, Nieto Y, Wang M, Wierda W, Keating M, Champlin R, Shpall EJ, Rezvani K. Use of CAR-Transduced Natural Killer Cells in CD19-Positive Lymphoid Tumors. N Engl J Med. 2020;382:545-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 920] [Cited by in RCA: 1461] [Article Influence: 292.2] [Reference Citation Analysis (0)] |
| 191. | Tang S, Qin C, Hu H, Liu T, He Y, Guo H, Yan H, Zhang J, Tang S, Zhou H. Immune Checkpoint Inhibitors in Non-Small Cell Lung Cancer: Progress, Challenges, and Prospects. Cells. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 117] [Article Influence: 39.0] [Reference Citation Analysis (0)] |
| 192. | Qin S, Xu L, Yi M, Yu S, Wu K, Luo S. Novel immune checkpoint targets: moving beyond PD-1 and CTLA-4. Mol Cancer. 2019;18:155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 429] [Cited by in RCA: 829] [Article Influence: 138.2] [Reference Citation Analysis (0)] |
| 193. | Yue P, Harper T, Bacot SM, Chowdhury M, Lee S, Akue A, Kukuruga MA, Wang T, Feldman GM. BRAF and MEK inhibitors differentially affect nivolumab-induced T cell activation by modulating the TCR and AKT signaling pathways. Oncoimmunology. 2019;8:e1512456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 194. | Lentz RW, Colton MD, Mitra SS, Messersmith WA. Innate Immune Checkpoint Inhibitors: The Next Breakthrough in Medical Oncology? Mol Cancer Ther. 2021;20:961-974. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 84] [Article Influence: 21.0] [Reference Citation Analysis (0)] |
| 195. | Paz-Ares L, Chen Y, Reinmuth N, Hotta K, Trukhin D, Statsenko G, Hochmair MJ, Özgüroğlu M, Ji JH, Garassino MC, Voitko O, Poltoratskiy A, Musso E, Havel L, Bondarenko I, Losonczy G, Conev N, Mann H, Dalvi TB, Jiang H, Goldman JW. Durvalumab, with or without tremelimumab, plus platinum-etoposide in first-line treatment of extensive-stage small-cell lung cancer: 3-year overall survival update from CASPIAN. ESMO Open. 2022;7:100408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 195] [Article Influence: 65.0] [Reference Citation Analysis (0)] |
| 196. | Zhu Y, Liu K, Qin Q, Zhu H. Serplulimab plus chemotherapy as first-line treatment for extensive-stage small-cell lung cancer: A cost-effectiveness analysis. Front Immunol. 2022;13:1044678. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 197. | Cheng Y, Han L, Wu L, Chen J, Sun H, Wen G, Ji Y, Dvorkin M, Shi J, Pan Z, Wang X, Bai Y, Melkadze T, Pan Y, Min X, Viguro M, Li X, Zhao Y, Yang J, Makharadze T, Arkania E, Kang W, Wang Q, Zhu J; ASTRUM-005 Study Group. Effect of First-Line Serplulimab vs Placebo Added to Chemotherapy on Survival in Patients With Extensive-Stage Small Cell Lung Cancer: The ASTRUM-005 Randomized Clinical Trial. JAMA. 2022;328:1223-1232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 288] [Article Influence: 96.0] [Reference Citation Analysis (0)] |
| 198. | Sharifi-Rad J, Quispe C, Patra JK, Singh YD, Panda MK, Das G, Adetunji CO, Michael OS, Sytar O, Polito L, Živković J, Cruz-Martins N, Klimek-Szczykutowicz M, Ekiert H, Choudhary MI, Ayatollahi SA, Tynybekov B, Kobarfard F, Muntean AC, Grozea I, Daştan SD, Butnariu M, Szopa A, Calina D. Paclitaxel: Application in Modern Oncology and Nanomedicine-Based Cancer Therapy. Oxid Med Cell Longev. 2021;2021:3687700. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 126] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 199. | Škubník J, Svobodová Pavlíčková V, Ruml T, Rimpelová S. Autophagy in cancer resistance to paclitaxel: Development of combination strategies. Biomed Pharmacother. 2023;161:114458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 28] [Reference Citation Analysis (0)] |
| 200. | Zhu L, Chen L. Progress in research on paclitaxel and tumor immunotherapy. Cell Mol Biol Lett. 2019;24:40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 339] [Article Influence: 56.5] [Reference Citation Analysis (0)] |
| 201. | Chen K, Shi W. Autophagy regulates resistance of non-small cell lung cancer cells to paclitaxel. Tumour Biol. 2016;37:10539-10544. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 64] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 202. | Fay N, Kouklovsky C, de la Torre A. Natural Product Synthesis: The Endless Quest for Unreachable Perfection. ACS Org Inorg Au. 2023;3:350-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 203. | Gallego-Jara J, Lozano-Terol G, Sola-Martínez RA, Cánovas-Díaz M, de Diego Puente T. A Compressive Review about Taxol(®): History and Future Challenges. Molecules. 2020;25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 200] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 204. | Miller DS, Filiaci VL, Mannel RS, Cohn DE, Matsumoto T, Tewari KS, DiSilvestro P, Pearl ML, Argenta PA, Powell MA, Zweizig SL, Warshal DP, Hanjani P, Carney ME, Huang H, Cella D, Zaino R, Fleming GF. Carboplatin and Paclitaxel for Advanced Endometrial Cancer: Final Overall Survival and Adverse Event Analysis of a Phase III Trial (NRG Oncology/GOG0209). J Clin Oncol. 2020;38:3841-3850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 212] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 205. | Ngoi NY, Syn NL, Goh RM, Goh BC, Huang RY, Soon YY, James E, Cook A, Clamp A, Tan DS. Weekly versus tri-weekly paclitaxel with carboplatin for first-line treatment in women with epithelial ovarian cancer. Cochrane Database Syst Rev. 2022;2:CD012007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 206. | Von Hoff DD, Ervin T, Arena FP, Chiorean EG, Infante J, Moore M, Seay T, Tjulandin SA, Ma WW, Saleh MN, Harris M, Reni M, Dowden S, Laheru D, Bahary N, Ramanathan RK, Tabernero J, Hidalgo M, Goldstein D, Van Cutsem E, Wei X, Iglesias J, Renschler MF. Increased survival in pancreatic cancer with nab-paclitaxel plus gemcitabine. N Engl J Med. 2013;369:1691-1703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4035] [Cited by in RCA: 4877] [Article Influence: 406.4] [Reference Citation Analysis (0)] |
| 207. | Chaturvedi VK, Singh A, Singh VK, Singh MP. Cancer Nanotechnology: A New Revolution for Cancer Diagnosis and Therapy. Curr Drug Metab. 2019;20:416-429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 198] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 208. | He M, Cao Y, Chi C, Zhao J, Chong E, Chin KXC, Tan NZV, Dmitry K, Yang G, Yang X, Hu K, Enikeev M. Unleashing novel horizons in advanced prostate cancer treatment: investigating the potential of prostate specific membrane antigen-targeted nanomedicine-based combination therapy. Front Immunol. 2023;14:1265751. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 10] [Reference Citation Analysis (1)] |
| 209. | Wu J. The Enhanced Permeability and Retention (EPR) Effect: The Significance of the Concept and Methods to Enhance Its Application. J Pers Med. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 414] [Article Influence: 103.5] [Reference Citation Analysis (0)] |
| 210. | Kamaly N, Xiao Z, Valencia PM, Radovic-Moreno AF, Farokhzad OC. Targeted polymeric therapeutic nanoparticles: design, development and clinical translation. Chem Soc Rev. 2012;41:2971-3010. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1402] [Cited by in RCA: 1195] [Article Influence: 91.9] [Reference Citation Analysis (1)] |
| 211. | Yao Y, Zhou Y, Liu L, Xu Y, Chen Q, Wang Y, Wu S, Deng Y, Zhang J, Shao A. Nanoparticle-Based Drug Delivery in Cancer Therapy and Its Role in Overcoming Drug Resistance. Front Mol Biosci. 2020;7:193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 629] [Article Influence: 125.8] [Reference Citation Analysis (0)] |
| 212. | Yoneshima Y, Morita S, Ando M, Nakamura A, Iwasawa S, Yoshioka H, Goto Y, Takeshita M, Harada T, Hirano K, Oguri T, Kondo M, Miura S, Hosomi Y, Kato T, Kubo T, Kishimoto J, Yamamoto N, Nakanishi Y, Okamoto I. Phase 3 Trial Comparing Nanoparticle Albumin-Bound Paclitaxel With Docetaxel for Previously Treated Advanced NSCLC. J Thorac Oncol. 2021;16:1523-1532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 213. | Schmid P, Adams S, Rugo HS, Schneeweiss A, Barrios CH, Iwata H, Diéras V, Hegg R, Im SA, Shaw Wright G, Henschel V, Molinero L, Chui SY, Funke R, Husain A, Winer EP, Loi S, Emens LA; IMpassion130 Trial Investigators. Atezolizumab and Nab-Paclitaxel in Advanced Triple-Negative Breast Cancer. N Engl J Med. 2018;379:2108-2121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2999] [Article Influence: 428.4] [Reference Citation Analysis (0)] |
| 214. | Lee H, Park S, Kang JE, Lee HM, Kim SA, Rhie SJ. Efficacy and safety of nanoparticle-albumin-bound paclitaxel compared with solvent-based taxanes for metastatic breast cancer: A meta-analysis. Sci Rep. 2020;10:530. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 30] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 215. | Shitara K, Takashima A, Fujitani K, Koeda K, Hara H, Nakayama N, Hironaka S, Nishikawa K, Makari Y, Amagai K, Ueda S, Yoshida K, Shimodaira H, Nishina T, Tsuda M, Kurokawa Y, Tamura T, Sasaki Y, Morita S, Koizumi W. Nab-paclitaxel versus solvent-based paclitaxel in patients with previously treated advanced gastric cancer (ABSOLUTE): an open-label, randomised, non-inferiority, phase 3 trial. Lancet Gastroenterol Hepatol. 2017;2:277-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 143] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 216. | Kogure Y, Iwasawa S, Saka H, Hamamoto Y, Kada A, Hashimoto H, Atagi S, Takiguchi Y, Ebi N, Inoue A, Kurata T, Okamoto I, Yamaguchi M, Harada T, Seike M, Ando M, Saito AM, Kubota K, Takenoyama M, Seto T, Yamamoto N, Gemma A. Efficacy and safety of carboplatin with nab-paclitaxel versus docetaxel in older patients with squamous non-small-cell lung cancer (CAPITAL): a randomised, multicentre, open-label, phase 3 trial. Lancet Healthy Longev. 2021;2:e791-e800. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 217. | Moser JC, Salvador E, Deniz K, Swanson K, Tuszynski J, Carlson KW, Karanam NK, Patel CB, Story M, Lou E, Hagemann C. The Mechanisms of Action of Tumor Treating Fields. Cancer Res. 2022;82:3650-3658. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 57] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 218. | Leal T, Kotecha R, Ramlau R, Zhang L, Milanowski J, Cobo M, Roubec J, Petruzelka L, Havel L, Kalmadi S, Ward J, Andric Z, Berghmans T, Gerber DE, Kloecker G, Panikkar R, Aerts J, Delmonte A, Pless M, Greil R, Rolfo C, Akerley W, Eaton M, Iqbal M, Langer C; LUNAR Study Investigators. Tumor Treating Fields therapy with standard systemic therapy versus standard systemic therapy alone in metastatic non-small-cell lung cancer following progression on or after platinum-based therapy (LUNAR): a randomised, open-label, pivotal phase 3 study. Lancet Oncol. 2023;24:1002-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 48] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 219. | Fisher JP, Adamson DC. Current FDA-Approved Therapies for High-Grade Malignant Gliomas. Biomedicines. 2021;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 186] [Article Influence: 46.5] [Reference Citation Analysis (0)] |
| 220. | Ceresoli GL, Aerts JG, Dziadziuszko R, Ramlau R, Cedres S, van Meerbeeck JP, Mencoboni M, Planchard D, Chella A, Crinò L, Krzakowski M, Rüssel J, Maconi A, Gianoncelli L, Grosso F. Tumour Treating Fields in combination with pemetrexed and cisplatin or carboplatin as first-line treatment for unresectable malignant pleural mesothelioma (STELLAR): a multicentre, single-arm phase 2 trial. Lancet Oncol. 2019;20:1702-1709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 110] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 221. | Virgil H. FDA Accepts PMA Application for TTFields in Non-Small Cell Lung Cancer. CancerNetwork®, home of the journal ONCOLOGY®. Jan 19, 2024. [cited 13 February 2024]. Available from: https://www.cancernetwork.com/view/fda-accepts-pma-application-for-ttfields-in-non-small-cell-lung-cancer. |
| 222. | Ghiaseddin AP, Shin D, Melnick K, Tran DD. Tumor Treating Fields in the Management of Patients with Malignant Gliomas. Curr Treat Options Oncol. 2020;21:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 71] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 223. | Shteingauz A, Porat Y, Voloshin T, Schneiderman RS, Munster M, Zeevi E, Kaynan N, Gotlib K, Giladi M, Kirson ED, Weinberg U, Kinzel A, Palti Y. AMPK-dependent autophagy upregulation serves as a survival mechanism in response to Tumor Treating Fields (TTFields). Cell Death Dis. 2018;9:1074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 76] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 224. | Silginer M, Weller M, Stupp R, Roth P. Biological activity of tumor-treating fields in preclinical glioma models. Cell Death Dis. 2017;8:e2753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 85] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 225. | Kim CY, Paek SH, Nam DH, Chang JH, Hong YK, Kim JH, Kim OL, Kim SH. Tumor treating fields plus temozolomide for newly diagnosed glioblastoma: a sub-group analysis of Korean patients in the EF-14 phase 3 trial. J Neurooncol. 2020;146:399-406. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 226. | Liu WT, Wang HT, Yeh YH, Wong TW. An Update on Recent Advances of Photodynamic Therapy for Primary Cutaneous Lymphomas. Pharmaceutics. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 227. | Kim EJ, Mangold AR, DeSimone JA, Wong HK, Seminario-Vidal L, Guitart J, Appel J, Geskin L, Lain E, Korman NJ, Zeitouni N, Nikbakht N, Dawes K, Akilov O, Carter J, Shinohara M, Kuzel TM, Piette W, Bhatia N, Musiek A, Pariser D, Kim YH, Elston D, Boh E, Duvic M, Huen A, Pacheco T, Zwerner JP, Lee ST, Girardi M, Querfeld C, Bohjanen K, Olsen E, Wood GS, Rumage A, Donini O, Haulenbeek A, Schaber CJ, Straube R, Pullion C, Rook AH, Poligone B. Efficacy and Safety of Topical Hypericin Photodynamic Therapy for Early-Stage Cutaneous T-Cell Lymphoma (Mycosis Fungoides): The FLASH Phase 3 Randomized Clinical Trial. JAMA Dermatol. 2022;158:1031-1039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 23] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 228. | Dong X, Zeng Y, Zhang Z, Fu J, You L, He Y, Hao Y, Gu Z, Yu Z, Qu C, Yin X, Ni J, Cruz LJ. Hypericin-mediated photodynamic therapy for the treatment of cancer: a review. J Pharm Pharmacol. 2021;73:425-436. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 48] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 229. | van Delft LCJ, Nelemans PJ, Kessels JPHM, Kreukels H, Roozeboom MH, de Rooij MJM, Mosterd K, de Haas ERM, Kelleners-Smeets NWJ. Long-Term Efficacy of Photodynamic Therapy with Fractionated 5-Aminolevulinic Acid 20% versus Conventional Two-Stage Topical Methyl Aminolevulinate for Superficial Basal-Cell Carcinoma. Dermatology. 2022;238:1044-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 230. | Agostinis P, Berg K, Cengel KA, Foster TH, Girotti AW, Gollnick SO, Hahn SM, Hamblin MR, Juzeniene A, Kessel D, Korbelik M, Moan J, Mroz P, Nowis D, Piette J, Wilson BC, Golab J. Photodynamic therapy of cancer: an update. CA Cancer J Clin. 2011;61:250-281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4122] [Cited by in RCA: 3567] [Article Influence: 254.8] [Reference Citation Analysis (0)] |
| 231. | Kwiatkowski S, Knap B, Przystupski D, Saczko J, Kędzierska E, Knap-Czop K, Kotlińska J, Michel O, Kotowski K, Kulbacka J. Photodynamic therapy - mechanisms, photosensitizers and combinations. Biomed Pharmacother. 2018;106:1098-1107. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 681] [Cited by in RCA: 1239] [Article Influence: 177.0] [Reference Citation Analysis (0)] |
| 232. | Ji B, Wei M, Yang B. Recent advances in nanomedicines for photodynamic therapy (PDT)-driven cancer immunotherapy. Theranostics. 2022;12:434-458. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 214] [Cited by in RCA: 264] [Article Influence: 88.0] [Reference Citation Analysis (0)] |
| 233. | Lucky SS, Soo KC, Zhang Y. Nanoparticles in photodynamic therapy. Chem Rev. 2015;115:1990-2042. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1896] [Cited by in RCA: 1962] [Article Influence: 196.2] [Reference Citation Analysis (0)] |
| 234. | Chen J, Fan T, Xie Z, Zeng Q, Xue P, Zheng T, Chen Y, Luo X, Zhang H. Advances in nanomaterials for photodynamic therapy applications: Status and challenges. Biomaterials. 2020;237:119827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 424] [Article Influence: 84.8] [Reference Citation Analysis (0)] |