Published online Sep 20, 2023. doi: 10.5662/wjm.v13.i4.223
Peer-review started: March 27, 2023
First decision: May 25, 2023
Revised: June 6, 2023
Accepted: June 19, 2023
Article in press: June 19, 2023
Published online: September 20, 2023
Processing time: 176 Days and 21.7 Hours
Ras suppressor 1 (RSU1), a highly conserved protein, plays an important role in actin cytoskeleton remodeling and cell-extracellular matrix adhesion. Aberration of RSU1 activity can cause changes in cell adhesion and migration, thereby enhancing tumor proliferation and metastasis. However, the correlation between RSU1 and gastrointestinal cancers (GICs), as well as its prognostic role related to tumor-infiltrating immune cells (TIICs) remains unclear.
To shows RSU1 plays a potential promoting role in facilitating tumor immune escape in GIC.
Differential expression of RSU1 in different tumors and their corresponding normal tissues was evaluated by exploring the Gene Expression Profiling Inter
High RSU1 expression was associated with poor overall survival of gastric cancer patients, exhibiting a hazard ratio (HR) = 1.36, first progression HR = 1.53, and post progression survival HR = 1.6. Specifically, high RSU1 Levels were associated with prognosis of gastric cancer in females, T4 and N3 stages, and Her-2-negative subtypes. Regarding immune-infiltrating cells, RSU1 expression level was positively correlated with infiltration of CD4+ T cells, macrophages, neutrophils, and dendritic cells (DCs) in colorectal adenocarcinoma and stomach adenocarcinoma. RSU1 expression was also predicted to be strongly correlated with immune marker sets in M2 macro
In gastrointestinal cancers, RSU1 is increased in tumor tissues, and predicts poor survival of patients. Increased RSU1 may be involved in promoting macrophage polarization, DC infiltration, and T cell exhaustion, inducing tumor immune escape and the development of tumors in GICs. We suggest that RSU1 is a promising prognostic biomarker reflecting immune infiltration level of GICs, as well as a potential therapeutic target for precision treatment through improving the immune response.
Core Tip: Ras suppressor 1 (RSU1), is a highly conserved protein involved in actin cytoskeleton remodeling and cell-extracellular matrix adhesion. The current study provides a comprehensive analysis of RSU1 in gastrointestinal cancer and shows its potential promoting role in facilitating tumor immune escape.
- Citation: Xu Y, Hou YY, Wu Z, Fang ZX, Wu HT, Liu J. Comprehensive analysis of cell-extracellular matrix protein Ras suppressor-1 in function and prognosis of gastrointestinal cancers. World J Methodol 2023; 13(4): 223-237
- URL: https://www.wjgnet.com/2222-0682/full/v13/i4/223.htm
- DOI: https://dx.doi.org/10.5662/wjm.v13.i4.223
The International Agency for Research on Cancer has released new data on the global burden of cancer for 2020. China has the world's highest number of new cases and deaths. There were 4.57 million new cancer cases in China in 2020, the top four being lung, colorectal, stomach and breast cancers. At the same time, the top five cancer deaths in China in 2020 were recognized as lung, liver, stomach, esophageal and colorectal cancers[1], with the incidence and mortality of gastrointestinal cancers (GICs) showing a clearly increasing pattern[2]. Currently, the prognosis of GIC patients has been slowly improving with the continuous progress of therapeutic strategies and extensive application of surgery, chemotherapy, biological targeted therapy, immunotherapy and other therapeutic methods[3]. However, poor clinical therapeutic effects remain for a subpopulation of GIC patients, showing the need for more specific and sensitive therapeutic targets for GIC patients. Development of new targeted drugs is a promising method in the treatment of GICs.
On the other hand, immune checkpoint inhibitors have emerged as promising antitumor drugs for multiple tumor types, either as single agents or in combination with chemotherapy[4]. For example, the KEYNOTE-062 and CheckMate-649 phase III clinical trials achieved positive results for immune checkpoint inhibitors in advanced first-line treatment of patients with gastric cancers (GCs)[5,6]. Therefore, clinical guidelines in many countries recommend immunotherapy combined with chemotherapy as the first-line treatment for metastatic GCs[7]. However, it is not clear how to select GIC patients who will benefit most from checkpoint inhibitors, necessitating further investigation for novel biomarkers to predict outcomes and immunotherapeutic response.
Ras suppressor 1 (RSU1) was originally found to suppress RAS-dependent oncogenic transformation, and localizes to human chromosome 10p13[8]. Later, it was found that RSU1 binds with high affinity, via its LRR domain, to the LIM5 domain of PINCH1, thereby forming an IPPR complex with ILK-PINCH-Parvin to regulate cell adhesion at sites of focal adhesion[9,10], and participate in physiological and pathological processes of local adhesion and tumor metastasis[11,12]. RSU1 has long been associated with cancers, but its expression in various cancer types or its role in metastasis has been unclear. It has been suggested that RSU1 is involved in the regulation of migration and invasion of breast cancer, liver cancer and brain cancer cells, and has the function of enhancing metastasis in a cell type-dependent manner[11,13-15]. RSU1 could also serve as a therapeutic anti-metastatic target in liver and breast cancer[16]. To evaluate the potential role of RSU1 in GIC, the correlation of RSU1 expression with prognosis in GIC patients with various clinicopathological factors was investigated, as well as the function of RSU1 in the occurrence of GIC and the correlation between RSU1 and immune infiltration.
This platform includes RNA sequencing data from 9736 tumor tissues and 8587 normal tissues from The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) databases. The main functions of GEPIA (http://gepia.cancer-pku.cn/) include gene expression analysis, gene correlation analysis, survival analysis, similar gene prediction, and dimension reduction analysis. The expression of RSU1 in tumors and normal tissues was analyzed via GEPIA.
Kaplan-Meier plotter (http://kmplot.com/analysis/) contains 10461 cancer samples for assessing the impact of 54675 genes on survival. Patients with cancers were divided into two groups (high- vs low-expression) to analyze the poorer overall survival (OS), poorer first progression (FP), progression free survival (PFS), and poorer post-progression survival (PPS) by means of HRs, 95% confidence intervals and log-rank P values. To predict the prognostic values of RSU1, GEO datasets were analyzed through the PrognoScan databases (http://dna00.bio.kyutech.ac.jp/PrognoScan/index.html) based on Cox P values.
DAVID (https://david.ncifcrf.gov/conversion.jsp) is a biological information database that integrates biological data, using analytical tools, to provide systematic and comprehensive annotation of biological functions for large-scale gene or protein lists, and extract biological information of interesting genes. RSU1-related genes were analyzed, regarding the top 10 highly enriched Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways and Gene Ontology (GO) items.
TIMER 2.0 (http://timer.cistrome.org) uses state-of-the-art algorithms to provide more reliable estimates of immune infiltration levels for TCGA or user-provided tumor profiles. TIMER 2.0 provides three modules, Immune Association, Cancer Exploration and Immune Estimation, for investigating associations between immune infiltration and genetic or clinical characteristics. TIMER was applied to analyze the expression of RSU1 in GIC patients and its relationship with 6 types of infiltrating immune cells (B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and DCs). The abundance of T cells, neutrophils, macrophages, and DCs, as well as tumor purity are important factors affecting immune infiltration in tumor samples analyzed by genomics.
ESTIMATE was conducted in the open-source R software to analyze the gene expression profiling of GICs for StromaScore, ImmuneScore and ESTIMATEScore. The datasets were downloaded from the GEO database, GSE17536, which had 177 patients with colorectal cancer (CRC), and GSE62254, which had 300 patients with GC. All patients were divided into two groups, high RSU1 vs low RSU1 based on the median RSU1 expression level.
The survival results from Kaplan-Meier plotter and GEPIA2 are displayed with HRs and P or Cox P values based on the log-rank test. Spearman correlation and statistical significance were used to evaluate the correlation of gene expression with immune biomarkers in GICs. Differences with P < 0.05 were considered significant.
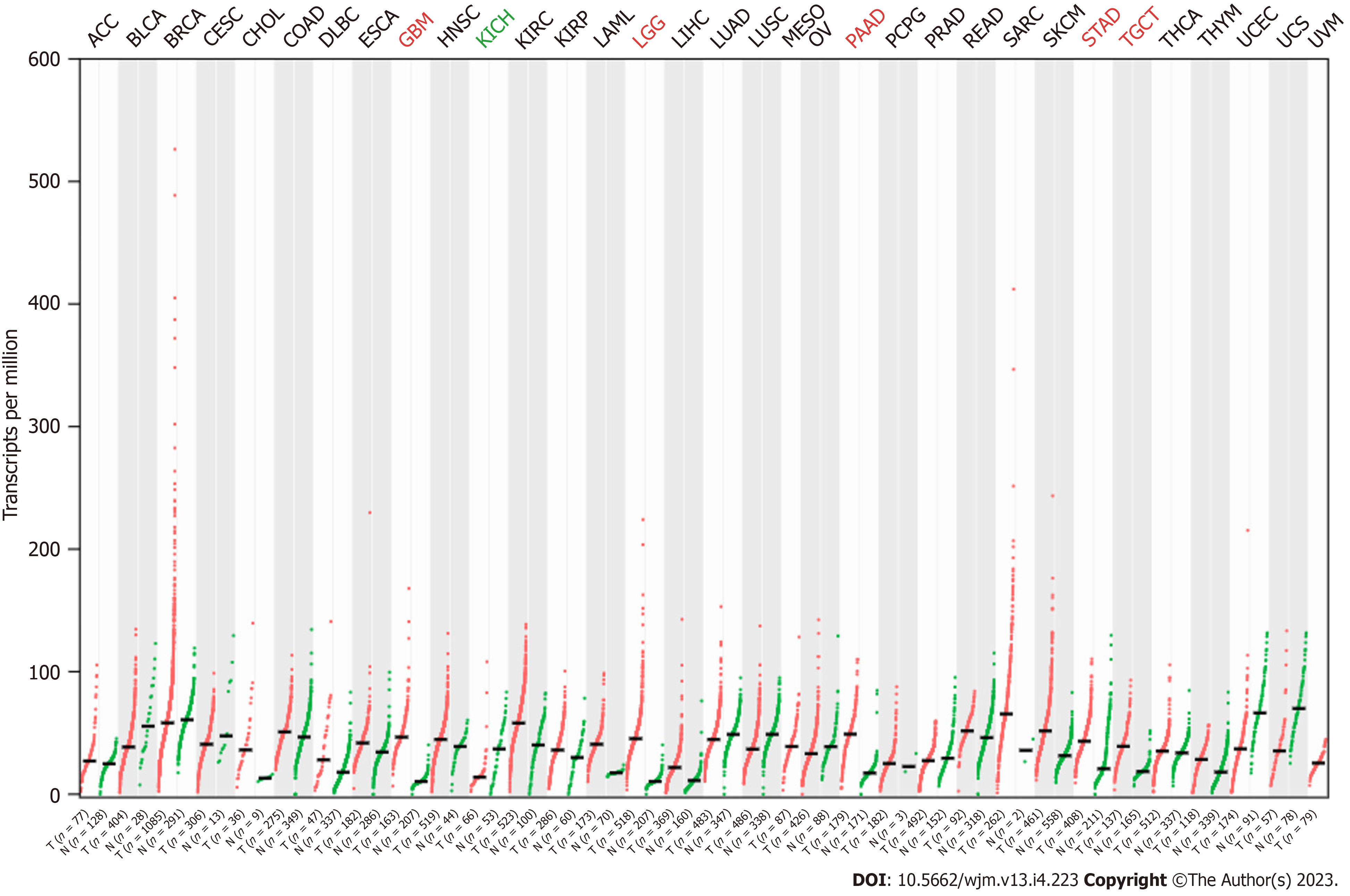
The expression pattern of RSU1 in different cancers was analyzed by GEPIA based on TCGA and GTEx datasets. It was found that, compared with normal tissues, RSU1 was highly expressed in glioblastoma multiforme (GBM), brain lower grade glioma (LGG), pancreatic adenocarcinoma (PAAD), stomach adenocarcinoma (STAD), and testicular germ cell tumors (TGCT) tissues, whereas a low level of RSU1 was found in kidney chromophobe (KICH) tissues (Figure 1A). However, there was no significant difference in RSU1 expression in colon adenocarcinoma (COAD), although the level of RSU1 in COAD tissues tended to be slightly increased.
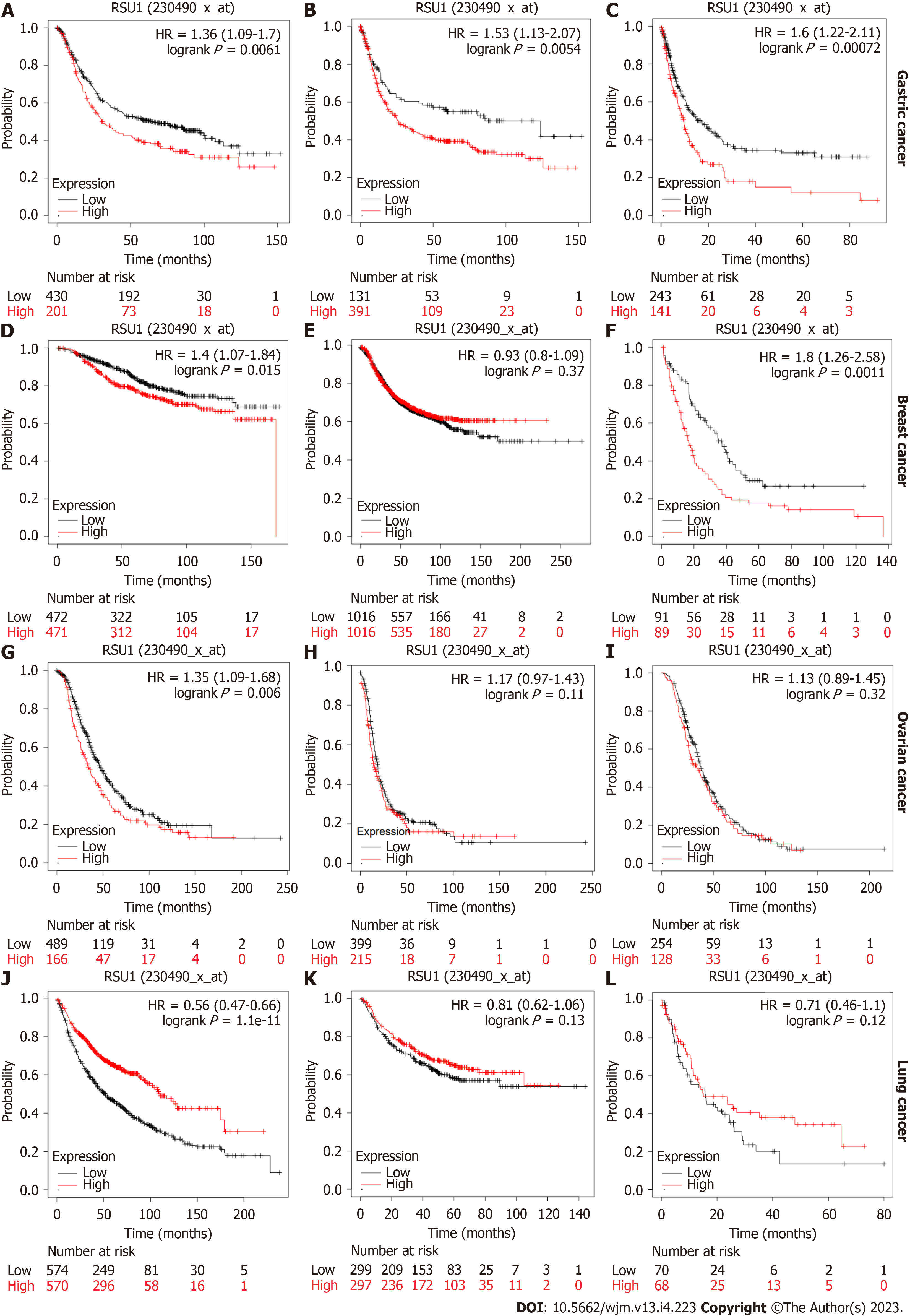
Further investigation focused on the prognostic role of RSU1 in different types of cancer. Based on the Kaplan-Meier plotter database, survival information was analyzed in patients with GC, breast cancer, ovarian cancer and lung cancer. A high expression level of RSU1 in patients with GC predicted poor OS, with hazard ratios (HR) = 1.36 (1.09-1.7), first progression HR = 1.53 (1.13-2.07) and post-progression survival HR = 1.6 (1.22-2.11) (Figure 2A-C). Regarding breast cancer, the expression level of RSU1 did not affect either the OS or PFS of patients with breast cancer, although significantly poor PPS was found in breast cancer patients with high RSU1 expression (HR = 1.8 (1.26-2.58), Figure 2D-F). In patients with ovarian cancer, the association between RSU1 level and survival was only found in OS [HR = 1.35 (1.09-1.68)], while PFS and PPS were not associated with RSU1 expression (Figure 2G-I). However, a high level of RSU1 predicted good OS in patients with lung cancer, with an HR = 0.56 (0.47-0.66), but no relationship with FP and PPS (Figure 2J-L). The above survival results in different types of cancer suggested that RSU1 may have distinct function in GCs, even in GICs.
To better understand the relevance and underlying mechanisms of RSU1 expression in GC, the relationship between the RSU1 expression and clinical characteristics of GC patients in the Kaplan-Meier plotter database was analyzed through six GC cohorts (GSE14210, GSE15459, GSE22377, GSE29272, GSE51105, GSE62254). Interestingly, in female patients with GC, the expression level of RSU1 was associated with OS, FP and PPS (P < 0.05, Table 1). Among different tumor stage parameters, the expression of RSU1 was correlated with large tumor size (T) and predicted poor survival of GC patients with the highest HR = 4.86. However, with or without lymph node metastasis, high RSU1 expression was associated with poor survival in GC, with HRs ranging from 1.35 to 10.98. Regarding distant metastasis, GC patients with increased RSU1 expression predicted shorter OS, and poor FP and PPS. Even HER2 status can affect the prognostic capability of RSU1, giving an HR = 1.46–1.79 in GC patients with negative HER2 status.
| OS (631) | FP (522) | PPS (384) | |||||||
| n | HR | P value | n | HR | P value | n | HR | P value | |
| Sex | |||||||||
| Female | 187 | 1.88 (1.22-2.89) | c | 179 | 1.65 (1.09-2.51) | a | 127 | 1.99 (1.21-3.25) | b |
| Male | 349 | 1.36 (0.94-1.97) | 0.098 | 341 | 1.4 (0.98-2.01) | 0.064 | 256 | 1.52 (1.09-2.14) | a |
| Stage | |||||||||
| I | 62 | 2.84 (0.63-12.88) | 0.16 | 60 | 2.7 (0.6-12.26) | 0.18 | 31 | 2.79 (0.53-14.68) | 0.21 |
| II | 135 | 1.47 (0.79-2.75) | 0.22 | 131 | 0.73 (0.4-1.32) | 0.29 | 105 | 2.01 (1.04-3.9) | a |
| III | 197 | 1.27 (0.87-1.87) | 0.22 | 186 | 1.51 (1.02-2.22) | a | 142 | 1.2 (0.77-1.86) | 0.42 |
| IV | 140 | 1.53 (0.95-2.46) | 0.078 | 141 | 1.17 (0.74-1.84) | 0.51 | 104 | 1.79 (1.13-2.83) | a |
| T | |||||||||
| T2 | 241 | 1.64 (0.98-2.73) | 0.055 | 239 | 1.42 (0.88-2.29) | 0.15 | 196 | 1.57 (0.9-2.72) | 0.11 |
| T3 | 204 | 1.3 (0.92-1.83) | 0.14 | 204 | 1.36 (0.95-1.94) | 0.091 | 150 | 1.36 (0.93-1.99) | 0.12 |
| T4 | 38 | 3.33 (1.12-9.96) | a | 39 | 2.12 (0.85-5.3) | 0.1 | 29 | 4.68 (1.34-16.32) | b |
| N | |||||||||
| N0 | 74 | 2.77 (1.19-6.43) | a | 72 | 2.69 (1.16-6.23) | a | 41 | 10.98 (2.7-44.62) | c |
| N1-3 | 422 | 1.54 (1.18-2.01) | b | 423 | 1.35 (1.04-1.74) | a | 337 | 1.64 (1.22-2.19) | c |
| M | |||||||||
| M0 | 444 | 1.72 (1.2-2.47) | b | 443 | 1.63 (1.16-2.31) | b | 342 | 1.77 (1.31-2.4) | c |
| M1 | 56 | 1.84 (1.02-3.29) | a | 56 | 1.44 (0.79-2.6) | 0.23 | 56 | 2.09 (0.98-4.45) | 0.052 |
| HER2 status | |||||||||
| Negative | 429 | 1.46 (1.12-1.91) | b | 429 | 1.55 (1.06-2.26) | a | 283 | 1.79 (1.28-2.5) | c |
| Positive | 202 | 1.41 (0.95-2.09) | 0.086 | 166 | 1.46 (0.9-2.38) | 0.13 | 101 | 1.46 (0.89-2.4) | 0.14 |
| Treatment | |||||||||
| Surgery alone | 380 | 1.44 (1.07-1.93) | a | 375 | 1.36 (0.97-1.9) | 0.071 | 277 | 1.82 (1.31-2.52) | c |
| 5-FU-based adjuvant | 34 | 1.77 (0.69-4.52) | 0.23 | 34 | 1.8 (0.77-4.25) | 0.17 | 21 | 0.39 (0.13-1.18) | 0.085 |
| Other adjuvant | 76 | 2.65 (1.09-6.4) | a | 80 | 3.46 (1.03-11.67) | a | 74 | 2.5 (0.98-6.41) | a |
To investigate the prognostic value of RSU1 in other GICs, GEO datasets were recruited for further analysis, as no survival information of CRC was found in Kaplan-Meier Plotter. Interestingly, both in GSE17536 and GSE17537, CRC patients with high RSU1 expression tended to have short OS, disease-free survival (DFS), and disease-specific survival (DSS). However, as the sample sizes were limited, statistical significance was only found in GSE17536 in regard to DSS in CRC patients (HR = 1.86, Figure 3).
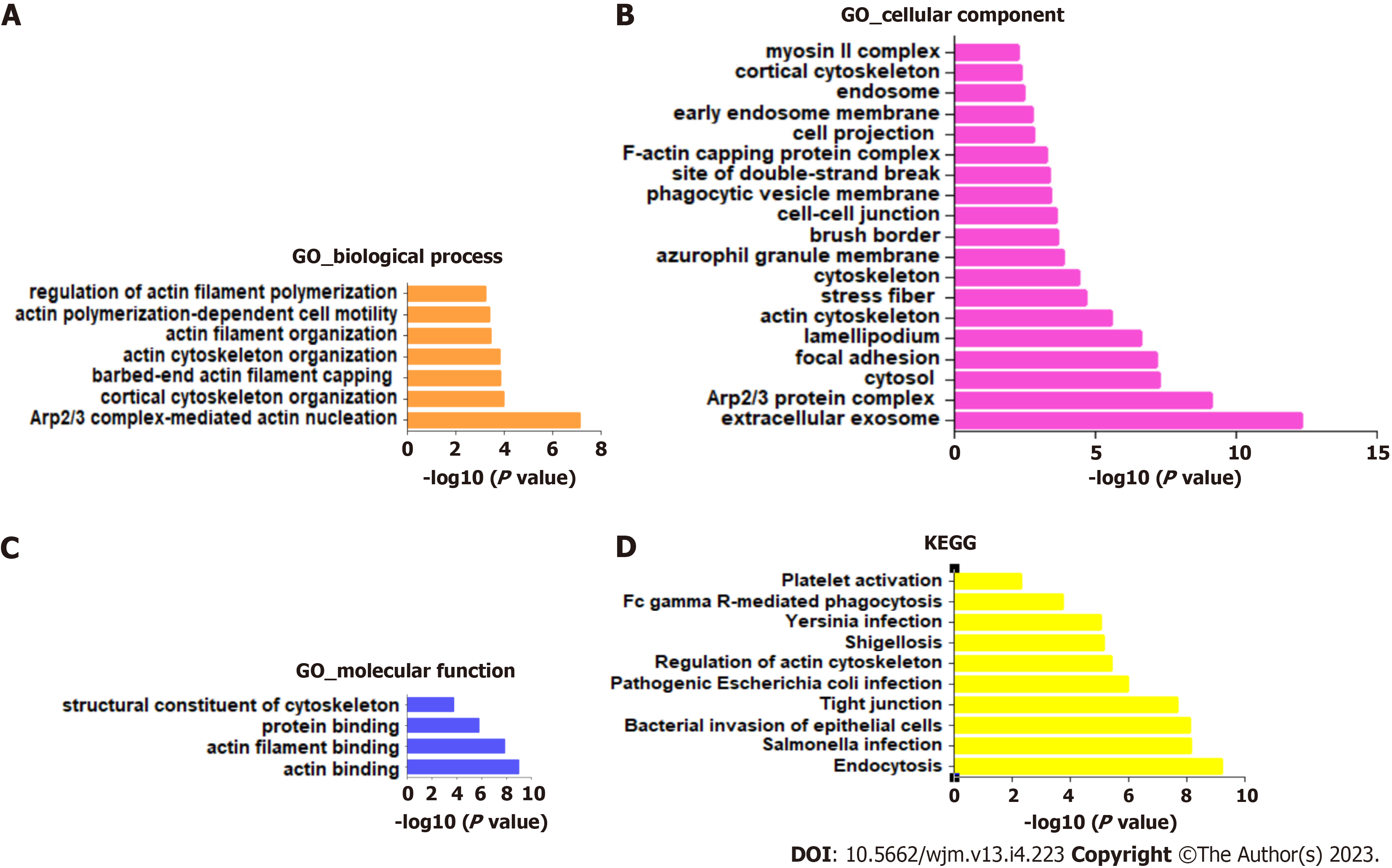
To investigate the potential signaling pathways involved in RSU1-regulated development of GIC, the top 100 RSU1-related genes were collected and analyzed with the KEGG pathways and GO projects, and mapped onto GO for biological process (BP), cellular component (CC), and molecular function (MF) analysis (Figure 4).
The highest enrichment of BP, CC and MF included actin cytoskeleton organization, focal adhesion, extracellular exosome, protein binding and actin binding (Figure 4A-C). In addition, among the top 10 KEGG pathways, endocytosis, Salmonella infection, bacterial invasion of epithelial cells, and tight junction were significantly enriched for RSU1 and RSU1-related genes (Figure 4D). It is reported that the actin cytoskeleton is essential for maintaining cell shape and promoting movement, and plays a key role in tumor invasion and metastasis[17], suggesting that RSU1 may be linked to the tumorigenesis and progression of cancers.
Tumor immune escape is a major obstacle limiting the efficacy of current immunotherapy. It has been reported that actin cytoskeletal remodeling can protect tumor cells from natural killer-mediated cytotoxicity[18]. Thus, RSU1 could affect immune infiltration in GICs based on its association with actin cytoskeletal remodeling.
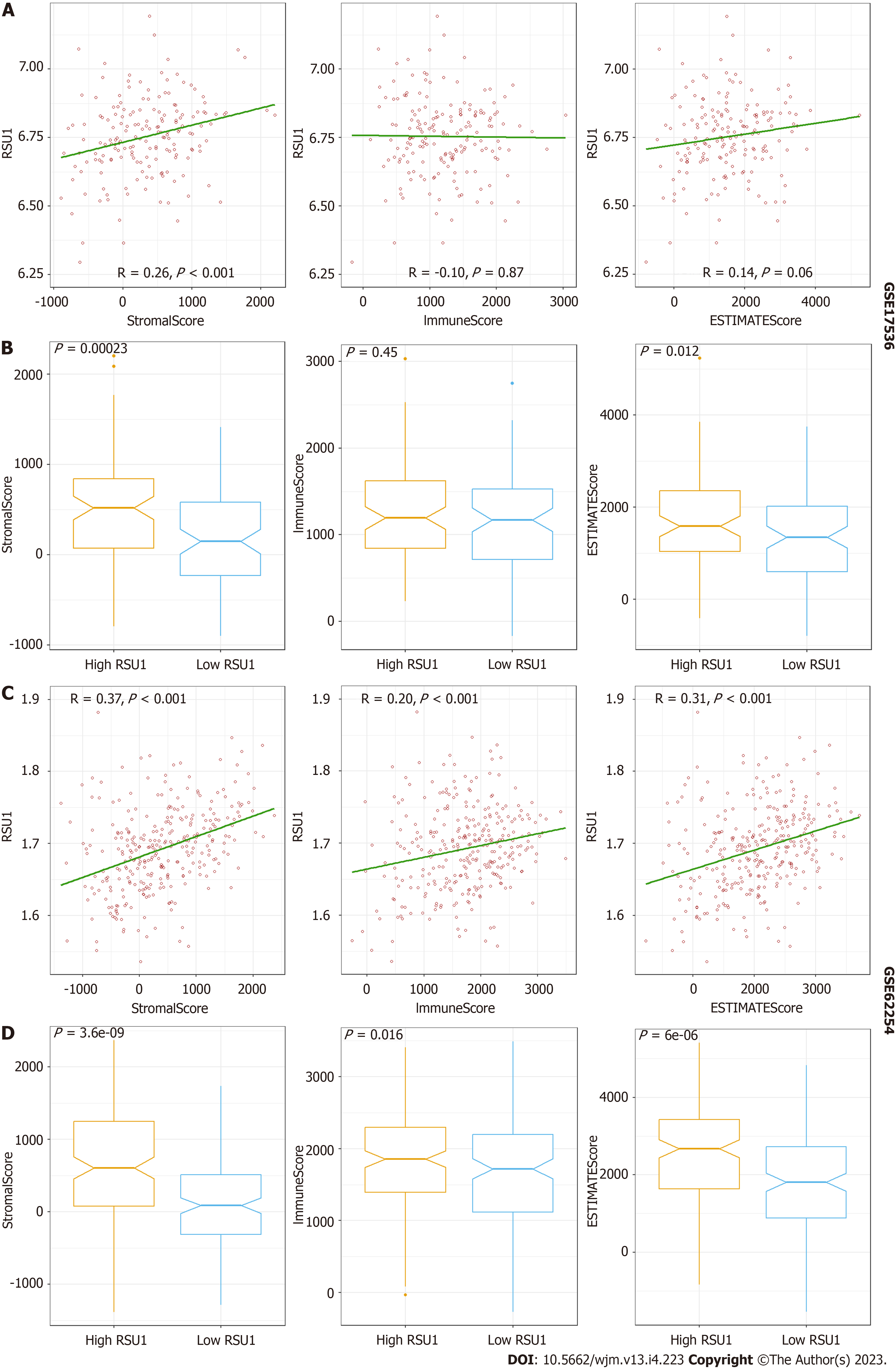
To explore the potential function of RSU1 in GICs, two datasets related to GICs were downloaded and analyzed through ESTIMATE in R software. It is found that along with the increased RSU1 expression level, the StromaScore and ESTIMATEScore were elevated gradually with P < 0.05 in CRC and GC (Figure 5A and C). After dividing the patients into high RSU1 vs low RSU1 groups, similar results were found in StromaScore and ESTIMATEScore. In the group with high RSU1 levels, StromaScore and ESTIMATEScore were significantly higher compared with the low RSU1 group (Figure 5B and D).
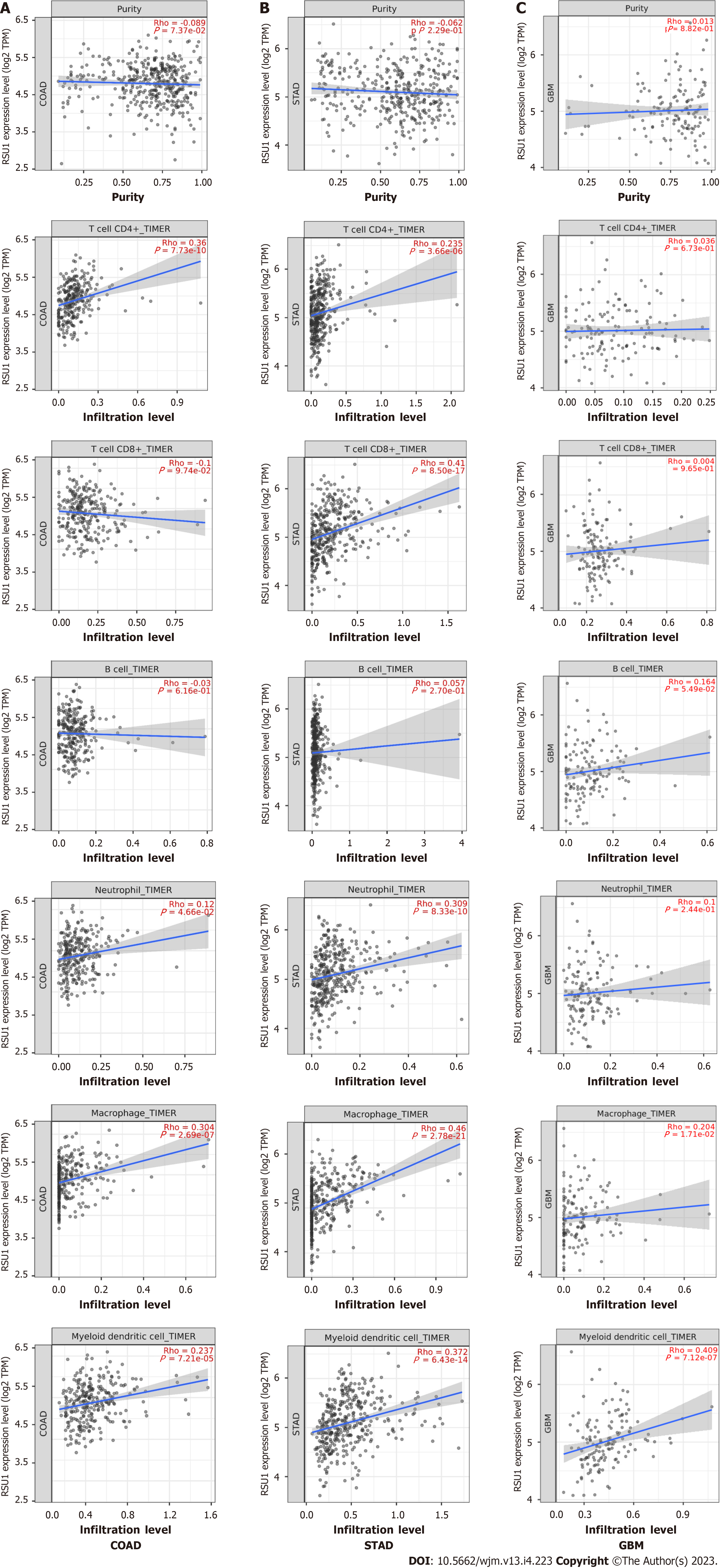
The relationship between RSU1 expression and immune infiltration levels in GIC was explored, revealing that in COAD, RSU1 expression was not significantly correlated with tumor purity (r = -0.089, P = 7.37e-02), CD8+ T cells (r = -0.1, P = 9.74e-02) or B cells (r = -0.03, P = 6.16e-01), but was positively correlated with the infiltration levels of CD4+ T cells (r = 0.36, P = 7.73e-10), macrophages (r = 0.304, P = 2.69e-07), neutrophils (r = 0.12, P = 4.66e-02) and DCs (r = 0.237, P = 7.21e-05) (n = 458) (Figure 6A). Interestingly, in STAD, RSU1 expression was found to be associated with the changed immune infiltration, which was positively correlated with infiltration levels of CD8+ T cells (r = 0.41, P = 8.50e-17), CD4+ T cells (r = 0.235, P = 3.66e-06), macrophages (r = 0.46, P = 2.78e-21), neutrophils (r = 0.309, P = 8.33e-10), and DCs (r = 0.372, P = 6.43e-14) (n = 415), but not for tumor purity (r = -0.062, P = 2.29e-01) and B cells (r = 0.057, P = 2.70e-01) (Figure 6B).
The clinical trials or vaccine therapies based on immune checkpoint blockade against GBM have not been successful, mainly due to its highly immunosuppressive environment and multiple resistance mechanisms[19]. To compare these findings, GBM was analyzed as control and found that except for macrophage and DC infiltration, expression of RSU1 was not associated with most immune cell infiltration (P > 0.05) (Figure 6C). These findings indicate that RSU1 may play a specific role in immune infiltration in the development of GIC, especially through regulating the infiltration of CD4+ T cells, macrophages and DCs.
As a relationship between RSU1 Level and immune infiltration was found, biomarkers of different immune cells were characterized to explore the potential function and mechanism of RSU1-related immune infiltration in GIC. The immune cells recruited included CD8+ T cells, T cells, B cells, monocytes, Tumor-associated macrophage (TAMs), M1 and M2 macrophages, neutrophils, natural killer (NK) cells, and DCs, as well as functionally different T cells, such as Th1, Th2, Tfh, Th17, Treg, and exhausted T cells (Table 2). After adjustment for purity, RSU1 expression levels were significantly correlated with 33 out of 57 immune cell biomarkers in COAD, and 48 out of 57 in STAD (Table 2).
| Immune cell | Biomarker | COAD | STAD | GBM | |||||||||
| None | Purity | None | Purity | None | Purity | ||||||||
| Cor | P value | Cor | P value | Cor | P value | Cor | P value | Cor | P value | Cor | P value | ||
| CD8+ T cell | CD8A | 0.052 | 2.64E-01 | 0.008 | 8.73E-01 | 0.306 | c | 0.305 | c | -0.056 | 4.90E-01 | -0.026 | 7.62E-01 |
| CD8B | -0.043 | 3.60E-01 | -0.076 | 1.25E-01 | 0.158 | b | 0.151 | b | -0.157 | 5.30E-02 | -0.124 | 1.50E-01 | |
| T cell | CD3D | 0.034 | 4.72E-01 | -0.009 | 8.63E-01 | 0.23 | c | 0.228 | c | -0.140 | 8.37E-02 | -0.114 | 1.86E-01 |
| CD3E | 0.123 | b | 0.099 | a | 0.248 | c | 0.245 | c | -0.042 | 6.06E-01 | 0.007 | 9.38E-01 | |
| CD2 | 0.176 | c | 0.168 | c | 0.304 | c | 0.307 | c | -0.074 | 3.61E-01 | -0.034 | 6.96E-01 | |
| B cell | CD19 | 0.116 | a | 0.099 | a | 0.183 | c | 0.161 | b | 0.019 | 8.14E-01 | -0.047 | 5.86E-01 |
| CD79A | 0.197 | c | 0.18 | c | 0.18 | c | 0.155 | b | 0.093 | 2.55E-01 | 0.133 | 1.21E-01 | |
| Monocyte | CD86 | 0.255 | c | 0.258 | c | 0.361 | c | 0.375 | c | 0.024 | 7.72E-01 | 0.104 | 2.28E-01 |
| CD115 (CSF1R) | 0.189 | c | 0.176 | c | 0.439 | c | 0.44 | c | 0.200 | a | 0.302 | c | |
| TAM | CCL2 | 0.352 | c | 0.337 | c | 0.348 | c | 0.355 | c | 0.034 | 6.74E-01 | 0.083 | 3.35E-01 |
| CD68 | 0.096 | a | 0.094 | 5.76E-02 | 0.17 | c | 0.169 | c | 0.169 | a | 0.275 | b | |
| IL10 | 0.21 | c | 0.204 | c | 0.384 | c | 0.406 | c | -0.043 | 5.98E-01 | 0.036 | 6.78E-01 | |
| M1 Macrophage | INOS (NOS2) | -0.196 | c | -0.196 | c | 0.035 | 4.75E-01 | 0.061 | 2.36E-01 | 0.049 | 5.50E-01 | 0.082 | 3.39E-01 |
| IRF5 | 0.216 | c | 0.219 | c | 0.108 | a | 0.093 | 7.03E-02 | 0.040 | 6.26E-01 | 0.094 | 2.74E-01 | |
| COX2 (PTGS2) | 0.084 | 7.40E-02 | 0.039 | 4.31E-01 | 0.186 | c | 0.183 | c | 0.198 | a | 0.254 | b | |
| M2 Macrophage | CD163 | 0.187 | c | 0.168 | c | 0.395 | c | 0.397 | c | 0.155 | 5.51E-02 | 0.213 | a |
| VSIG4 | 0.18 | c | 0.158 | b | 0.401 | c | 0.421 | c | 0.018 | 8.25E-01 | 0.101 | 2.39E-01 | |
| MS4A4A | 0.221 | c | 0.204 | c | 0.421 | c | 0.431 | c | 0.060 | 4.58E-01 | 0.166 | 5.21E-02 | |
| Neutrophils | CD66B (CEACAM8) | -0.024 | 6.15E-01 | -0.013 | 7.98E-01 | 0.029 | 5.57E-01 | 0.034 | 5.08E-01 | 0.033 | 6.87E-01 | 0.044 | 6.11E-01 |
| CD11B (ITGAM) | 0.136 | b | 0.13 | b | 0.392 | c | 0.392 | c | 0.233 | b | 0.319 | c | |
| CCR7 | 0.17 | c | 0.16 | b | 0.322 | c | 0.325 | c | 0.226 | b | 0.254 | b | |
| Natural killer cell | KIR2DL1 | -0.024 | 6.08E-01 | -0.057 | 2.51E-01 | 0.143 | b | 0.145 | b | -0.025 | 7.62E-01 | -0.027 | 7.52E-01 |
| KIR2DL3 | -0.076 | 1.06E-01 | -0.091 | 6.78E-02 | 0.094 | 5.45E-02 | 0.083 | 1.07E-01 | -0.022 | 7.88E-01 | -0.052 | 5.48E-01 | |
| KIR2DL4 | -0.072 | 1.23E-01 | -0.11 | a | 0.005 | 9.25E-01 | 0.003 | 9.61E-01 | -0.009 | 9.11E-01 | 0.009 | 9.13E-01 | |
| KIR3DL1 | -0.049 | 2.92E-01 | -0.073 | 1.40E-01 | 0.142 | b | 0.127 | a | 0.042 | 6.06E-01 | 0.018 | 8.34E-01 | |
| KIR3DL2 | -0.01 | 8.31E-01 | -0.063 | 2.04E-01 | 0.146 | b | 0.147 | b | -0.086 | 2.93E-01 | -0.060 | 4.86E-01 | |
| KIR3DL3 | -0.086 | 6.63E-02 | -0.093 | 6.04E-02 | -0.078 | 1.12E-01 | -0.064 | 2.16E-01 | -0.118 | 1.48E-01 | -0.107 | 2.12E-01 | |
| KIR2DS4 | -0.015 | 7.48E-01 | -0.026 | 5.99E-01 | 0.051 | 3.03E-01 | 0.044 | 3.95E-01 | -0.001 | 9.88E-01 | 0.003 | 9.72E-01 | |
| Dendritic cell | HLA-DPB1 | 0.051 | 2.73E-01 | 0.024 | 6.27E-01 | 0.287 | c | 0.291 | c | 0.032 | 6.91E-01 | 0.108 | 2.08E-01 |
| HLA-DQB1 | 0.056 | 2.29E-01 | 0.037 | 4.55E-01 | 0.152 | b | 0.153 | b | 0.152 | 6.11E-02 | 0.234 | b | |
| HLA-DRA | 0.125 | b | 0.103 | a | 0.268 | c | 0.27 | c | -0.049 | 5.51E-01 | 0.014 | 8.73E-01 | |
| HLA-DPA1 | 0.16 | c | 0.142 | b | 0.255 | c | 0.256 | c | 0.032 | 6.91E-01 | 0.108 | 2.08E-01 | |
| BDCA-1 (CD1C) | 0.29 | c | 0.291 | c | 0.303 | c | 0.302 | c | -0.030 | 7.10E-01 | 0.025 | 7.70E-01 | |
| BDCA-4 (NRP1) | 0.348 | c | 0.37 | c | 0.523 | c | 0.52 | c | 0.376 | c | 0.462 | c | |
| CD11c (ITGAX) | 0.156 | c | 0.15 | b | 0.341 | c | 0.348 | c | 0.081 | 3.19E-01 | 0.099 | 2.49E-01 | |
| Th1 | T-bet (TBX21) | 0.069 | 1.39E-01 | 0.06 | 2.27E-01 | 0.266 | c | 0.264 | c | 0.237 | b | 0.258 | b |
| STAT4 | 0.159 | c | 0.148 | b | 0.301 | c | 0.294 | c | -0.091 | 2.63E-01 | -0.043 | 6.18E-01 | |
| STAT1 | 0.197 | c | 0.182 | c | 0.189 | c | 0.177 | c | 0.078 | 3.38E-01 | 0.075 | 3.84E-01 | |
| IFN-γ (IFNG) | 0.023 | 6.28E-01 | 0.009 | 8.53E-01 | 0.098 | a | 0.091 | 7.73E-02 | 0.120 | 1.40E-01 | 0.130 | 1.30E-01 | |
| TNF-α (TNF) | 0.118 | a | 0.098 | a | 0.104 | a | 0.071 | 1.67E-01 | 0.022 | 7.83E-01 | 0.026 | 7.67E-01 | |
| Th2 | GATA3 | 0.176 | c | 0.183 | c | 0.305 | c | 0.304 | c | 0.125 | 1.24E-01 | 0.143 | 9.67E-02 |
| STAT6 | 0.006 | 8.92E-01 | 0.01 | 8.44E-01 | 0.224 | c | 0.219 | c | 0.289 | c | 0.353 | c | |
| STAT5A | 0.155 | c | 0.172 | c | 0.431 | c | 0.426 | c | 0.071 | 3.81E-01 | 0.104 | 2.24E-01 | |
| IL13 | 0.124 | b | 0.099 | a | 0.151 | b | 0.153 | b | 0.021 | 7.93E-01 | 0.029 | 7.39E-01 | |
| Tfh | BCL6 | 0.145 | b | 0.143 | b | 0.388 | c | 0.367 | c | 0.238 | b | 0.232 | b |
| IL21 | 0.074 | 1.13E-01 | 0.06 | 2.31E-01 | 0.133 | b | 0.12 | a | 0.023 | 7.73E-01 | -0.026 | 7.67E-01 | |
| Th17 | STAT3 | 0.318 | c | 0.327 | c | 0.426 | c | 0.411 | c | 0.404 | c | 0.394 | c |
| IL17A | 0.055 | 2.42E-01 | 0.057 | 2.51E-01 | -0.124 | a | -0.133 | b | -0.077 | 3.42E-01 | -0.082 | 3.42E-01 | |
| Treg | FOXP3 | 0.218 | c | 0.214 | c | 0.224 | c | 0.223 | c | 0.065 | 4.24E-01 | 0.073 | 3.98E-01 |
| CCR8 | 0.317 | c | 0.327 | c | 0.341 | c | 0.336 | c | 0.052 | 5.26E-01 | 0.082 | 3.38E-01 | |
| STAT5B | 0.401 | c | 0.413 | c | 0.499 | c | 0.491 | c | 0.218 | b | 0.179 | a | |
| TGFβ (TGFB1) | 0.114 | a | 0.106 | 3.28E-02 | 0.418 | c | 0.406 | c | 0.325 | c | 0.387 | c | |
| T cell (exhausted) | PD1 (PDCD1) | -0.016 | 7.28E-01 | -0.05 | 3.16E-01 | 0.205 | c | 0.202 | c | 0.085 | 2.96E-01 | 0.132 | 1.23E-01 |
| CTLA4 | 0.1 | a | 0.092 | 6.43E-02 | 0.17 | c | 0.164 | b | 0.030 | 7.12E-01 | 0.060 | 4.89E-01 | |
| LAG3 | -0.001 | 9.91E-01 | -0.041 | 4.04E-01 | 0.202 | c | 0.196 | c | 0.116 | 1.52E-01 | 0.126 | 1.42E-01 | |
| TIM-3 (HAVCR2) | 0.223 | c | 0.22 | c | 0.38 | c | 0.391 | c | 0.046 | 5.76E-01 | 0.106 | 2.16E-01 | |
| GZMB | 0.028 | 5.53E-01 | 0.007 | 8.89E-01 | 0.134 | b | 0.132 | b | -0.009 | 9.15E-01 | 0.043 | 6.20E-01 | |
Interestingly, the expression levels of biomarkers in most monocyte, TAM, and M2 macrophage marker groups were strongly correlated with RSU1 expression in GIC. It has been reported that the polarization of macrophages from M1 (anti-tumor macrophages) to M2 (pro-tumor macrophages) triggers apoptosis of CD8 T cells, and restricts T cell receptor clustering, all of which contribute to immune escape and promote tumor progression[20]. Specifically, the chemokines of TAMs (CCL-2 and IL10), and M2 (CD163, VSIG4, and MS4A4A) were positively correlated with RSU1 expression in GICs (P < 0.001), suggesting a potential role of RSU1 in modulating macrophage polarization in GICs.
High expression of RSU1 was also associated with high infiltration of DCs in GIC, based on its positive association with DC markers, such as HLA-DRA, HLA-DPA1, BDCA-1, BDCA-4 and CD11c (P < 0.05). For T cells with different functions in GIC, RSU1 was positively correlated with FOXP3, CCR8, and STAT5B in Treg cells, and associated with CTLA and TIM-3 in T cell exhaustion, indicating the potential role of RSU1 in regulating Treg and T cell exhaustion.
For comparison, GBM was also analyzed for the relationship between RSU1 level and immune cell biomarkers. Only 12 out of 57 immune cell biomarkers were found to be associated with high RSU1 expression in GBM, those being CD115 in monocytes, CD68 in TAMs, COX2 in M1 macrophages, CD11B and CCR7 in neutrophils, BDCA-4 and CD11c in DCs, STAT6 in Th2 T cells, BCL6 in Tfh T cells, STAT3 in Th17 T cells, and STAT5B and TGFβ in Tregs. These findings confirm a potential role of RSU1 in immune escape in GIC through regulating macrophage polarization, DC infiltration and T cell exhaustion, resulting in promotion of tumor progression.
The actin cytoskeleton mediates many essential biological functions in all eukaryotic cells. In addition to providing the structural framework necessary to determine cell shape and polarity, the actin cytoskeleton, especially its dynamics, is also closely related to cell movement, division, adhesion, and phagocytosis[21]. The actin cytoskeleton also mediates many pathological functions, such as playing a key role in tumor cell invasion and metastasis[22,23]. Among diverse different regulating factors, RSU1 participates in actin cytoskeletal remodeling, which is essential for the development of cancers. Current research reported the potential role of RSU1 in regulating immune escape in GICs through mediating the actin cytoskeleton.
It is found that RSU1 is involved in tumor progression in various cancer cell lines in vitro, but in vivo studies of the role of RSU1 in cancer is still missing. In this study, the expression pattern of RSU1 in different cancers was examined. Although at the mRNA level, RUS1 Level is associated only with GBM, LGG, PAAD, STAD, and TGCT, in patients with GICs, the high level of RSU1 is predicted to promote the development of gastric and colorectal cancers. In breast cancer studies, Vasaturo et al[24] demonstrated that overexpression of RSU1 in Michigan Cancer Foundation-7 (MCF-7) breast cancer cells induced p21 activation and reduced cancer cell proliferation by inhibiting cyclin-dependent kinase, suggest
Recent studies have shown that actin cytoskeletal remodeling is also closely related to the function of various immune cells (including T cells, B cells, and macrophages), which are involved in the formation of immune synapses and the development and maturation of T cells[27,28]. Actin cytoskeletal remodeling is also involved in the function of B cells and chemotaxis and phagocytosis in macrophages, through B cell antigen presentation by regulating BCR signaling[29,30]. Interestingly, actin is increased in resistant tumor cells, and the actin response is related to the protection of cancer cells against NK cell attack[18]. However, the regulatory function of tumor cell RSU1 on immune cells has not been studied yet. To elaborate the relationship between RSU1 and different immune cells, we found RSU1 expression is positively correlated with infiltration levels of CD4+ T cells, macrophages, neutrophils, and DCs in STAD. RSU1 plays a crucial role in immune escape of STAD microenvironments through macrophage polarization, DC infiltration, and T cell exhaustion, resulting in promotion of tumor progression.
The results of two clinical studies, CheckMate-649 and KEYNOTE-062, predicted that the response to immunotherapy cannot be judged solely by traditional indications, such as PD-L1 or tumor mutational burden. Since then, a large number of tumor patients with PD-L1 negative or low TMB have produced unusually long-lasting responses. Therefore, the development of new markers to predict immunotherapeutic response needs further investigation. PD-1, PD-L1 and CTLA4 are the main targets of immunotherapy. The present study shows that RSU1 is positively correlated with the expression of PD1 and CTLA4 in GC, and therefore RUS1 may be a potential prognostic factor for predicting the efficacy of immunotherapy.
In conclusion, our study provides insights into the potential role of RSU1 in tumor immunology and its prognostic value. RSU1 is associated with prognosis and TIICs in patients with different types of cancer, especially COAD and STAD, including CD4+ T cells, macrophages, neutrophils, and DCs. Higher RSU1 expression is associated with poor prognosis of GC patients and RSU1 plays a crucial role in immune escape of STAD microenvironments through macrophage polarization, DCs infiltration, and T cell exhaustion, and promotes tumor progression. RSU1 mRNA level is correlated with prognosis and immune infiltration level in GIC, suggesting that RSU1 can be used as a biomarker of prognosis.
At present, gastrointestinal cancers (GICs) in clinical immunotherapy effect is not significant, this study found that Ras suppressor 1 (RSU1) participation in the immune escape process may affect the efficacy of immunotherapy, providing a new target for improving the efficacy of immunotherapy.
The main topic of this study is to explore the role of RSU1 in GICs. Currently, GICs have no significant effect on immunotherapy, and this study found that RSU1 can affect the efficacy of immunotherapy through immune escape, providing a way to improve the efficacy of immunotherapy.
RSU1 is a promising prognostic biomarker reflecting the level of GICs immunoinfiltration and a potential therapeutic target by improving the immune response. At present, this study needs clinical data for verification.
We evaluated differential expression of RSU1 in different tumors and their corresponding normal tissues by exploring Gene Expression Profiling Interactive Analysis datasets. The prognostic relationship between RSU1 expression and GICs patients was evaluated using Kaplan-Meier plotter and PrognoScan. Then, RSU1 related genes were screened and functional enrichment was performed by DAVID. Tumor Immune Estimation Resource (TIMER) was used to further characterize the correlation between RSU1 and tumor-infiltrating immune cells. In addition, the correlation between RSU1 and immune cell surface molecules was analyzed by TIMER.
Our study reveals the potential role of RSU1 in tumor immunology and its prognostic value. RSU1 is involved in immune escape of tumor microenvironment through macrophage polarization, dendritic cells infiltration, T cell depletion and other pathways to promote tumor progression. RSU1 can be used as a prognostic biomarker to provide therapeutic target for improving the efficacy of immunotherapy.
The new theories that this study proposes is RSU1 plays a crucial role in immune escape and thus the efficacy of immunotherapy in gastrointestinal cancers. The new methods that this study proposed is the immune status was expressed by analyzing the expression of immune-related factors in the online database.
This is the first comprehensive analysis of the potential role and prognostic value of the RSU1 in diverse gastrointestinal cancers. However, the current research is limited, and future studies need to further explore the molecular mechanism of RSU1 in regulating the oncogenesis and development of GICs.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Oncology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: de Souza HSP, Brazil; Gkretsi V, Cyprus; Huang LQ S-Editor: Liu JH L-Editor: A P-Editor: Xu ZH
| 1. | Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71:209-249. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75126] [Cited by in RCA: 64637] [Article Influence: 16159.3] [Reference Citation Analysis (176)] |
| 2. | Yu YY, Zhu YJ, Xiao ZZ, Chen YD, Chang XS, Liu YH, Tang Q, Zhang HB. The pivotal application of patient-derived organoid biobanks for personalized treatment of gastrointestinal cancers. Biomark Res. 2022;10:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (77)] |
| 3. | Liu HN, Yao C, Wang XF, Zhang NP, Chen YJ, Pan D, Zhao GP, Shen XZ, Wu H, Liu TT. Diagnostic and economic value of carcinoembryonic antigen, carbohydrate antigen 19-9, and carbohydrate antigen 72-4 in gastrointestinal cancers. World J Gastroenterol. 2023;29:706-730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (1)] |
| 4. | Eefsen RL, Larsen JS, Klarskov LL, Altaf R, Høgdall E, Ingeholm P, Lykke J, Nielsen DL, Pfeiffer P, Poulsen LØ, Qvortrup C, Schou JV, Mau-Sørensen M, Østerlind K, Jensen BV. Therapy with pembrolizumab in treatment-naïve patients with nonmetastatic, mismatch repair deficient colorectal cancer. Int J Cancer. 2023;152:2145-2152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 9] [Reference Citation Analysis (1)] |
| 5. | Shitara K, Van Cutsem E, Bang YJ, Fuchs C, Wyrwicz L, Lee KW, Kudaba I, Garrido M, Chung HC, Lee J, Castro HR, Mansoor W, Braghiroli MI, Karaseva N, Caglevic C, Villanueva L, Goekkurt E, Satake H, Enzinger P, Alsina M, Benson A, Chao J, Ko AH, Wainberg ZA, Kher U, Shah S, Kang SP, Tabernero J. Efficacy and Safety of Pembrolizumab or Pembrolizumab Plus Chemotherapy vs Chemotherapy Alone for Patients With First-line, Advanced Gastric Cancer: The KEYNOTE-062 Phase 3 Randomized Clinical Trial. JAMA Oncol. 2020;6:1571-1580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 821] [Cited by in RCA: 867] [Article Influence: 173.4] [Reference Citation Analysis (1)] |
| 6. | Janjigian YY, Shitara K, Moehler M, Garrido M, Salman P, Shen L, Wyrwicz L, Yamaguchi K, Skoczylas T, Campos Bragagnoli A, Liu T, Schenker M, Yanez P, Tehfe M, Kowalyszyn R, Karamouzis MV, Bruges R, Zander T, Pazo-Cid R, Hitre E, Feeney K, Cleary JM, Poulart V, Cullen D, Lei M, Xiao H, Kondo K, Li M, Ajani JA. First-line nivolumab plus chemotherapy versus chemotherapy alone for advanced gastric, gastro-oesophageal junction, and oesophageal adenocarcinoma (CheckMate 649): a randomised, open-label, phase 3 trial. Lancet. 2021;398:27-40. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1962] [Cited by in RCA: 1898] [Article Influence: 474.5] [Reference Citation Analysis (1)] |
| 7. | Naito Y, Nishida T, Doi T. Current status of and future prospects for the treatment of unresectable or metastatic gastrointestinal stromal tumours. Gastric Cancer. 2023;26:339-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (1)] |
| 8. | Cutler ML, Bassin RH, Zanoni L, Talbot N. Isolation of rsp-1, a novel cDNA capable of suppressing v-Ras transformation. Mol Cell Biol. 1992;12:3750-3756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 38] [Article Influence: 1.2] [Reference Citation Analysis (1)] |
| 9. | Kadrmas JL, Smith MA, Clark KA, Pronovost SM, Muster N, Yates JR 3rd, Beckerle MC. The integrin effector PINCH regulates JNK activity and epithelial migration in concert with Ras suppressor 1. J Cell Biol. 2004;167:1019-1024. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 72] [Article Influence: 3.4] [Reference Citation Analysis (1)] |
| 10. | Fukuda K, Lu F, Qin J. Molecular basis for Ras suppressor-1 binding to PINCH-1 in focal adhesion assembly. J Biol Chem. 2021;296:100685. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 11. | Louca M, Stylianou A, Minia A, Pliaka V, Alexopoulos LG, Gkretsi V, Stylianopoulos T. Ras suppressor-1 (RSU-1) promotes cell invasion in aggressive glioma cells and inhibits it in non-aggressive cells through STAT6 phospho-regulation. Sci Rep. 2019;9:7782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (1)] |
| 12. | Zacharia LC, Stylianopoulos T, Gkretsi V. Ras Suppressor-1 (RSU-1) in Cancer Cell Metastasis: Friend or Foe? Crit Rev Oncog. 2017;22:249-253. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 13. | Gkretsi V, Bogdanos DP. Elimination of Ras Suppressor-1 from hepatocellular carcinoma cells hinders their in vitro metastatic properties. Anticancer Res. 2015;35:1509-1512. [PubMed] |
| 14. | Louca M, Gkretsi V, Stylianopoulos T. Coordinated Expression of Ras Suppressor 1 (RSU-1) and Growth Differentiation Factor 15 (GDF15) Affects Glioma Cell Invasion. Cancers (Basel). 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 1.7] [Reference Citation Analysis (1)] |
| 15. | Gkretsi V, Stylianou A, Louca M, Stylianopoulos T. Identification of Ras suppressor-1 (RSU-1) as a potential breast cancer metastasis biomarker using a three-dimensional in vitro approach. Oncotarget. 2017;8:27364-27379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (1)] |
| 16. | Louca M, Stylianopoulos T, Gkretsi V. Ras Suppressor-1 (RSU1) in Cancer Cell Metastasis: A Tale of a Tumor Suppressor. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (1)] |
| 17. | Blanquie O, Bradke F. Cytoskeleton dynamics in axon regeneration. Curr Opin Neurobiol. 2018;51:60-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 94] [Cited by in RCA: 131] [Article Influence: 18.7] [Reference Citation Analysis (1)] |
| 18. | Al Absi A, Wurzer H, Guerin C, Hoffmann C, Moreau F, Mao X, Brown-Clay J, Petrolli R, Casellas CP, Dieterle M, Thiery JP, Chouaib S, Berchem G, Janji B, Thomas C. Actin Cytoskeleton Remodeling Drives Breast Cancer Cell Escape from Natural Killer-Mediated Cytotoxicity. Cancer Res. 2018;78:5631-5643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 110] [Article Influence: 15.7] [Reference Citation Analysis (1)] |
| 19. | Hsieh HT, Huang HC, Chung CW, Chiang CC, Hsia T, Wu HF, Huang RL, Chiang CS, Wang J, Lu TT, Chen Y. CXCR4-targeted nitric oxide nanoparticles deliver PD-L1 siRNA for immunotherapy against glioblastoma. J Control Release. 2022;352:920-930. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 32] [Reference Citation Analysis (1)] |
| 20. | Farhad M, Rolig AS, Redmond WL. The role of Galectin-3 in modulating tumor growth and immunosuppression within the tumor microenvironment. Oncoimmunology. 2018;7:e1434467. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 79] [Cited by in RCA: 157] [Article Influence: 22.4] [Reference Citation Analysis (1)] |
| 21. | Pollard TD, Cooper JA. Actin, a central player in cell shape and movement. Science. 2009;326:1208-1212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1579] [Cited by in RCA: 1498] [Article Influence: 93.6] [Reference Citation Analysis (1)] |
| 22. | Gerasimcik N, Dahlberg CI, Baptista MA, Massaad MJ, Geha RS, Westerberg LS, Severinson E. The Rho GTPase Cdc42 Is Essential for the Activation and Function of Mature B Cells. J Immunol. 2015;194:4750-4758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (1)] |
| 23. | Yoshida T, Ozawa Y, Kimura T, Sato Y, Kuznetsov G, Xu S, Uesugi M, Agoulnik S, Taylor N, Funahashi Y, Matsui J. Eribulin mesilate suppresses experimental metastasis of breast cancer cells by reversing phenotype from epithelial-mesenchymal transition (EMT) to mesenchymal-epithelial transition (MET) states. Br J Cancer. 2014;110:1497-1505. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 208] [Cited by in RCA: 272] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 24. | Vasaturo F, Dougherty GW, Cutler ML. Ectopic expression of Rsu-1 results in elevation of p21CIP and inhibits anchorage-independent growth of MCF7 breast cancer cells. Breast Cancer Res Treat. 2000;61:69-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 25. | Giotopoulou N, Valiakou V, Papanikolaou V, Dubos S, Athanassiou E, Tsezou A, Zacharia LC, Gkretsi V. Ras suppressor-1 promotes apoptosis in breast cancer cells by inhibiting PINCH-1 and activating p53-upregulated-modulator of apoptosis (PUMA); verification from metastatic breast cancer human samples. Clin Exp Metastasis. 2015;32:255-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Christou C, Christodoulou MI, Zaravinos A, Gkretsi V. Ras suppressor 1 long form (RSU1L) silencing promotes apoptosis in invasive breast cancer cells. Cell Signal. 2023;101:110522. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 3] [Article Influence: 1.5] [Reference Citation Analysis (1)] |
| 27. | Na BR, Jun CD. TAGLN2-mediated actin stabilization at the immunological synapse: implication for cytotoxic T cell control of target cells. BMB Rep. 2015;48:369-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 18] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Phee H, Au-Yeung BB, Pryshchep O, O'Hagan KL, Fairbairn SG, Radu M, Kosoff R, Mollenauer M, Cheng D, Chernoff J, Weiss A. Pak2 is required for actin cytoskeleton remodeling, TCR signaling, and normal thymocyte development and maturation. Elife. 2014;3:e02270. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 47] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 29. | Burbage M, Keppler SJ. Shaping the humoral immune response: Actin regulators modulate antigen presentation and influence B-T interactions. Mol Immunol. 2018;101:370-376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Davidson AJ, Millard TH, Evans IR, Wood W. Ena orchestrates remodelling within the actin cytoskeleton to drive robust Drosophila macrophage chemotaxis. J Cell Sci. 2019;132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |