Copyright
©The Author(s) 2023.
World J Methodol. Dec 20, 2023; 13(5): 484-491
Published online Dec 20, 2023. doi: 10.5662/wjm.v13.i5.484
Published online Dec 20, 2023. doi: 10.5662/wjm.v13.i5.484
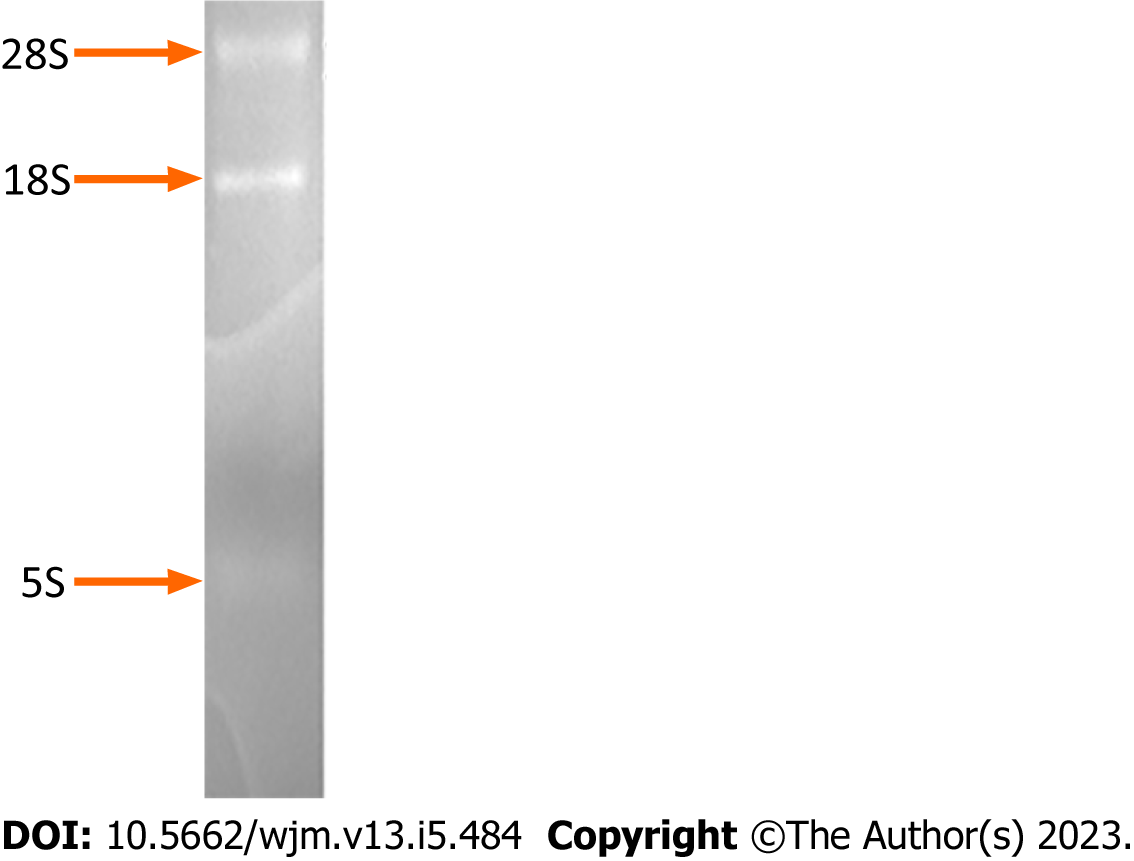
Figure 1 Determination of the integrity of the total RNA isolated from the cultured HepG2 cells.
Electrophoresis of the total RNA on a formaldehyde-denaturing agarose gel and subsequent staining with 0.5 mg/mL ethidium bromide revealed discrete bands corresponding to 28S, 18S, and 5S RNA molecules.
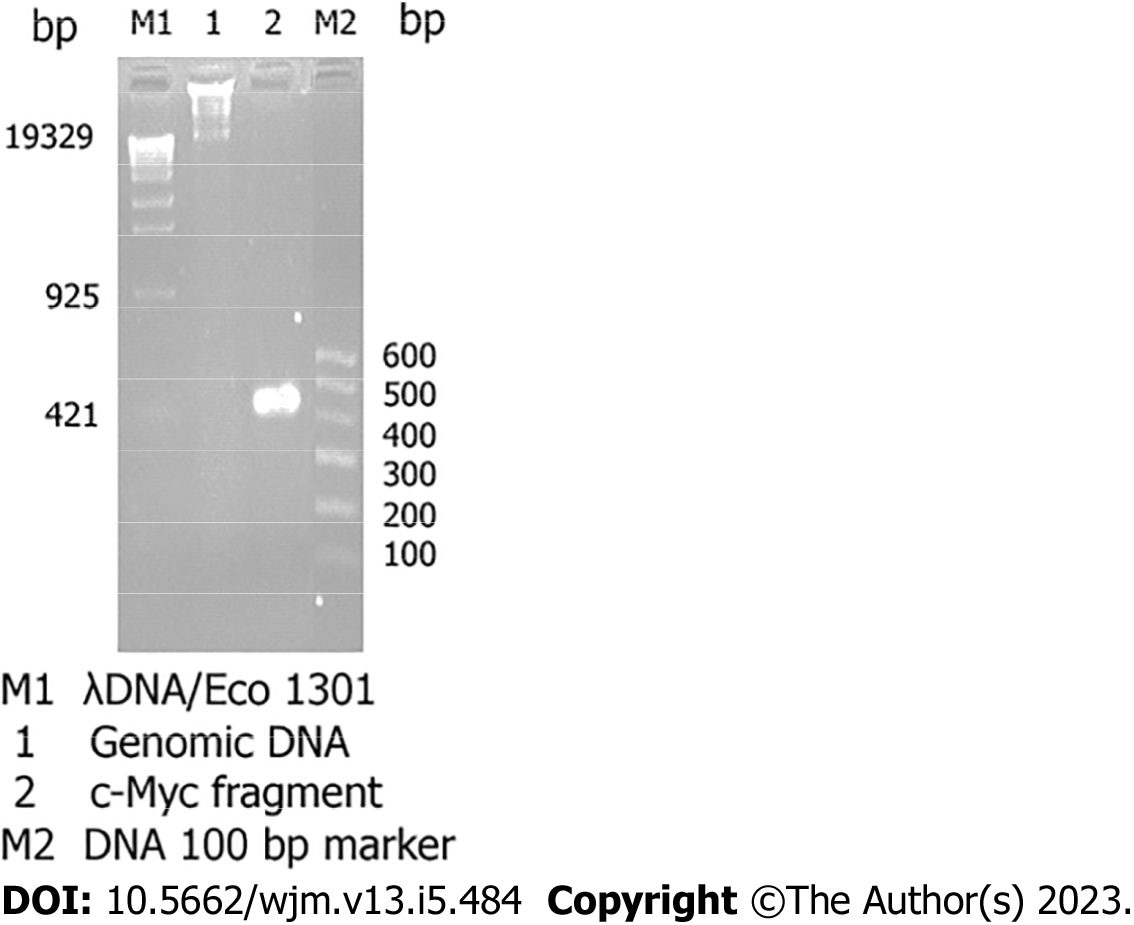
Figure 2 Identification of genomic DNA and determination of the quality of the total RNA isolated from cultured HepG2 cells.
Electrophoresis of the genomic DNA and products of reverse transcription (RT)-polymerase chain reaction (PCR) amplification of the c-Myc fragment on a 2.0% agarose gel and subsequent staining with 0.5 mg/mL ethidium bromide revealed the presence of 1.0-20 kb genomic DNA fragments (Lane 1), and a 500 bp c-Myc fragment that was amplified by RT-PCR using the total RNA as the template (Lane 2).
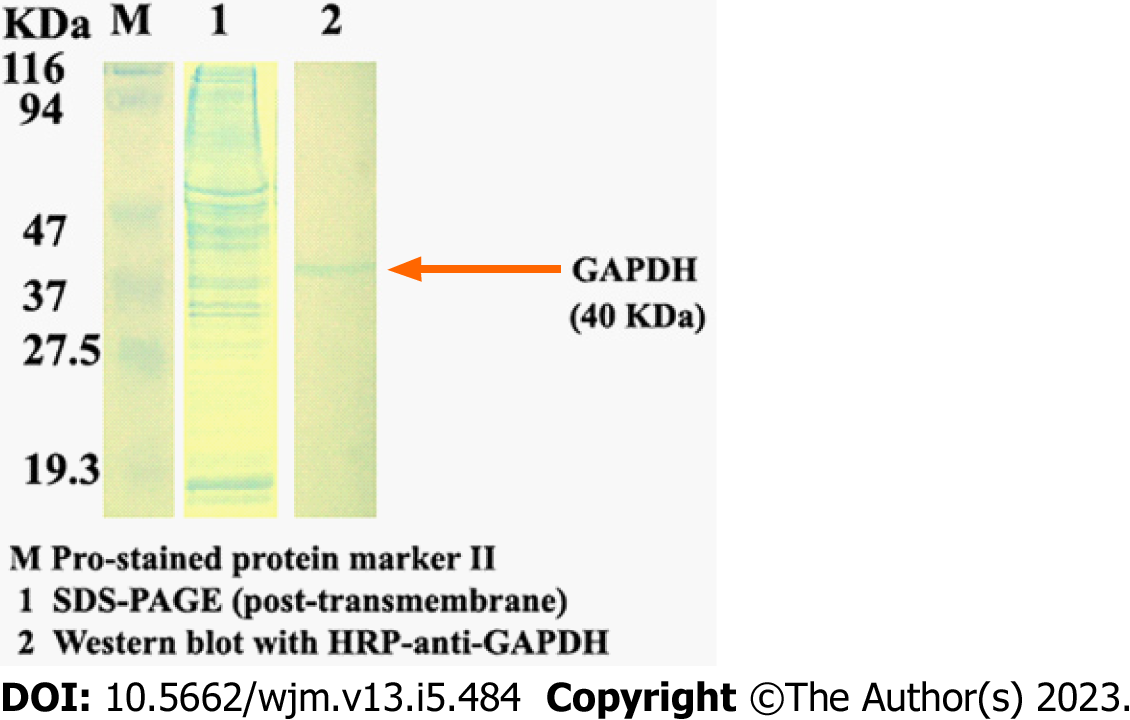
Figure 3 Identification of the quality of the total proteins isolated from the same cultured HepG2 cells.
The total proteins were separated on 12% gels by SDS-PAGE (Lane 1). Following transfer onto a polyvinylidene fluoride membrane, the 40-kDa GAPDH-specific band was blotted with HRP-anti-GAPDH and observed using sediment type mono-ingredient TMB substrate solution (Lane 2).
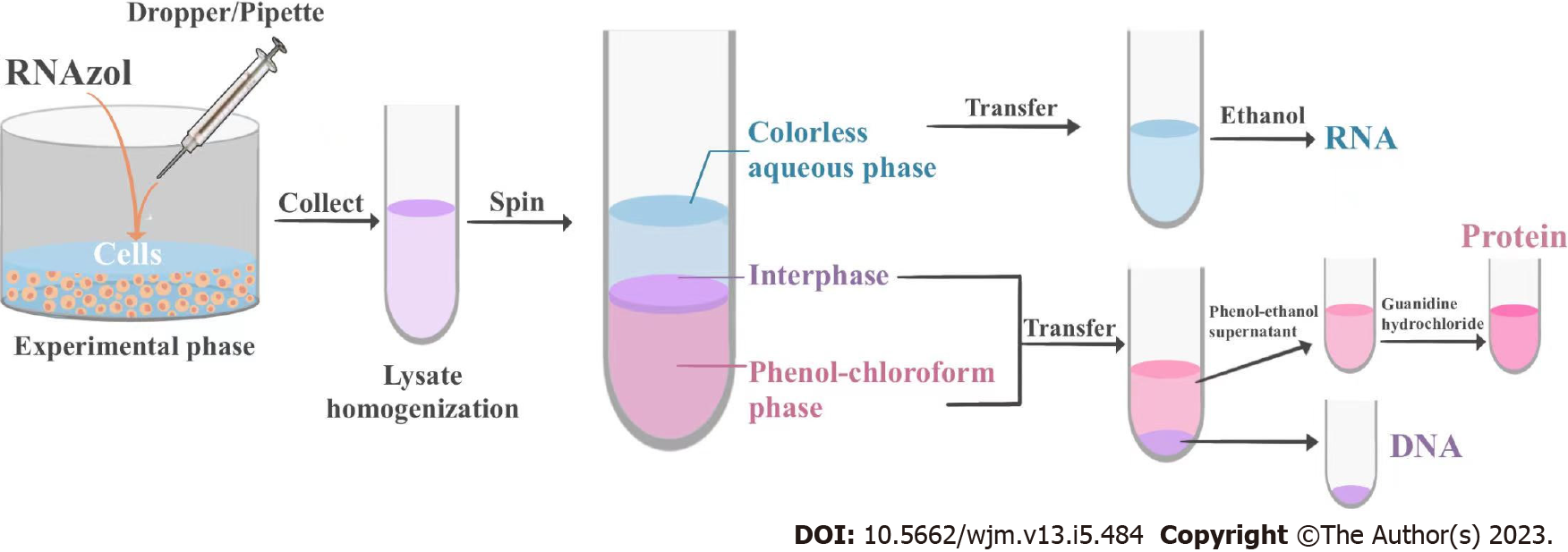
Figure 4 Flow chart of ENAP protocol.
- Citation: Cui YY. Sequential extraction of RNA, DNA and protein from cultured cells of the same group. World J Methodol 2023; 13(5): 484-491
- URL: https://www.wjgnet.com/2222-0682/full/v13/i5/484.htm
- DOI: https://dx.doi.org/10.5662/wjm.v13.i5.484