Published online Jul 20, 2021. doi: 10.5662/wjm.v11.i4.130
Peer-review started: January 29, 2021
First decision: May 6, 2021
Revised: May 11, 2021
Accepted: June 16, 2021
Article in press: June 16, 2021
Published online: July 20, 2021
Processing time: 170 Days and 23.5 Hours
The gastrointestinal microbiota plays a pivotal role in health and has been linked to many diseases. With the rapid accumulation of pyrosequencing data of the bacterial composition, the causal-effect relationship between specific dysbiosis features and diseases is now being explored. The aim of this review is to describe the key functional bacterial proteins and antigens in the context of dysbiosis related-diseases. We subjectively classify the key functional proteins into two categories: Primary key proteins and secondary key proteins. The primary key proteins mainly act by themselves and include biofilm inhibitors, toxin degraders, oncogene degraders, adipose metabolism modulators, anti-inflammatory peptides, bacteriocins, host cell regulators, adhesion and invasion molecules, and intestinal barrier regulators. The secondary key proteins mainly act by eliciting host immune responses and include flagellin, outer membrane proteins, and other autoantibody-related antigens. Knowledge of key bacterial proteins is limited compared to the rich microbiome data. Understanding and focusing on these key proteins will pave the way for future mechanistic level cause-effect studies of gut dysbiosis and diseases.
Core Tip: Revealing the causal-effect relationship between specific dysbiosis features and diseases requires understanding the roles of key bacterial proteins that are involved in dysbiosis. Some bacterial proteins may affect the microbiome by their inherent functions. Others shape the microbiome mainly by eliciting host immune responses. These key proteins warrant attention in future bioinformatic analyses and mechanistic studies.
- Citation: Zeng XY, Li M. Looking into key bacterial proteins involved in gut dysbiosis. World J Methodol 2021; 11(4): 130-143
- URL: https://www.wjgnet.com/2222-0682/full/v11/i4/130.htm
- DOI: https://dx.doi.org/10.5662/wjm.v11.i4.130
The gastrointestinal microbiota is linked to numerous diseases, including inflammatory bowel disease (IBD), irritable bowel syndrome (IBS), colorectal cancer, cirrhosis, and many others. Thanks to the rapid decrease in the cost of pyrosequencing, the gut microbiota, often represented by the fecal bacteria composition, is now easy to profile by 16S rDNA sequencing and shotgun metagenomic sequencing. With the accumulation of known microbiome-disease correlations in many descriptive studies, the mechanisms of known dysbiosis features in the pathogenesis of related diseases have become a new frontier to be explored. Understanding these mechanisms is a prerequisite to developing the precise intervention methods targeting the gut microbiome. Thus, it is necessary to review the key microbial proteins involved in gut dysbiosis.
The gut microbiome produces numerous products for itself and the host. The collection of small molecules produced by the gut microbiota, termed the metabolome, represents promising targets for investigation and translation. The methodology and findings of studies of the gut metabolome have been reviewed elsewhere[1,2]. In addition, the gut microbiota produces exosomes, which have been reviewed by other excellent reviews[3]. The virome[4,5], parasitome[6], helminths, and protozoa-omics[6] are also recognized by omic-approaches but with less well documented mechanisms. In this review, we will focus on the key peptides, proteins, and antigens produced by bacteria and fungi in the context of dysbiosis and diseases.
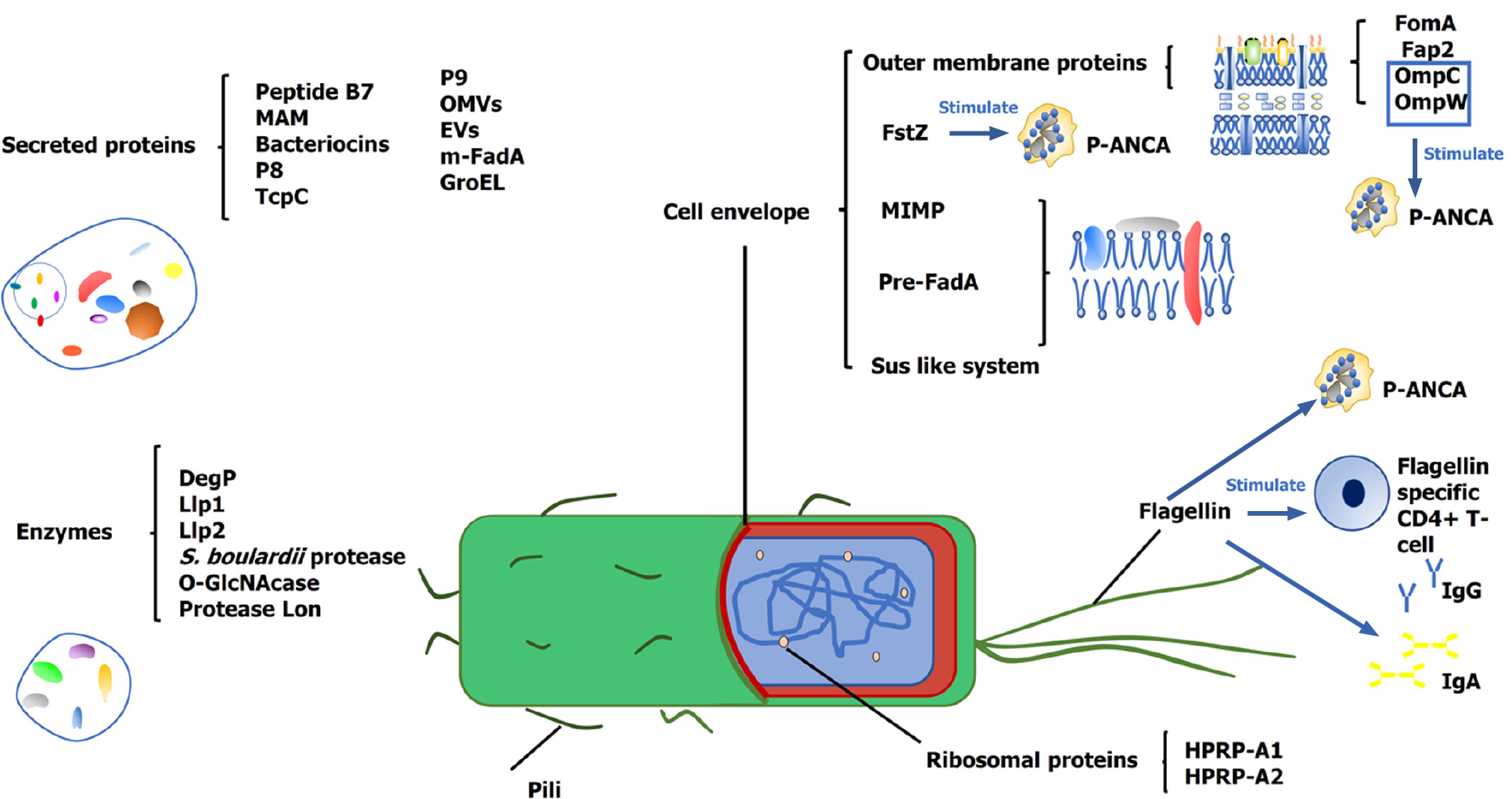
To organize the review, we categorize the bacterial proteins into two groups: (1) The primary key proteins, whose action mainly depends on their inherent properties (Table 1); and (2) The secondary key proteins, whose action mainly depends on the host response to them (Figure 1, Table 2). This classification mainly depends on the current knowledge and is relative. Often, the bacteria-host interaction is bilateral. Thus, this classification is subjective and only helps navigate the mechanisms. For each group, we organize the key proteins according to their functions to assist in navigating this field rapidly.
| Classification | Name | Function | Ref. |
| Enzyme | DegP | Inhibiting EHEC biofilms. | [11] |
| Enzyme | Llp1, Llp2 | Inhibiting biofilm formation of pathogen. | [12] |
| Enzyme | Protease of S. boulardii | Digesting both C. difficile toxin A and its receptor binding sites. | [15] |
| Enzyme | Lon protease | Degrading the oncogene c-MYC. | [19] |
| Secreted protein | P9 | Inducing the secretion of GLP-1. Inducing the secretion of IL-6 in macrophages. | [21,22] |
| Secreted protein | Peptide B7 | Reducing CCR2 expression on all APCs from health people. | [25] |
| Secreted protein | MAM | Inhibiting the NF-κB pathway and several cell immune responses. Inducing expression of TGF β. | [26-28] |
| Surface layer protein | MIMP | Inducing the secretion of anti- inflammatory cytokines and inhibiting inflammatory cytokines. Enhancing the intestinal barrier. | [29] |
| Enzyme | OGA | Hydrolysing O-GlcNAcylated NF-κB-p65 and IKKβ to inhibit NF-κB signaling. | [30] |
| Bacteriocins | PediocinPA-1/AcHnisin Z | Reducing colonization of VRE in vivo. | [38] |
| Microcin | limiting the expansion of pathogens. | [39,40] | |
| Bacteriocins | Enterocins | Inhibiting a wide spectrum of Gram-positive bacteria. Inhibiting the growth of cancer cells. | [42,43] |
| Bacteriocins | Bacteriocin A, B | Degrading pathogenic biofilm and having antibacterial potential. | [36] |
| Bacteriocins | Nisin A | Changing the integrity of the cancer cell membrane. | [44] |
| Secreted protein | P8 | Inducing host cell growth arrest at the G2 phase. | [46,47] |
| Ribosomal proteins | HPRP-A1; HPRP-A2 | Resisting infection. Arresting the cancer cells cycle at the G0/G1 phase and G2/M phase. | [48-53] |
| Innermembrane protein | Pre-FadA | Binding host epithelial cells. | [54] |
| Secreted protein | m-FadA | Inducing the invasion of host cells. | [54] |
| Outer membrane protein | Fap2 | Leading to colonization of Fn. Facilitating tumor immunity evasion. Binding to and activating TIGIT. Inducing host lymphocyte apoptosis. | [55-57] |
| Secreted proteins | OMVs of Fn | Inducing the colonization of host epithelial cells. | [58-60] |
| Cell envelope-associated multiprotein systems | Sus-like systems | Inducing the colonization of host epithelial cells. | [64] |
| Pili | SpaCBA | Inducing the adhesion of mucus. | [66,67] |
| Secreted proteins | EVs | Inducing the expression of the TJ protein-encoding genes and regulating the intestinal barrier. Inducing the expression of PPARα and PPARγ genes and ANGPTL4 gene. Inhibiting blood lipase lipoproteins in the bloodstream. | [71,72] |
| Secreted proteins | TcpC OMVs of EcN | Enhancing epithelial barrier. | [77-81] |
| Classification | Name | Function | Ref. |
| Flagellin | Inducing the secretion of proinflammatory cytokines. | [87] | |
| Flagellin | Recuiting flagellin specific CD4+ T-cells. | [85] | |
| Flagellin | Inducing the secretion of flagellin antibodies. | [88,89] | |
| Flagellin | Inducing the secretion of AMPs. | [90] | |
| Flagellin | Inducing the secretion of human β-defensin 2. | [91] | |
| Flagellin | Inducing the expression of lncRNA (HIF1A-AS2) and suppressing NF-kB signaling pathway activation. | [92] | |
| Outer membrane protein | OmpC, OmpW | Adhesion and invasion of the CD-associated Escherichia coli in intestinal epithelial cells. Cross-reactive bacterial proteins. | [95-97] |
| Outer membrane protein | FomA | Inducing upregulation of CD86, MHC II, and primary B cells. Inducing secretion of antigen-specific antibody IgA and IgG. | [99,100] |
| Bacterial division protein | FtsZ | Cross-reacting with TBB-5 and mediating the secretion of p-ANCA. | [102,103] |
| Bacterial heat shock protein | GroEL | Cross-reacting with Hsp60 and inducing antibodies. | [110,111] |
Biofilm formation is a process of extracellular synthesis by bacteria, and it has adverse effects on the immune response of the host[7], resulting in dysbiosis[8]. Bacteria are found in the intestinal mucosa of humans and clinical observations have revealed bacterial biofilms associated with mucosal colonization in patients with IBD[7]. Many infections also involve pathogens forming biofilms, including enterohemorrhagic Escherichia coli (EHEC)[9]. Probiotics have been documented to produce enzymes degrading biofilms of other species. Escherichia coli (E. coli) Nissle 1917 (EcN), a probiotic capable of alleviating inflammation, can produce its own biofilm and outcompete that of other intestinal pathogens[10]. Fang et al[11] found that DegP, a bifunctional (protease and chaperone) periplasmic protein secreted by EcN, contributes to the inhibition of EHEC biofilms by directly interacting with the EHEC cell surface while not affecting its own biofilm. Another probiotic, Lactobacillus rhamnosus GG (LGG), could also disrupt the biofilm formation of pathogenic E. coli and Salmonella[12]. This effect is mediated by its lectin like proteins, termed Llp1 (lectin-like protein 1) and Llp2[12]. Llp2, which is more active than Llp1, showed inhibitory activity against biofilm formation by various pathogens, including clinical Salmonella species and uropathogenic E. coli (UPEC) [12]. Thus, biofilm production and inhibition might represent key bacterial events in microbiome evolution, as well as promising targets to manage dysbiosis.
Probiotics may degrade pathogenic toxins and thus contribute to the homeostasis of gut microbiota. Clostridium difficile (C. difficile) mediates intestinal inflammation and mucosal damage by releasing two potent exotoxins, toxin A and toxin B[13], while the fungal probiotic Saccharomyces boulardii (S. boulardii) is known as the most efficient probiotic to prevent intestinal inflammation and mucosal damage associated with C. difficile infection[14]. The protective effect of S. boulardii is dependent on a 54 kDa protease, which digests both toxin A and its receptor binding sites[15]. Several human studies demonstrated that treatment with S. boulardii CNCM I-745 in dysbiosis leads to faster reestablishment of a healthy microbiome[16].
Oncogene c-MYC is associated with oncogenic transcription in malignant tumor driven by chronic bacterial infections[17], and the up-regulated c-MYC also indicates a poor prognosis in some human cancers[18]. The Lon protease from UPEC shows potential for therapeutic targeting of c-MYC in cancers, the degradation of c-MYC is dependent on both direct Lon protease cleavage and Hly-dependent activation of CK1α1, and UPEC represses transcriptional MYC regulators to inhibit c-MYC expression[19]. In mice, the recombinant Lon (rLon) protease without major toxicity delayed tumor development and increased survival in MYC-dependent bladder and colon cancer models[19]. These results indicate that probiotics may block tumor proliferation by degrading the oncogene.
Akkermansia muciniphila (A. muciniphila), one of the gut microbiota, is connected with metabolic disorders, and it reduces the energy absorption under cold conditions in the intestine epithelium[20]. P9 is an 84 kDa protein, which is secreted by A. muciniphila. P9 increases the glucagon-like peptide-1 (GLP-1) secretion in a calcium-dependent manner and specifically promotes interscapular brown adipose tissue (iBAT) non-shivering thermogenesis in the gut hormone-releasing L cells and HFD mice[21]. The ligand–receptor capture (LRC)-TriCEPS technology shows that the P9 interacts with intercellular adhesion molecule 2 (ICAM-2), and ICAM-2 reduces the secretion of the P9-induced GLP-1 in a dose-dependent manner[21]. Moreover, P9 induced the secretion of interleukin-6 (IL-6) in macrophages[21], and IL-6 can stimulate GLP-1 secretion by intestinal L cells[22].
The mucosal immune response plays an important role in IBD pathogenesis, and perturbations of the gut microbiota are a key element[23]. Probiotics can modulate the intestinal cytokine milieu to treat IBD[24] and other diseases. Peptide B7 from the probiotic Bifidobacterium longum decreases CCR2 expression on all antigen presenting cells from healthy controls but not from active IBD patients[25]. Although this bioactive peptide is useless for the treatment of active IBD patients, we cannot ignore its potential to prevent inflammation flares in the quiescent phase[25]. Another probiotic, Faecalibacterium prausnitzii (F. prausnitzii), one of the most abundant species in the human gut microbiota, possesses a 15 kDa protein with anti-inflammatory properties, termed a microbial anti-inflammatory molecule (MAM)[26]. The inflammatory suppressive role of MAMs from F. prausnitzii may be related to their effects on the inhibition of the NF-κB pathway, several cell immune responses such as Th1, Th2, and Th17 cells, and the expression of TGF-β[27,28]. The micro integral membrane protein (MIMP) identified from Lactobacillus plantarum was found to decrease proinflammatory cytokines (IFN-γ, IL-17 and IL-23), increase anti-inflammatory cytokines (IL-4 and IL-10), and fortify the intestinal barrier in a dextran sulphate sodium induced colitis model[29]. Probiotics have been documented to produce enzymes hydrolyzing key proteins in the NF-κB pathway[30]. O-GlcNAcase (OGA) is rich in Bacteroidetes and Firmicutes, the major probiotics distributed in the human gut, and reduced expression of bacterial OGA genes has been found in ulcerative colitis (UC)[30]. Bacterial OGAs are an advanced therapeutic strategy in UC that act by hydrolyzing O-GlcNAcylated NF-κB-p65 and IKKβ to inhibit NF-κB signaling in both immune cells and intestinal epithelial cells[30].
Bacteriocins are ribosomally synthesized bactericidal or bacteriostatic peptides[31,32]. Bacteriocins from probiotics maintain the microbial population-level and community-level dynamics and inhibit other strains[33]. Bacteriocins are mainly divided into two classes: Posttranslationally modified class I and unmodified class II[34,35]. In a previous study, pediocin, enterocin-A, and enterocin-B were regarded as class II bacteriocins[31], and nisin belonged to class I bacteriocins[36,37]. Pediocin PA-1/AcH secreted by Pediococcus acidilactici (P. acidilactici) MM33 and nisin Z secreted by Lactococcus lactis (L. lactis) MM19, have been proven to reduce colonization of vancomycin-resistant enterococci (VRE) in vivo[38]. Microcin-producing EcN limits the expansion of competing Enterobacteriaceae, including commensal E. coli, adherent-invasive E. coli, and Salmonella enterica in the inflamed gut[39] by utilizing catecholate siderophores[40]. Enterococcus faecium produces two synergistic bacteriocins, enterocin-A (a pediocin-like bacteriocin) and enterocin-B. Although the inhibitory spectra of enterocins A and B have small differences, both enterocins from Enterococcus faecium TI36 inhibit a wide spectrum of Gram-positive bacteria but not Gram-negative bacteria[41]. With a similar inhibitory spectrum, enterocin A has lower minimum inhibitory concentration (MIC) values than enterocin B[41].
Furthermore, the bactericidal effect is drastically increased when a mixture of the two bacteriocins is used[41]. The findings of a previous study suggested that the heterodimer of bacteriocin A and B from Enterococcus faecium por1 had antibacterial, pathogenic biofilm degradation potential but did not result in haemolysis of human red blood cells[42]. A cancer cell growth inhibitory potential of enterocins has been demonstrated, and apoptotic makers were observed in enterocin treated cancer cells including HeLa, HT-29, and AGS cells[42]. The mechanism of their effects on cancer is that cancer cells have more microvilli on their surface, which allows the membrane of cancer cells to bind large quantities of bacteriocins[43]; thus, Nisin A from L. lactis changes the integrity of the cancer cell membrane and obstructs the rearrangement of phospholipids, resulting in increased ion permeability[44]. These bacteriocins enable probiotics to treat enterobacterial infections in the gut and even some cancers.
Some oral bacteria disseminate into the colon and alter the composition of the microbiota in the colon, resulting in intestinal dysbiosis and possibly leading to colorectal cancer (CRC)[8]. FadA from Fusobacterium nucleatum drives CRC proliferation through E-cadherin and increases the expression of transcription factors and inflammatory genes via activation of β-catenin signaling[45]. Some bacterial proteins provide new strategies to treat cancer. An 8 kDa protein called p8 was isolated from Lactobacillus rhamnosus (LR) KCTC 12202BP, which regulates the p53-p21-Cyclin B1/Cdk1 signaling pathway and causes cell growth arrest at the G2 phase in a dose-dependent manner[46]. Bacterial drug delivery systems are being applied to treat CRC. The p8 protein from Pediococcus pentosaceus SL4 (PP-p8) showed antiproliferative activity in a mouse CRC model[47]. Moreover, endogenous p8 expression was much more effective than exogenous recombinant- p8 expression. This makes gene therapy possible[47].
HPRP-A1 and its enantiomer HPRP-A2 are derived from ribosomal protein L1 (RpL1) of Helicobacter pylori[48]. These proteins can resist infection including fungi, bacteria, and parasites[49,50]. Moreover, they have anticancer potential, and both peptides lead to apoptosis via caspase-3-, caspase-8-, and caspase-9-dependent pathways and inhibit cancer cell growth by arresting the cell cycle at the G0/G1 phase and G2/M phase. HPRP-A1 and its enantiomer HPRP-A2 play an important role in the inhibition of gastrointestinal cancer[51-53].
Fusobacterium nucleatum (Fn) is associated with CRC and promotes tumor formation. Fn is able to adhere to and invade intestinal endothelial cells by binding to adhesin FadA, a virulence factor from Fn[54]. FadA from E. coli enhances the connection between host epithelial cells and bacteria. FadA has two forms, anchored form (pre-FadA) and secreted form (mature FadA), thus the pre FadA-mFadA complex is regarded as a unique adhesin/invasin[54]. Fusobacterial lectin (Fap2) might mediate the binding of Fn to the host factor Gal-GalNAc in CRC, and Gal-GalNAc is highly expressed in human colorectal adenocarcinoma and metastases[55]. Other findings support that Fap2 of Fn not only leads to colonization but also facilitates tumor immunity evasion[55]. Fap2 directly binds to and activates TIGIT (an inhibitory receptor on human natural killer cells and different T cells), and the interaction between these two molecules inhibits the cytotoxicity of NK cells and the activities of cytotoxic T lymphocytes and T helper cells, increasing the immune evasion of tumor cells[56]. Fap2, as an apoptosis-inducing protein, also induces host lymphocyte apoptosis and destroys the host immune response, facilitating Fn survival[57]. Liu et al[58] identified outer membrane vesicles (OMVs) in Fn by LC/MS/MS analysis and identified several pathogenic proteins in OMVs, including FadA, Fap2, MORN2, YadA (Yersinia adhesin)-like protein, and autotransporter proteins[58]. The MORN2 domains of Fn may contribute to adhesion and active invasion[59]. Two YadA-like proteins exist in OMVs and outer membrane fractions, which reveal great adhesion ability[58]; therefore, YadA-like proteins are involved in resisting host immune defenses dependent on resisting serum killing activity and phagocytosis[60]. OMVs provide new insight into the research and development of vaccines against Fn[58].
Bacteroidetes is one of the most numerous Gram-negative bacteria in the mammalian gastrointestinal tract[61]. Cell envelope-associated multiprotein systems, namely, Sus (starch utilization system)-like systems[62], are abundant in Bacteroides. Polysaccharide utilization loci (PULs) in Sus-like systems are not only used to bind to and degrade dietary sugar[63], but they also encode a unique pathway, the ccfA–E genes, called commensal colonization factors (CCF systems) for species-specific saturable niche colonization[64]. Moreover, the CCF system is medicated by B. fragilis colonization during infection with Citrobacter rodentium and antibiotic treatment[64].
LGG has a very good mucus adhesive capacity compared to another Lactobacillus strains[65].The LGG-specific SpaCBA pili are long and thin proteinaceous protrusions on bacterial surface, which involved in three pilin monomers: SpaA , SpaB, and SpaC[66]. The SpaCBA pili mediate adhesive capacity to mucus and contribute to biofilm formation[67]. Moreover, the SpaCBA pili may also regular immune response. The spaCBA knockout LGG had twofold increased IL-8 and some pro-inflammatory markers in Caco-2 cells compared to wild-type[67].
Under dysbiosis, increased permeability of the intestinal epithelium leads to low-grade inflammation and metabolic dysfunctions[68]. However, according to the leaky gut hypothesis, if only the F. prausnitzii is present as a probiotic, it will not beneficial to the intestine health and dysbiosis-induced diseases but enter the bloodstream by passing though the gut barrier and may cause systemic consequences because of obesity and a high-fat diet (HFD)[69,70]. Moosavi et al[71] show that F. prausnitzii–derived extracellular vesicles (EVs) contain different proteins with a molecular weight of 11 to 245 kDa. Compared with F. prausnitzii, its EVs in the Caco-2 cell line significantly regulate the intestinal barrier permeability due to increasing the expression of the tight junction (TJ) protein encoding genes ZO1 and OCLN, as well as PPARα and PPARγ genes and their targeted gene ANGPTL4 at the mRNA level[71]. TJ proteins connect the adjacent epithelial cells and block the paracellular space in order to obstruct pathogens[72]. ANGPTL4 inhibits blood lipase lipoproteins in the bloodstream, which reduces the intake of free fatty acids and cholesterol into the tissues[73-76].
TcpC from EcN enhanced the intestinal barrier function by increasing the expre
Flagellin is a common conserved component of bacteria, and it induces both innate and specific immunity, showing a close relationship between dysbiosis and IBD[85], but flagellin of some probiotics has anti-inflammatory effects[86]. Flagellin is regarded as the major antigen in pathogenic bacteria. Flagellin binds with the pattern-recognition receptor Toll-like receptor 5 (TLR5), inducing the secretion of proinflammatory cytokines[87]. Compared with healthy controls, both Crohn’s disease (CD) and UC patients have a relative increase in the proportion of flagellin specific CD4+ T-cells. Cook et al[85] found a positive correlation between the relative abundance of bacteria [Escherichia/Shigella and (Ruminococcus) gnavus group] in IBD patients and high concentrations of flagellin antibodies, including anti-Fla2 IgG and anti-Fla2 IgA[88]. Specifically, CBir1 flagellin has been associated with complicated CD, and enzyme-linked immunosorbent assays proved that anti-CBir1 IgG is independently associated with CD[89]. Flagellin may provide a clinically novel approach to prevent pathogen infections, including vancomycin-resistant Enterococcus (VRE). Intestinal epithelial cells and Paneth cells secrete the antimicrobial protein (AMP) RegIIIγ to kill microorganisms and directly respond to flagellin via the Toll-like receptor (TLR)–myeloid differentiation factor 88–mediated pathway[90]. Flagellin of EcN stimulates intestinal epithelial cells to produce human β-defensin 2 via three main MAP kinase pathways, including ERK1/2, JNK, and p38[91]. Bacterial flagellin also induces negative regulation of inflammation. Roseburia intestinalis (R. intestinalis), a dominant symbiotic microbiota in the intestine, suppresses inflammation by inducing Treg cells and upregulating anti-inflammatory cytokines. However, R. intestinalis is significantly reduced in CD patients[86]. Flagellin in R. intestinalis induces the expression of lncRNA (HIF1A-AS2) in a dose- and time-dependent manner via p38 STAT1 activation, and HIF1A-AS2 inhibits the expression of inflammatory genes by suppressing NF-kB signaling pathway activation[92].
Some evidence linking intestinal dysbiosis with autoimmune diseases has shown that they are both associated with increased inflammation[93,94]. Bacterial outer membra
Primary sclerosing cholangitis (PSC) and autoimmune hepatitis (AIH) are frequently associated with chronic IBD, including UC and CD[101]. The immune reaction in PSCs is mediated by autoantibodies, including pANCA, that recognize both β-tubulin isotype 5 (TBB-5) and the bacterial antigen cell division protein FtsZ[102]. Human TBB-5 and FtsZ share a high degree of structural homology in evolutionarily conserved epitopes[103]. Moreover, B cells respond directly to microbial constituents in PSCs and AIH[104].
Helicobacter hepaticus (Hh) can induce intestinal inflammation in DC-LMP1/CD40 mice[105]. These immunodeficient mice lost intestinal CD103+ DCs and IL-10+ Helios−induced Tregs (iTregs) but had increased IL17+ IFNγ+ Th17/Th1 cells and pathogenic IFNγ+ Th1 cells[106,107]. They developed fetal colitis similar to human IBD, because CD40-CD40L interactions are connected with the pathogenesis of IBD[108,109].A 60 kDa Hh-protein, GroEL, as the main antigen recognized by antibodies in an iTreg-free setting, triggers fatal colitis[110]. The bacterial GroEL and human heat shock protein 60 (Hsp60) share a high similarity and molecular mimicry[111,112], hence the antibodies cross-react with Hsp60 and GroEL, which contribute to IBD and autoimmune diseases[110].
Understanding the key bacterial proteins is significant to both the diagnosis and management of dysbiosis related diseases. For the incendiary proteins involved in autoimmune diseases and tumors, the presence of the specific marker in the microbiota or its specific antibodies might indicate the prognosis of diseases. The therapeutic value of targeting these markers would also be tempting. Knowing the key elements of microbiota could provide much more specific target than generally modulating the microbiota, which is super-high dimensional in taxonomy.
The current mainstream microbiome manipulation approaches are intensively investigated, including supplement of probiotics and prebiotics, and fecal microbiota transplantation (FMT). However, the probiotics should be strain-defined to gain standardized safety, dose, and effect; the adverse events associated with FMT have been found recently. The transplanted probiotics met indigenous microbiome-mediated mucosal colonization resistance in mice and even a specific colonization resistance in a person-, strain-, and region- dependent manner in humans[113]. Our recent mathematical model studies also suggested intriguing behavior of microbiome in response to probiotic supplement[114]. For FMT, the risk of unknown infections is still inevitable even after rigorous tests on the donors. The specific microbial proteins are easier to be cloned, purified, tested, optimized, and standardized, which is crucial for the pharmacology. Furthermore, the natural beneficial bacterial proteins can be artificially engineered and optimized to maximum their mechanism. This review summarizes the pathogenic and therapeutic mechanisms of some bioactive microbial proteins. This field is cutting edge, and there is a need for further studies to explore the role of the key gut microbial proteins in dysbiosis associated diseases.
Manuscript source: Invited manuscript
Specialty type: Gastroenterology and hepatology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Biondi A S-Editor: Ma YJ L-Editor: Wang TQ P-Editor: Zhang YL
| 1. | Chen MX, Wang SY, Kuo CH, Tsai IL. Metabolome analysis for investigating host-gut microbiota interactions. J Formos Med Assoc. 2019;118 Suppl 1:S10-S22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 121] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 2. | De Angelis M, Garruti G, Minervini F, Bonfrate L, Portincasa P, Gobbetti M. The Food-gut Human Axis: The Effects of Diet on Gut Microbiota and Metabolome. Curr Med Chem. 2019;26:3567-3583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 77] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 3. | Dagnelie MA, Corvec S, Khammari A, Dréno B. Bacterial extracellular vesicles: A new way to decipher host-microbiota communications in inflammatory dermatoses. Exp Dermatol. 2020;29:22-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 33] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 4. | Lopetuso LR, Ianiro G, Scaldaferri F, Cammarota G, Gasbarrini A. Gut Virome and Inflammatory Bowel Disease. Inflamm Bowel Dis. 2016;22:1708-1712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Zuo T, Lu XJ, Zhang Y, Cheung CP, Lam S, Zhang F, Tang W, Ching JYL, Zhao R, Chan PKS, Sung JJY, Yu J, Chan FKL, Cao Q, Sheng JQ, Ng SC. Gut mucosal virome alterations in ulcerative colitis. Gut. 2019;68:1169-1179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 196] [Cited by in RCA: 285] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 6. | Marzano V, Mancinelli L, Bracaglia G, Del Chierico F, Vernocchi P, Di Girolamo F, Garrone S, Tchidjou Kuekou H, D'Argenio P, Dallapiccola B, Urbani A, Putignani L. "Omic" investigations of protozoa and worms for a deeper understanding of the human gut "parasitome". PLoS Negl Trop Dis. 2017;11:e0005916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 34] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 7. | Srivastava A, Gupta J, Kumar S, Kumar A. Gut biofilm forming bacteria in inflammatory bowel disease. Microb Pathog. 2017;112:5-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 8. | Koliarakis I, Messaritakis I, Nikolouzakis TK, Hamilos G, Souglakos J, Tsiaoussis J. Oral Bacteria and Intestinal Dysbiosis in Colorectal Cancer. Int J Mol Sci. 2019;20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 171] [Article Influence: 28.5] [Reference Citation Analysis (0)] |
| 9. | Sharma G, Sharma S, Sharma P, Chandola D, Dang S, Gupta S, Gabrani R. Escherichia coli biofilm: development and therapeutic strategies. J Appl Microbiol. 2016;121:309-319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 252] [Article Influence: 28.0] [Reference Citation Analysis (0)] |
| 10. | Hancock V, Dahl M, Klemm P. Probiotic Escherichia coli strain Nissle 1917 outcompetes intestinal pathogens during biofilm formation. J Med Microbiol. 2010;59:392-399. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 92] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 11. | Fang K, Jin X, Hong SH. Probiotic Escherichia coli inhibits biofilm formation of pathogenic E. coli via extracellular activity of DegP. Sci Rep. 2018;8:4939. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 61] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 12. | Petrova MI, Imholz NC, Verhoeven TL, Balzarini J, Van Damme EJ, Schols D, Vanderleyden J, Lebeer S. Lectin-Like Molecules of Lactobacillus rhamnosus GG Inhibit Pathogenic Escherichia coli and Salmonella Biofilm Formation. PLoS One. 2016;11:e0161337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 66] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 13. | Pothoulakis C, Lamont JT. Microbes and microbial toxins: paradigms for microbial-mucosal interactions II. The integrated response of the intestine to Clostridium difficile toxins. Am J Physiol Gastrointest Liver Physiol. 2001;280:G178-G183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 111] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 14. | McFarland LV. Meta-analysis of probiotics for the prevention of antibiotic associated diarrhea and the treatment of Clostridium difficile disease. Am J Gastroenterol. 2006;101:812-822. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 563] [Cited by in RCA: 492] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 15. | Castagliuolo I, LaMont JT, Nikulasson ST, Pothoulakis C. Saccharomyces boulardii protease inhibits Clostridium difficile toxin A effects in the rat ileum. Infect Immun. 1996;64:5225-5232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 166] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 16. | Moré MI, Swidsinski A. Saccharomyces boulardii CNCM I-745 supports regeneration of the intestinal microbiota after diarrheic dysbiosis - a review. Clin Exp Gastroenterol. 2015;8:237-255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 17. | Wierstra I, Alves J. Cyclin E/Cdk2, P/CAF, and E1A regulate the transactivation of the c-myc promoter by FOXM1. Biochem Biophys Res Commun. 2008;368:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 18. | Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 351] [Cited by in RCA: 580] [Article Influence: 82.9] [Reference Citation Analysis (0)] |
| 19. | Butler DSC, Cafaro C, Putze J, Wan MLY, Tran TH, Ambite I, Ahmadi S, Kjellström S, Welinder C, Chao SM, Dobrindt U, Svanborg C. A bacterial protease depletes c-MYC and increases survival in mouse models of bladder and colon cancer. Nat Biotechnol. 2021;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 20. | Chevalier C, Stojanović O, Colin DJ, Suarez-Zamorano N, Tarallo V, Veyrat-Durebex C, Rigo D, Fabbiano S, Stevanović A, Hagemann S, Montet X, Seimbille Y, Zamboni N, Hapfelmeier S, Trajkovski M. Gut Microbiota Orchestrates Energy Homeostasis during Cold. Cell. 2015;163:1360-1374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 419] [Cited by in RCA: 553] [Article Influence: 61.4] [Reference Citation Analysis (0)] |
| 21. | Yoon HS, Cho CH, Yun MS, Jang SJ, You HJ, Kim JH, Han D, Cha KH, Moon SH, Lee K, Kim YJ, Lee SJ, Nam TW, Ko G. Akkermansia muciniphila secretes a glucagon-like peptide-1-inducing protein that improves glucose homeostasis and ameliorates metabolic disease in mice. Nat Microbiol. 2021;6:563-573. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 326] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 22. | Ellingsgaard H, Hauselmann I, Schuler B, Habib AM, Baggio LL, Meier DT, Eppler E, Bouzakri K, Wueest S, Muller YD, Hansen AM, Reinecke M, Konrad D, Gassmann M, Reimann F, Halban PA, Gromada J, Drucker DJ, Gribble FM, Ehses JA, Donath MY. Interleukin-6 enhances insulin secretion by increasing glucagon-like peptide-1 secretion from L cells and alpha cells. Nat Med. 2011;17:1481-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 687] [Cited by in RCA: 684] [Article Influence: 48.9] [Reference Citation Analysis (0)] |
| 23. | Mowat AM. To respond or not to respond - a personal perspective of intestinal tolerance. Nat Rev Immunol. 2018;18:405-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 125] [Article Influence: 20.8] [Reference Citation Analysis (0)] |
| 24. | Fernández-Tomé S, Montalban-Arques A, Díaz-Guerra A, Galvan-Roman JM, Marin AC, Mora-Gutiérrez I, Ortega Moreno L, Santander C, Sánchez B, Chaparro M, Gisbert JP, Bernardo D. Peptides encrypted in the human intestinal microbial-exoproteome as novel biomarkers and immunomodulatory compounds in the gastrointestinal tract. J Funct Foods. 2019;52:459-468. [RCA] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 3] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 25. | Fernández-Tomé S, Marin AC, Ortega Moreno L, Baldan-Martin M, Mora-Gutiérrez I, Lanas-Gimeno A, Moreno-Monteagudo JA, Santander C, Sánchez B, Chaparro M, Gisbert JP, Bernardo D. Immunomodulatory Effect of Gut Microbiota-Derived Bioactive Peptides on Human Immune System from Healthy Controls and Patients with Inflammatory Bowel Disease. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 29] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 26. | Quévrain E, Maubert MA, Michon C, Chain F, Marquant R, Tailhades J, Miquel S, Carlier L, Bermúdez-Humarán LG, Pigneur B, Lequin O, Kharrat P, Thomas G, Rainteau D, Aubry C, Breyner N, Afonso C, Lavielle S, Grill JP, Chassaing G, Chatel JM, Trugnan G, Xavier R, Langella P, Sokol H, Seksik P. Identification of an anti-inflammatory protein from Faecalibacterium prausnitzii, a commensal bacterium deficient in Crohn's disease. Gut. 2016;65:415-425. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 450] [Cited by in RCA: 570] [Article Influence: 63.3] [Reference Citation Analysis (0)] |
| 27. | Breyner NM, Michon C, de Sousa CS, Vilas Boas PB, Chain F, Azevedo VA, Langella P, Chatel JM. Microbial Anti-Inflammatory Molecule (MAM) from Faecalibacterium prausnitzii Shows a Protective Effect on DNBS and DSS-Induced Colitis Model in Mice through Inhibition of NF-κB Pathway. Front Microbiol. 2017;8:114. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 147] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 28. | Quévrain E, Maubert MA, Sokol H, Devreese B, Seksik P. The presence of the anti-inflammatory protein MAM, from Faecalibacterium prausnitzii, in the intestinal ecosystem. Gut. 2016;65:882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Yin M, Yan X, Weng W, Yang Y, Gao R, Liu M, Pan C, Zhu Q, Li H, Wei Q, Shen T, Ma Y, Qin H. Micro Integral Membrane Protein (MIMP), a Newly Discovered Anti-Inflammatory Protein of Lactobacillus Plantarum, Enhances the Gut Barrier and Modulates Microbiota and Inflammatory Cytokines. Cell Physiol Biochem. 2018;45:474-490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 42] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 30. | He X, Gao J, Peng L, Hu T, Wan Y, Zhou M, Zhen P, Cao H. Bacterial O-GlcNAcase genes abundance decreases in ulcerative colitis patients and its administration ameliorates colitis in mice. Gut. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 31. | Martinez FA, Balciunas EM, Converti A, Cotter PD, de Souza Oliveira RP. Bacteriocin production by Bifidobacterium spp. A review. Biotechnol Adv. 2013;31:482-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 147] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 32. | Klaenhammer TR. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev. 1993;12:39-85. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 892] [Cited by in RCA: 943] [Article Influence: 29.5] [Reference Citation Analysis (0)] |
| 33. | Riley MA, Wertz JE. Bacteriocin diversity: ecological and evolutionary perspectives. Biochimie. 2002;84:357-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 194] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 34. | Collado MC, Hernández M, Sanz Y. Production of bacteriocin-like inhibitory compounds by human fecal Bifidobacterium strains. J Food Prot. 2005;68:1034-1040. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 35. | Riley MA, Wertz JE. Bacteriocins: evolution, ecology, and application. Annu Rev Microbiol. 2002;56:117-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 817] [Cited by in RCA: 738] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 36. | Lee H, Kim HY. Lantibiotics, class I bacteriocins from the genus Bacillus. J Microbiol Biotechnol. 2011;21:229-235. [PubMed] |
| 37. | Sahl HG, Bierbaum G. Lantibiotics: biosynthesis and biological activities of uniquely modified peptides from gram-positive bacteria. Annu Rev Microbiol. 1998;52:41-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 328] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 38. | Millette M, Cornut G, Dupont C, Shareck F, Archambault D, Lacroix M. Capacity of human nisin- and pediocin-producing lactic Acid bacteria to reduce intestinal colonization by vancomycin-resistant enterococci. Appl Environ Microbiol. 2008;74:1997-2003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 116] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 39. | Sassone-Corsi M, Nuccio SP, Liu H, Hernandez D, Vu CT, Takahashi AA, Edwards RA, Raffatellu M. Microcins mediate competition among Enterobacteriaceae in the inflamed gut. Nature. 2016;540:280-283. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 396] [Article Influence: 44.0] [Reference Citation Analysis (0)] |
| 40. | Patzer SI, Baquero MR, Bravo D, Moreno F, Hantke K. The colicin G, H and X determinants encode microcins M and H47, which might utilize the catecholate siderophore receptors FepA, Cir, Fiu and IroN. Microbiology (Reading). 2003;149:2557-2570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 176] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 41. | Casaus P, Nilsen T, Cintas LM, Nes IF, Hernández PE, Holo H. Enterocin B, a new bacteriocin from Enterococcus faecium T136 which can act synergistically with enterocin A. Microbiology (Reading). 1997;143 ( Pt 7):2287-2294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 258] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 42. | Ankaiah D, Palanichamy E, Antonyraj CB, Ayyanna R, Perumal V, Ahamed SIB, Arul V. Cloning, overexpression, purification of bacteriocin enterocin-B and structural analysis, interaction determination of enterocin-A, B against pathogenic bacteria and human cancer cells. Int J Biol Macromol. 2018;116:502-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 54] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 43. | Chan SC, Hui L, Chen HM. Enhancement of the cytolytic effect of anti-bacterial cecropin by the microvilli of cancer cells. Anticancer Res. 1998;18:4467-4474. [PubMed] |
| 44. | Joo NE, Ritchie K, Kamarajan P, Miao D, Kapila YL. Nisin, an apoptogenic bacteriocin and food preservative, attenuates HNSCC tumorigenesis via CHAC1. Cancer Med. 2012;1:295-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 45. | Rubinstein MR, Wang X, Liu W, Hao Y, Cai G, Han YW. Fusobacterium nucleatum promotes colorectal carcinogenesis by modulating E-cadherin/β-catenin signaling via its FadA adhesin. Cell Host Microbe. 2013;14:195-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1511] [Cited by in RCA: 1662] [Article Influence: 138.5] [Reference Citation Analysis (1)] |
| 46. | An BC, Hong S, Park HJ, Kim BK, Ahn JY, Ryu Y, An JH, Chung MJ. Anti-Colorectal Cancer Effects of Probiotic-Derived p8 Protein. Genes (Basel). 2019;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 27] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 47. | An BC, Ryu Y, Yoon YS, Choi O, Park HJ, Kim TY, Kim SI, Kim BK, Chung MJ. Colorectal Cancer Therapy Using a Pediococcus pentosaceus SL4 Drug Delivery System Secreting Lactic Acid Bacteria-Derived Protein p8. Mol Cells. 2019;42:755-762. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 48. | Pütsep K, Brändén CI, Boman HG, Normark S. Antibacterial peptide from H. pylori. Nature. 1999;398:671-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 145] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 49. | Liu R, Ni Y, Song J, Xu Z, Qiu J, Wang L, Zhu Y, Huang Y, Ji M, Chen Y. Research on the effect and mechanism of antimicrobial peptides HPRP-A1/A2 work against Toxoplasma gondii infection. Parasite Immunol. 2019;41:e12619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 50. | Zhu J, Huang Y, Chen M, Hu C, Chen Y. Functional Synergy Of Antimicrobial Peptides And Chlorhexidine Acetate Against Gram-Negative/Gram-Positive Bacteria And A Fungus In Vitro And In Vivo. Infect Drug Resist. 2019;12:3227-3239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 51. | Cho E, Lee JK, Park E, Seo CH, Luchian T, Park Y. Antitumor activity of HPA3P through RIPK3-dependent regulated necrotic cell death in colon cancer. Oncotarget. 2018;9:7902-7917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 52. | Hu C, Chen X, Huang Y, Chen Y. Synergistic effect of the pro-apoptosis peptide kla-TAT and the cationic anticancer peptide HPRP-A1. Apoptosis. 2018;23:132-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 29] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 53. | Hao W, Hu C, Huang Y, Chen Y. Coadministration of kla peptide with HPRP-A1 to enhance anticancer activity. PLoS One. 2019;14:e0223738. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 54. | Xu M, Yamada M, Li M, Liu H, Chen SG, Han YW. FadA from Fusobacterium nucleatum utilizes both secreted and nonsecreted forms for functional oligomerization for attachment and invasion of host cells. J Biol Chem. 2007;282:25000-25009. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 145] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 55. | Abed J, Emgård JE, Zamir G, Faroja M, Almogy G, Grenov A, Sol A, Naor R, Pikarsky E, Atlan KA, Mellul A, Chaushu S, Manson AL, Earl AM, Ou N, Brennan CA, Garrett WS, Bachrach G. Fap2 Mediates Fusobacterium nucleatum Colorectal Adenocarcinoma Enrichment by Binding to Tumor-Expressed Gal-GalNAc. Cell Host Microbe. 2016;20:215-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 579] [Article Influence: 72.4] [Reference Citation Analysis (0)] |
| 56. | Gur C, Ibrahim Y, Isaacson B, Yamin R, Abed J, Gamliel M, Enk J, Bar-On Y, Stanietsky-Kaynan N, Coppenhagen-Glazer S, Shussman N, Almogy G, Cuapio A, Hofer E, Mevorach D, Tabib A, Ortenberg R, Markel G, Miklić K, Jonjic S, Brennan CA, Garrett WS, Bachrach G, Mandelboim O. Binding of the Fap2 protein of Fusobacterium nucleatum to human inhibitory receptor TIGIT protects tumors from immune cell attack. Immunity. 2015;42:344-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1015] [Cited by in RCA: 975] [Article Influence: 97.5] [Reference Citation Analysis (0)] |
| 57. | Kaplan CW, Lux R, Huynh T, Jewett A, Shi W, Haake SK. Fusobacterium nucleatum apoptosis-inducing outer membrane protein. J Dent Res. 2005;84:700-704. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 58. | Liu J, Hsieh CL, Gelincik O, Devolder B, Sei S, Zhang S, Lipkin SM, Chang YF. Proteomic characterization of outer membrane vesicles from gut mucosa-derived fusobacterium nucleatum. J Proteomics. 2019;195:125-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 48] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 59. | Manson McGuire A, Cochrane K, Griggs AD, Haas BJ, Abeel T, Zeng Q, Nice JB, MacDonald H, Birren BW, Berger BW, Allen-Vercoe E, Earl AM. Evolution of invasion in a diverse set of Fusobacterium species. mBio. 2014;5:e01864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 82] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 60. | Heise T, Dersch P. Identification of a domain in Yersinia virulence factor YadA that is crucial for extracellular matrix-specific cell adhesion and uptake. Proc Natl Acad Sci USA. 2006;103:3375-3380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 61. | Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635-1638. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5700] [Cited by in RCA: 5587] [Article Influence: 279.4] [Reference Citation Analysis (2)] |
| 62. | Martens EC, Koropatkin NM, Smith TJ, Gordon JI. Complex glycan catabolism by the human gut microbiota: the Bacteroidetes Sus-like paradigm. J Biol Chem. 2009;284:24673-24677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 434] [Cited by in RCA: 476] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 63. | Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012;10:323-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 838] [Cited by in RCA: 1035] [Article Influence: 79.6] [Reference Citation Analysis (0)] |
| 64. | Lee SM, Donaldson GP, Mikulski Z, Boyajian S, Ley K, Mazmanian SK. Bacterial colonization factors control specificity and stability of the gut microbiota. Nature. 2013;501:426-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 381] [Cited by in RCA: 460] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 65. | Tuomola EM, Ouwehand AC, Salminen SJ. The effect of probiotic bacteria on the adhesion of pathogens to human intestinal mucus. FEMS Immunol Med Microbiol. 1999;26:137-142. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 135] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 66. | Segers ME, Lebeer S. Towards a better understanding of Lactobacillus rhamnosus GG--host interactions. Microb Cell Fact. 2014;13 Suppl 1:S7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 216] [Cited by in RCA: 299] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 67. | Lebeer S, Claes I, Tytgat HL, Verhoeven TL, Marien E, von Ossowski I, Reunanen J, Palva A, Vos WM, Keersmaecker SC, Vanderleyden J. Functional analysis of Lactobacillus rhamnosus GG pili in relation to adhesion and immunomodulatory interactions with intestinal epithelial cells. Appl Environ Microbiol. 2012;78:185-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 249] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 68. | Ahmadi Badi S, Moshiri A, Fateh A, Rahimi Jamnani F, Sarshar M, Vaziri F, Siadat SD. Microbiota-Derived Extracellular Vesicles as New Systemic Regulators. Front Microbiol. 2017;8:1610. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 99] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 69. | Maguire M, Maguire G. Gut dysbiosis, leaky gut, and intestinal epithelial proliferation in neurological disorders: towards the development of a new therapeutic using amino acids, prebiotics, probiotics, and postbiotics. Rev Neurosci. 2019;30:179-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 54] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 70. | Camilleri M. Leaky gut: mechanisms, measurement and clinical implications in humans. Gut. 2019;68:1516-1526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 663] [Article Influence: 110.5] [Reference Citation Analysis (0)] |
| 71. | Moosavi SM, Akhavan Sepahi A, Mousavi SF, Vaziri F, Siadat SD. The effect of Faecalibacterium prausnitzii and its extracellular vesicles on the permeability of intestinal epithelial cells and expression of PPARs and ANGPTL4 in the Caco-2 cell culture model. J Diabetes Metab Disord. 2020;19:1061-1069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 72. | Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011;141:769-776. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 754] [Cited by in RCA: 855] [Article Influence: 61.1] [Reference Citation Analysis (1)] |
| 73. | Shan L, Yu XC, Liu Z, Hu Y, Sturgis LT, Miranda ML, Liu Q. The angiopoietin-like proteins ANGPTL3 and ANGPTL4 inhibit lipoprotein lipase activity through distinct mechanisms. J Biol Chem. 2009;284:1419-1424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 112] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 74. | Yoshida K, Shimizugawa T, Ono M, Furukawa H. Angiopoietin-like protein 4 is a potent hyperlipidemia-inducing factor in mice and inhibitor of lipoprotein lipase. J Lipid Res. 2002;43:1770-1772. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 270] [Cited by in RCA: 290] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 75. | Mandard S, Zandbergen F, van Straten E, Wahli W, Kuipers F, Müller M, Kersten S. The fasting-induced adipose factor/angiopoietin-like protein 4 is physically associated with lipoproteins and governs plasma lipid levels and adiposity. J Biol Chem. 2006;281:934-944. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 311] [Cited by in RCA: 339] [Article Influence: 17.0] [Reference Citation Analysis (0)] |
| 76. | Sukonina V, Lookene A, Olivecrona T, Olivecrona G. Angiopoietin-like protein 4 converts lipoprotein lipase to inactive monomers and modulates lipase activity in adipose tissue. Proc Natl Acad Sci USA. 2006;103:17450-17455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 320] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 77. | Hering NA, Richter JF, Fromm A, Wieser A, Hartmann S, Günzel D, Bücker R, Fromm M, Schulzke JD, Troeger H. TcpC protein from E. coli Nissle improves epithelial barrier function involving PKCζ and ERK1/2 signaling in HT-29/B6 cells. Mucosal Immunol. 2014;7:369-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 54] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Ukena SN, Singh A, Dringenberg U, Engelhardt R, Seidler U, Hansen W, Bleich A, Bruder D, Franzke A, Rogler G, Suerbaum S, Buer J, Gunzer F, Westendorf AM. Probiotic Escherichia coli Nissle 1917 inhibits leaky gut by enhancing mucosal integrity. PLoS One. 2007;2:e1308. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 310] [Cited by in RCA: 349] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 79. | Zyrek AA, Cichon C, Helms S, Enders C, Sonnenborn U, Schmidt MA. Molecular mechanisms underlying the probiotic effects of Escherichia coli Nissle 1917 involve ZO-2 and PKCzeta redistribution resulting in tight junction and epithelial barrier repair. Cell Microbiol. 2007;9:804-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 310] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 80. | Toloza L, Giménez R, Fábrega MJ, Alvarez CS, Aguilera L, Cañas MA, Martín-Venegas R, Badia J, Baldomà L. The secreted autotransporter toxin (Sat) does not act as a virulence factor in the probiotic Escherichia coli strain Nissle 1917. BMC Microbiol. 2015;15:250. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 81. | Alvarez CS, Badia J, Bosch M, Giménez R, Baldomà L. Outer Membrane Vesicles and Soluble Factors Released by Probiotic Escherichia coli Nissle 1917 and Commensal ECOR63 Enhance Barrier Function by Regulating Expression of Tight Junction Proteins in Intestinal Epithelial Cells. Front Microbiol. 2016;7:1981. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 145] [Article Influence: 16.1] [Reference Citation Analysis (0)] |
| 82. | Luettig J, Rosenthal R, Barmeyer C, Schulzke JD. Claudin-2 as a mediator of leaky gut barrier during intestinal inflammation. Tissue Barriers. 2015;3:e977176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 201] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 83. | Hering NA, Fromm M, Schulzke JD. Determinants of colonic barrier function in inflammatory bowel disease and potential therapeutics. J Physiol. 2012;590:1035-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 210] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 84. | Landy J, Ronde E, English N, Clark SK, Hart AL, Knight SC, Ciclitira PJ, Al-Hassi HO. Tight junctions in inflammatory bowel diseases and inflammatory bowel disease associated colorectal cancer. World J Gastroenterol. 2016;22:3117-3126. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 294] [Cited by in RCA: 369] [Article Influence: 41.0] [Reference Citation Analysis (4)] |
| 85. | Cook L, Lisko DJ, Wong MQ, Garcia RV, Himmel ME, Seidman EG, Bressler B, Levings MK, Steiner TS. Analysis of Flagellin-Specific Adaptive Immunity Reveals Links to Dysbiosis in Patients With Inflammatory Bowel Disease. Cell Mol Gastroenterol Hepatol. 2020;9:485-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 86. | Shen Z, Zhu C, Quan Y, Yang J, Yuan W, Yang Z, Wu S, Luo W, Tan B, Wang X. Insights into Roseburia intestinalis which alleviates experimental colitis pathology by inducing anti-inflammatory responses. J Gastroenterol Hepatol. 2018;33:1751-1760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 96] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 87. | Means TK, Hayashi F, Smith KD, Aderem A, Luster AD. The Toll-like receptor 5 stimulus bacterial flagellin induces maturation and chemokine production in human dendritic cells. J Immunol. 2003;170:5165-5175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 317] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 88. | Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296-1306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 298] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 89. | Targan SR, Landers CJ, Yang H, Lodes MJ, Cong Y, Papadakis KA, Vasiliauskas E, Elson CO, Hershberg RM. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn's disease. Gastroenterology. 2005;128:2020-2028. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 352] [Cited by in RCA: 351] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 90. | Kinnebrew MA, Ubeda C, Zenewicz LA, Smith N, Flavell RA, Pamer EG. Bacterial flagellin stimulates Toll-like receptor 5-dependent defense against vancomycin-resistant Enterococcus infection. J Infect Dis. 2010;201:534-543. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 196] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 91. | Schlee M, Wehkamp J, Altenhoefer A, Oelschlaeger TA, Stange EF, Fellermann K. Induction of human beta-defensin 2 by the probiotic Escherichia coli Nissle 1917 is mediated through flagellin. Infect Immun. 2007;75:2399-2407. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 225] [Cited by in RCA: 231] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 92. | Quan Y, Song K, Zhang Y, Zhu C, Shen Z, Wu S, Luo W, Tan B, Yang Z, Wang X. Roseburia intestinalis-derived flagellin is a negative regulator of intestinal inflammation. Biochem Biophys Res Commun. 2018;501:791-799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 93. | Wu HJ, Wu E. The role of gut microbiota in immune homeostasis and autoimmunity. Gut Microbes. 2012;3:4-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 622] [Cited by in RCA: 899] [Article Influence: 69.2] [Reference Citation Analysis (0)] |
| 94. | de Oliveira GLV, Leite AZ, Higuchi BS, Gonzaga MI, Mariano VS. Intestinal dysbiosis and probiotic applications in autoimmune diseases. Immunology. 2017;152:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 201] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 95. | Cohavy O, Bruckner D, Gordon LK, Misra R, Wei B, Eggena ME, Targan SR, Braun J. Colonic bacteria express an ulcerative colitis pANCA-related protein epitope. Infect Immun. 2000;68:1542-1548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 143] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 96. | Wei B, Dalwadi H, Gordon LK, Landers C, Bruckner D, Targan SR, Braun J. Molecular cloning of a Bacteroides caccae TonB-linked outer membrane protein identified by an inflammatory bowel disease marker antibody. Infect Immun. 2001;69:6044-6054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 57] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 97. | Rolhion N, Carvalho FA, Darfeuille-Michaud A. OmpC and the sigma(E) regulatory pathway are involved in adhesion and invasion of the Crohn's disease-associated Escherichia coli strain LF82. Mol Microbiol. 2007;63:1684-1700. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 106] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 98. | Bolstad AI, Jensen HB, Bakken V. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55-71. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 99. | Toussi DN, Liu X, Massari P. The FomA porin from Fusobacterium nucleatum is a Toll-like receptor 2 agonist with immune adjuvant activity. Clin Vaccine Immunol. 2012;19:1093-1101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 44] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 100. | Gu X, Song LJ, Li LX, Liu T, Zhang MM, Li Z, Wang P, Li M, Zuo XL. Fusobacterium nucleatum Causes Microbial Dysbiosis and Exacerbates Visceral Hypersensitivity in a Colonization-Independent Manner. Front Microbiol. 2020;11:1281. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 101. | Broomé U, Bergquist A. Primary sclerosing cholangitis, inflammatory bowel disease, and colon cancer. Semin Liver Dis. 2006;26:31-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 157] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 102. | Tripathi A, Debelius J, Brenner DA, Karin M, Loomba R, Schnabl B, Knight R. The gut-liver axis and the intersection with the microbiome. Nat Rev Gastroenterol Hepatol. 2018;15:397-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 872] [Cited by in RCA: 970] [Article Influence: 138.6] [Reference Citation Analysis (0)] |
| 103. | Terjung B, Söhne J, Lechtenberg B, Gottwein J, Muennich M, Herzog V, Mähler M, Sauerbruch T, Spengler U. p-ANCAs in autoimmune liver disorders recognise human beta-tubulin isotype 5 and cross-react with microbial protein FtsZ. Gut. 2010;59:808-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 107] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 104. | Terjung B, Spengler U. Atypical p-ANCA in PSC and AIH: a hint toward a "leaky gut"? Clin Rev Allergy Immunol. 2009;36:40-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 105. | Cahill RJ, Foltz CJ, Fox JG, Dangler CA, Powrie F, Schauer DB. Inflammatory bowel disease: an immunity-mediated condition triggered by bacterial infection with Helicobacter hepaticus. Infect Immun. 1997;65:3126-3131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 247] [Cited by in RCA: 232] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 106. | Barthels C, Ogrinc A, Steyer V, Meier S, Simon F, Wimmer M, Blutke A, Straub T, Zimber-Strobl U, Lutgens E, Marconi P, Ohnmacht C, Garzetti D, Stecher B, Brocker T. CD40-signalling abrogates induction of RORγt+ Treg cells by intestinal CD103+ DCs and causes fatal colitis. Nat Commun. 2017;8:14715. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 107. | Kusters P, Seijkens T, Bürger C, Legein B, Winkels H, Gijbels M, Barthels C, Bennett R, Beckers L, Atzler D, Biessen E, Brocker T, Weber C, Gerdes N, Lutgens E. Constitutive CD40 Signaling in Dendritic Cells Limits Atherosclerosis by Provoking Inflammatory Bowel Disease and Ensuing Cholesterol Malabsorption. Am J Pathol. 2017;187:2912-2919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 108. | Ludwiczek O, Kaser A, Tilg H. Plasma levels of soluble CD40 Ligand are elevated in inflammatory bowel diseases. Int J Colorectal Dis. 2003;18:142-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 109. | Danese S, Sans M, Fiocchi C. The CD40/CD40L costimulatory pathway in inflammatory bowel disease. Gut. 2004;53:1035-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 120] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 110. | Friedrich V, Forné I, Matzek D, Ring D, Popper B, Jochum L, Spriewald S, Straub T, Imhof A, Krug A, Stecher B, Brocker T. Helicobacter hepaticus is required for immune targeting of bacterial heat shock protein 60 and fatal colitis in mice. Gut Microbes. 2021;13:1-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 111. | Füst G, Uray K, Bene L, Hudecz F, Karádi I, Prohászka Z. Comparison of epitope specificity of anti-heat shock protein 60/65 IgG type antibodies in the sera of healthy subjects, patients with coronary heart disease and inflammatory bowel disease. Cell Stress Chaperones. 2012;17:215-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 112. | Bachmaier K, Penninger JM. Chlamydia and Antigenic Mimicry. In: Oldstone M, editor. Molecular Mimicry: Infection-Inducing Autoimmune Disease. Berlin, Heidelberg: Springer Berlin Heidelberg, 2005: 153-163. |
| 113. | Zmora N, Zilberman-Schapira G, Suez J, Mor U, Dori-Bachash M, Bashiardes S, Kotler E, Zur M, Regev-Lehavi D, Brik RB, Federici S, Cohen Y, Linevsky R, Rothschild D, Moor AE, Ben-Moshe S, Harmelin A, Itzkovitz S, Maharshak N, Shibolet O, Shapiro H, Pevsner-Fischer M, Sharon I, Halpern Z, Segal E, Elinav E. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388-1405.e21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 702] [Cited by in RCA: 971] [Article Influence: 138.7] [Reference Citation Analysis (0)] |
| 114. | Li M, Xu R, Li YQ. Sequential laxative-probiotic usage for treatment of irritable bowel syndrome: a novel method inspired by mathematical modelling of the microbiome. Sci Rep. 2020;10:19291. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |