Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
Peer-review started: August 9, 2014
First decision: October 14, 2014
Revised: February 19, 2015
Accepted: March 18, 2015
Article in press: March 20, 2015
Published online: April 12, 2015
Processing time: 249 Days and 1.5 Hours
Epigenetic is the study of those alterations regulating gene expression without altering DNA sequence and inherited by transmission through cell division. Mutational and epimutational events that alterate cellular growth and division are combined in carcinogenesis. Advances in genome and epigenome-wide analysis identify DNA hypomethylation, hypermethylation of tumor suppressor genes, aberrant histone modifications and/or specific miRNA expression profiles to contribute to tumor initiation and progression. The major challenge for cancer researchers is to enlighten the complex relationship between the epigenetic and genetic machinery in order to optimize combined therapies, reducing chemoresistance and minimizing adverse effects in cancer patients. In this review we will cover many distinct aspects of epigenetic phenomenon. Firstly, we will globally explain the most common epigenetic events and their effects on gene expression regulation. Secondly, we will review the evidence of the correlation between epigenetics and cancer progression, focusing in particular on the effect of aberrant hypo- and hyper-methylation. We will also consider the main methods currently used for methylation analysis, covering both locus-specific technologies and genome-wide analysis. Finally, we will discuss the introduction of novel epigenetic drugs in combination with conventional treatments in order to develop more effective cancer therapies. Such information could help in understanding the important role of epigenetics in cancer.
Core tip: Carcinogenesis occurs through a combination of mutational and epimutational alterations involving key pathways in cellular growth and division. Tumour cells exhibit two main differences from normal cells in DNA methylation: a global reduction in DNA methylation and the hypermethylation of specific sequences, mainly CpG islands, that cause the transcriptional silencing of tumour suppressor genes, thus directly driving the carcinogenic. In this review, we’ll focus on our current understanding of this process, aiming to discuss how the analysis of cancer methylomes and the re-expression of epigenetically silenced genes have potential uses in developing more effective cancer therapies.
- Citation: Lattanzio L, Lo Nigro C. Epigenetics and DNA methylation in cancer. World J Transl Med 2015; 4(1): 11-24
- URL: https://www.wjgnet.com/2220-6132/full/v4/i1/11.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v4.i1.11
In the early 1940s the word “epigenetics” was introduced in the biological vocabulary to describe those phenomena that traditional genetics could not completely explain. Conrad Waddington (1905-1975) defined epigenetics as “the branch of biology which studies the causal interactions between genes and their products which bring the phenotype into being”. Today the most common definition for “epigentics” is “the study of changes in gene function that are mitotically and/or meiotically heritable and that do not entail a change in DNA sequence”[1].
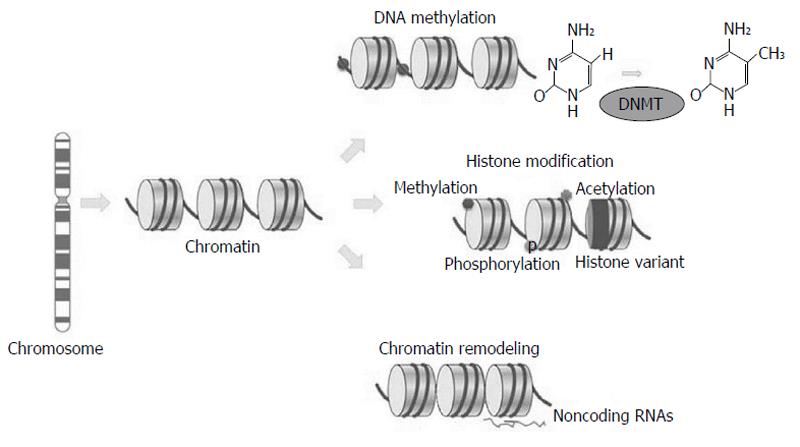
The epigenetic field covers chromatin-based events that regulate DNA-templated processes. Specific chromatin-modifying enzymes highly regulated modifications to both DNA and to histones, proteins involved in DNA packaging into structural units defined nucleosomes[2]. Table 1 describes the most well-known DNA and histone modifications and their correlated functions.
| Chromatin modification | Nomenclature | Chromatin-reader motif | Attributed function |
| DNA modification | |||
| 5-methylcytosine | 5mC | MBD domain | Transcription |
| 5-formylcytosine | 5fC | Unknown | Unknown |
| 5-hydroxymethylcytosine | 5hmC | Unknown | Transcription |
| 5-carboxylcytosine | 5caC | Unknown | Unknown |
| Histone modification | |||
| Acetylation | K-ac | BromodomainTandem PHD fingers | Transcription, repair, replication, and condensation |
| Methylation (lysine) | K-me1, K-me2, K-me3 | Chromodomain, tudor domain, MBT domain, PWWP domain, PHD fingers | Transcription and repair |
| Methylation (arginine) | R-me, R-me2s, R-me2a | Tudor domain | Transcription |
| Phosphorylation (serine and threonine) | S-ph, T-ph | 14-3-3. BRCT | Transcription, repair and condensation |
| Phosphorylation (tyrosine) | Y-ph | SH2 | Transcription and repair |
| Ubiquitylation | K-ub | UIM, IUIM | Transcription and repair |
| Sumoylation | K-su | SIM | Transcription and repair |
| ADP ribosylation | E-ar | Macro domain, PBZ domain | Transcription and repair |
| Deimination | R→Cit | Unknown | Transcription and decondensation |
| Propoline isomerisation | P-cis↔P-trans | Unknown | Transcription |
| Crotonylation | K-cr | Unknown | Transcription |
| Propionylation | K-pr | Unknown | Unknown |
| Butyrylation | K-bu | Unknown | Unknown |
| Formylation | K-fo | Unknown | Unknown |
| Hydroxylation | Y-oh | Unknown | Unknown |
| O-GIcNAcylation (serine and threonine) | S-GIcNAc; T-GIcNAc | Unknown | Transcription |
Epigenetic modifications play critical role in regulating DNA transcription, repair and duplication. Genomic alterations leading to deregulated expression patterns in chromatin regulators could be responsible for cancer induction and progression[2].
Earliest studies on gene expression and DNA methylation indicated the possible link of epigenetics to cancer, as detailed in the history of cancer epigenetics by Feinberg et al[3] and confirmed by recent results from the International Cancer Genome Consortium (ICGC). The analysis of genomes of various cancers allowed us to identify recurrent somatic mutations causing a loss or gain of function in tumour suppressor genes and in oncogenes, respectively. The so called “driver” mutations often present at a high prevalence, are recurrently found in various tumours[1]. Recent studies identified many of these mutations also in numerous epigenetic regulators. Feinberg hypothesised that epigenetic changes may induce genetic alterations causing cancer initiation and/or progression.
Differently from somatic mutations, epigenetic changes occur without changing DNA sequence; they include chromatin structure variations, due to methylation or histone variants, nucleosome remodelling and non-coding regulatory RNAs changes (Figure 1).
It is now well demonstrated that epigenetic events are hereditable changes in gene structures aimed to perpetuate altered activity states[4]. These alterations in the state of chromosomal regions are called epimutations and could play a significant role in carcinogenesis as they have been commonly found in epigenetic regulators.
The first epigenetic mark studied in correlation with cancer was aberrant DNA methylation causing deregulation in normal gene expression[5]. DNA methylation is a covalent chemical change that causes the addition of a methyl (CH3) group at the 5’ carbon position of a cytosine ring. The presence of methyl groups determines the turning off of gene transcription and thus the silencing of these genes. The methylation pattern is inherited by the daughter cells during mitosis, allowing maintenance of gene transcription regulation after replication and generating a stable gene silencing mechanism.
A family of DNA methyltransferases (DNMTs) regulate DNA methylation in the CpG dinucleotide by catalyzing the addition of CH3 groups from S-adenosyl-L-methionine to the 5’ position of cytosine. Methyl-binding domain (MBD) proteins (MeCP2, MBD1, MBD2, and MBD4) are able to bind to methylated CpGs, causing transcriptional silencing[1]. Aberrant methylation frequently occurs in cancer and the most common types of alteration are hypo-, hypermethylation and loss of imprinting (Table 2).
| DNA hypomethylation | Consequence |
| Global hymethylation | Reactivation of endoparasitic and repetitive genomic sequences Chromosomal and genomic instability |
| Hypomethylation of gene bodies | Activation of incorrect sites of transcription initiation |
| Loss of promoter methylation | Activation of metastasis and tumour promoting genes |
| DNA hypermethylation | Consequence |
| Promoter CpG island | Tumour-suppressor gene silencing |
| (CpGI) methylation | Inhibition of transcription factors suppressors |
| Loss of imprinting | Abnormal transcriptional inactivation Deregulation of imprinted genes |
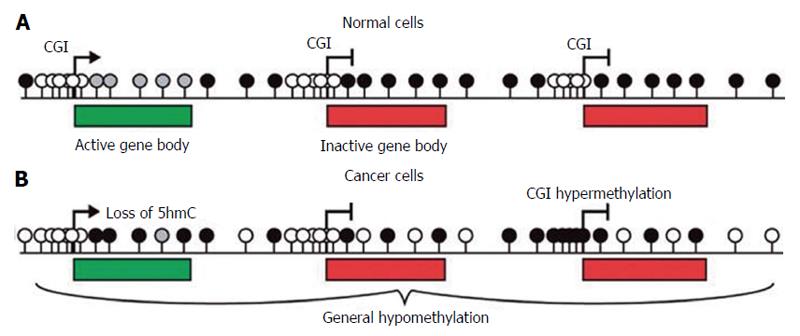
In somatic cells DNA methylation mainly occurs at cytosines usually concentrated in islands (CGIs) which frequently correspond to the promoters of tumour suppressor genes (TSGs) (Figure 2A), which are unmethylated in normal cells.
In cancer cells methylation levels are frequently reduced in specific repetitive elements or in target chromosomal regions (Figure 2B). LINE-1 elements hypomethylation has been described in colorectal, urothelial and hepatocellular cancers, disrupting normal patterns of gene expression. Moreover, Alu elements are hypomethylated with LINE-1 elements in prostate adenocarcinomas, pancreatic endocrine tumors, and carcinoid tumors[6]. Hypomethylation of these elements is strongly linked to tumorigenesis through insertional mutagenesis, genomic rearrangements, deletions or inversions causing genomic instability and gene activation[7]. Although hypomethylation has been clearly correlated to cancer development, the chemical process resulting in the removal of methyl groups (demethylation) and its role in gene regulation are still unclear. The family of enzymes Ten-eleven translocation [TET (TET1, TET2 and TET3)] has been identify to be active in initiating demethylation. They are 2-oxoglutarate-/Fe(II)-dependent oxygenases that convert the 5-methylcytosine (5mC) into 5-hydroxymethylcytosine with mechanisms still not well described[8].
DNA hypermethylation is the most well studied abnormality of DNA methylation. This method of gene inactivation is the most common mode used by cancer cells to silence TSGs, thus affecting DNA repair, apoptosis, angiogenesis, cell cycle regulation, and capability of invasion. TSGs that are cancer-specifically silenced by CpG island hypermethylation of their promoters are, for example, retinoblastoma, CDKN2A (p16), hMLH1, and VHL genes[6]. However, hypermethylation could also hit DNA repair genes and transcription factors indirectly affecting downstream targets, thus leading to genetic errors and tumorigenesis[7].
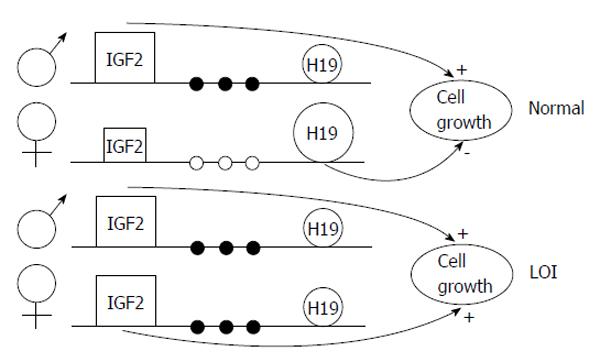
A less studied epigenetic event is the loss of parental allele specific monoallelic expression of genes, the so-called loss of imprinting (LOI); this may be caused by hypomethylation of one of the two parental alleles (Figure 3). Insulin-like growth factor 2 LOI has been associated with an increased risk of colorectal cancer[5,9] and other neoplasias. Data demonstrated that LOI can also cause tumour suppressor gene silencing; for example, ARHI, a candidate breast tumour gene, shows aberrant allele-specific silencing. Moreover, LIT1, an untranslated RNA, undergoes LOI in about half of patients with Beckwith-Wiedemann syndrome, determining downregulation of CDKN1C (which encodes KIP2, also known as p57)[3]. Table 3 shows some of the most well known genes epigenetically regulated in cancer.
| Cancer-associated pathway | Gene |
| Cell cycle | Rb, p16INK4a, p16INK4b, 14-3-3, cyclin E, p14ARF |
| Signal transduction | ErbB2, RASSF1, LKB1/STK11, APC |
| Apoptosis | DAPK gene, Caspase-8 gene |
| DNA repair | MGMT, MHL1, BRCA1, FNACF |
| Carcinogen metabolism | GSTP1 gene |
| Hormonal response | Oestrogen receptor gene, progesterone receptor gene, RAR-b2 gene |
| Senescence | TERT, TERG |
| Invasion/metastasis | TIMP-3 gene, E cadherin gene, VHL gene |
| Transcription | Runx3, Twist, Er α, Er β, PR, RAR, vitamin D receptor |
| Drug responsiveness | Glutatione S-transferase, thymidylate synthase |
The fact that aberrant hypermethylation in cancer causes TSG silencing is supported by three important evidences: (1) hypermethylation has been observed alongside inherited germ line mutations and could be the specific “hit” that completely disables TSG activity (i.e., CDNK2A-p16/ARF); (2) in sporadic cancers the tissue specificity of TSG hypermethylation causes predisposition in the specific tissues as the inherited mutations in these same genes. For example, MLH1 mutations and hypermethylation predispose to colorectal cancer, the latter been limited to colorectal tumoural tissues. Similarly, BRCA1 mutations predispose to breast and ovarian tumours and hypermethylation is limited to those tissues; and (3) the strongest evidence that aberrant DNA hypermethylation contributes to silencing of TSG in cancer is that demethylation of promoters is able to reactivate those genes. Many studies demonstrate the ability of 5-aza-2’-deoxycytidine to cause DNMT1 degradation and methyltransferase maintenance, leading to reactivation of hypermethylated gene promoters[4].
The mechanism(s) responsible for aberrant promoter hypermethylation in cancer are still unclear. However, genomewide analyses of normal and tumour cells demonstrate two principal processes: active mechanisms targeting specific factors to CGIs, or passive ones deriving from a loss of protection against de novo methylation.
Over-expression or increased activity of DNMTs might cause aberrant CGI hypermethylation. Many studies initially reported DNMTs to be increased, but more recently it has been attributed to cell cycle regulation or to an increased number of cycling cells.
Moreover, methylome analyses demonstrate that hypermethylation in not random, but hit specific set of genes. The promoters of such genes seem to be relatively poor of retrotransposons (transposable elements that duplicate via RNA intermediates and are reverse-transcribed and inserted at new genomic locations) compared with hypermethylation-resistant promoters[4].
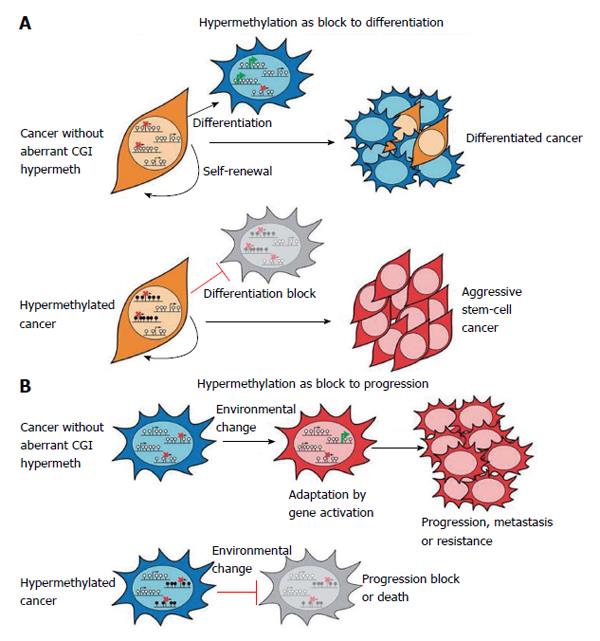
Many results have demonstrated epigenetic plasticity of cancer cells, suggesting that hypermethylation could act as a block to differentiation and to progression. This derives from studies reporting gene expression profiles of aggressive tumours to be similar to those of embryonic stem (ES) cells. Thus, it has been proposed that in cancer the hypermethylation in ES polycomb repressive complexes targets might impact differentiation and maintain stem-cell-like state (Figure 4A). Moreover, hypermethylation could act in cancer progression (Figure 4B)[4]. In fact, dissemination of tumour from the primary site requires the re-modelling of gene expression profiles. Moreover, drug resistance might result from secondary activating mutations and/or epigenetic alterations. Hypermethylation causing gene repression might provide a protection to these events and favour cancer progression[4].
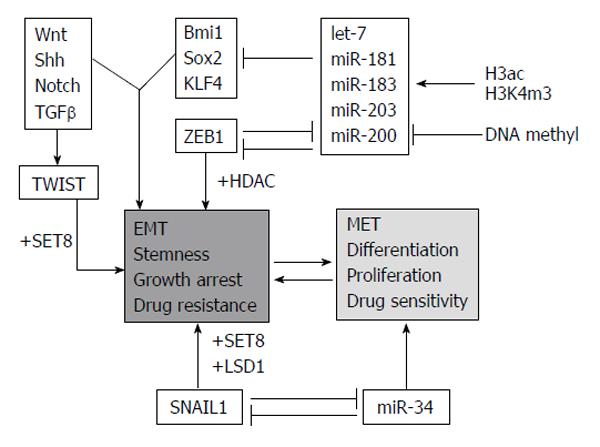
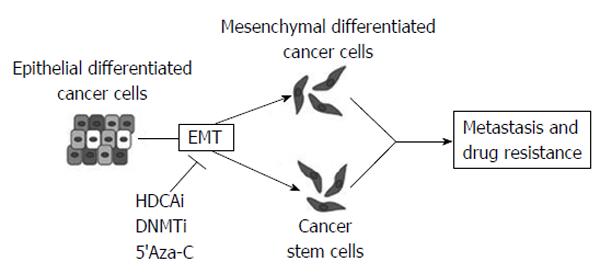
Epithelial-mesenchymal transition (EMT) and the otherway process, mesenchymal-epithelial transition, (MET) are important during cellular growth and in physiological tissue repair (wound healing) but they also play a crucial role in carcinogenesis. In normal tissues, many intercellular junctions (desmosomes, adherens, tight junctions) ensure tissue homeostasis and stability, linking epithelial cells together and to the extracellular matrix. In particular conditions, such as physiological circadian changes or tissue loss or damage, epithelial cells can acquire a mesenchymal phenotype, including an intermediate stem cell phenotype, such us in embryogenesis. Recently, many studies have aimed at understanding the role of EMT and MET in cancer progression and, in particular, in the initial processes of tissue invasion and extravasation. EMT, in fact, is minutely regulated by networks of activating/deactivating signalling pathways and also by epigenetic alterations (DNA methylation, histone modifications and by miRNAs). For this reason, anomalies in regulating those mechanisms might cause cancer initiation and progression, depending on the capability of cells to react to external and internal stimuli.
One of the main mechanisms used by epithelial tumor cells to convert into de-differentiated, mesenchymal cells is by silencing epithelial genes, such as E-cadherin, and loosing cell-cell contacts. Loss of E-cadherin happens in early tumor progression, so that the EMT process is strictly related to metastatic invasion. The replacement of E-cadherin by N-cadherin (cadherin switching)[10] depends on multiple cellular signaling mechanisms [Hedgehog, Wnt, Notch, transforming growth factor β, fibroblast growth factor (FGF), epidermal growth factor and platelet-derived growth factor]. Moreover, many epigenetic events are involved in the EMT program and are responsible of the silencing of specific epithelial markers, leading the epithelial cells to be aggressive and invasive (Figure 5)[11].
During tumorigenesis, cells acquire metastatic potential following angiogenesis, induction of cell surface metalloproteases, decrease in the expression of cell-cell adhesion molecules, and increased expression of cell surface receptors that aid in motility. E-cadherin and alpha-4 integrins, two of the most common cell adhesion receptors, are silenced by methylation in several cancers[12]. Similarly, intracellular basement membrane proteins (i.e., NID1 and NID2) are also silenced by methylation in cancer. Therefore, it is evident that epigenetics could also play a critical role in the metastatic process[13]. The phenomenon of metastasis is a complex process involving several distinct steps: tumor cells, supported by angiogenesis, infiltrate the basement membrane.
Aberrant methylation of metastasis initiation genes could be responsible of tumor invasiveness (for a detailed review, refer to Cock-Rada and Weitzman, 2013[8]).
Several genes have been identified which regulate the metastatic process, can predict prognosis and metastasis and are used in daily clinical practice[8]. These genes are usually involved in regulation of extracellular matrix (ECM) and angiogenesis, regulation of cell adhesion and invasion, and repressive and activating histone modifications.
In particular, in the beginning of tumour progression, cellular matrix metalloproteinases (MMPs) are able to degradate the ECM for angiogenesis. The loss of MMP regulation and release of angiogenic stimuli (FGF-2 and vascular endothelial growth factor) contribute to this process[14]. Tissue inhibitor of metalloproteinase 2 (TIMP-2) is a MMP inhibitor suppressed in some solid and lymphoid tumours by CpGI hyper-methylation[15,16]. TIMP-3 was also found to be silenced by DNA methylation in gastric and oesophageal cancers and to correlate with poor survival[17].
Cells migrate throught the ECM, invade adjacent structures and traverse into lymphatic or blood vessels, so that are able to disseminate to distant sites, form micrometastases and eventually colonise the new organ with macrometastases (Figure 6).
Exposure to ionizing radiation (IR) could cause alteration in gene expression, deregulation of cell cycle, and apoptosis. Nonetheless, although several studies demonstrate that IR could also alterate DNA methylation[6], the epigenetic events following IR still need to be defined at the molecular level.
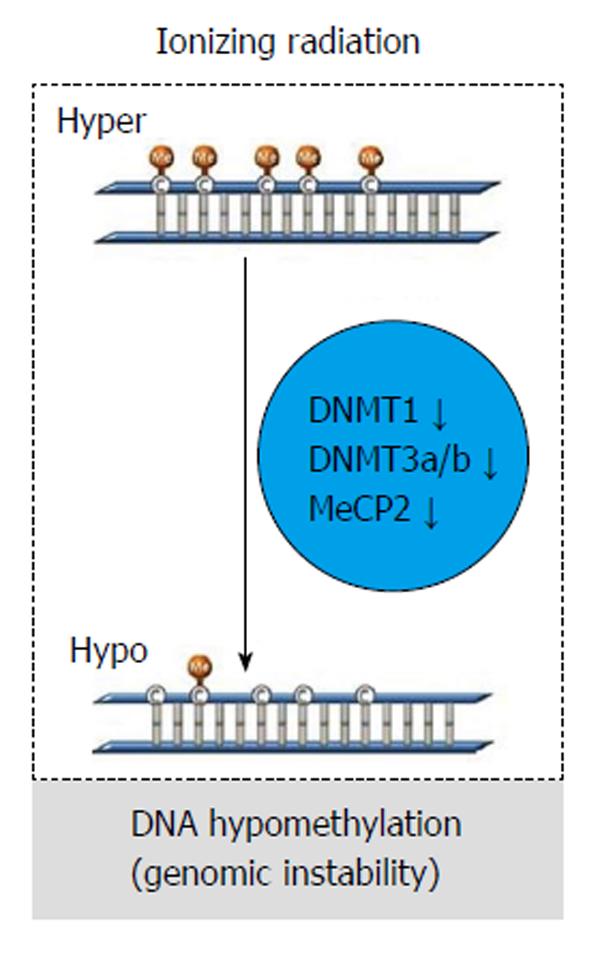
In vivo studies show that IR causes a dose-dependent and sex/tissue-specific global hypomethylation, together with a decrease in methyltransferases (DNMTs; DNMT1, DNMY3a, and DNMT3b) and methyl CpG-binding protein (MeCP2) level (Figure 7). Thus, radiation exposure could be strictly correlated to DNA hypomethylation patterns resulting in genomic instability[6].
Interestingly, recent studies in colon cancer report a correlation between the DNA methyltransferase inhibitor 5-aza and radio-sensitivity[18]. In breast cancer cells, fractionated IR caused DNA methylation alterations at specific loci (TRAPP9, FOXC1, and LINE1)[19]. More recently, it has been shown that radiosensitive and radioresistant cancer cells present differential DNA methylation alterations[20]. Nevertheless, the epigenetic mechanisms at the basis of those alterations and site-specificity in DNA methylation, is still not clear and requires further studies, with the final aim of identifying useful methylation target for developing cancer targeting therapies[6].
Currently, many studies are focusing on the role of microRNAs (miRNAs) in cancer development and metastasis. Here we refer the reader to several excellent recent reviews[21-25].
miRNAs are small, non-coding RNAs involved in post-transcriptional gene expression regulation through binding to complementary sequences in the 3’-untranslated region of messenger RNAs (mRNA). This interaction leads to mRNA cleavage or inhibited protein synthesis, thus reducing protein expression of the targeted gene. When affecting expression of oncogenes and tumor suppressor genes, common breakpoints and fragile sites (preferential sites of chromatid exchange, deletion, translocation, amplification, or integration of plasmid DNA and tumor-associated viruses), the up- or down-regulation of miRNAs could be critical for tumorigenesis and cancer progression. Indeed, a large set of aberrantly regulated miRNAs have been already identified in several tumor entities, although the biological mechanisms at the basis of miRNA regulation are still poorly studied[21].
Recent studies demonstrated that many miRNAs could act as TSGs, but others are frequently overexpressed in human tumors possibly exerting a tumorigenic function. For example, miR-17-92 cluster shows an oncogene function, is transactivated by the c-MYC oncogene, and accelerates lymphomagenesis in murine models[26]. Moreover, miR-155 has been shown to induce leukemia in transgenic murine models and plays a critical role in inflammation and immune response[27]. miR-21 has been found in several tumor types as a regulator of important TSGs such as PTEN1 and PDCD4[25].
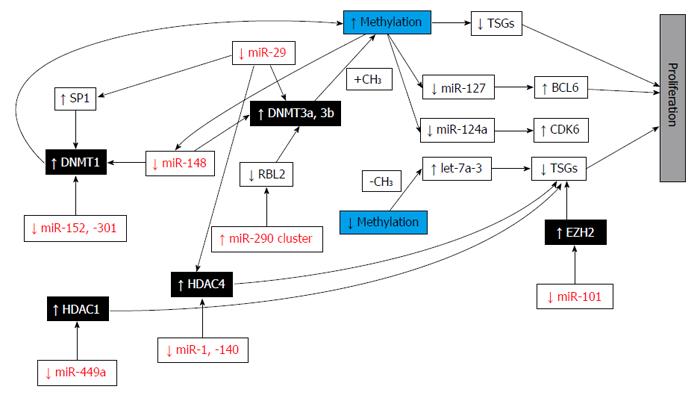
On the basis of their correlation with cancer, miRNAs are divided as: oncogenic, tumor-suppressive, and “context-dependent” miRNAs[1]. In cancer cells, the loss of miRNA regulation could activate oncogenes or repress target tumor suppressor genes. Moreover, mutations could occur also in miRNA sequences, leading to lack of recognition of its binding target and thus to oncogene activation and/or tumor suppressor repression. miR-155, miR-21, and miR-17 to -92 are, for example, oncogenic miRNAs and their expression has been found to be amplified in several tumor types; furthermore, tumor-suppressive miRNAs (miR-146, -15 and -16) appear to be down-regulated in cancers. miRNA mutations are also known to target epigenetically modifying enzymes, such as EZH2 and DNMT3. Alterations of miRNA expression, including miR-101 and miR-29, may cause extensive alterations in histone acetylation or DNA methylation of other miRNAs that target oncogenes and TSG. Since a correlation between miRNA expression and tumorigenesis has been demonstrated, miRNA might be useful therapeutics, replacing tumor-suppressive miRNA or targeting the oncogenic ones[1].
The study of the influence of DNA methylation on miRNA transcription on a genome-wide level has been hampered by poor miRNA promoter annotation. Recently, large collaborations (ICGC and The Cancer Genome Atlas), have created extensive data sets of genetic, epigenetic, and transcriptome profiles of different tumor entities and cell lines. Furthermore, the Encyclopedia of DNA Elements (ENCODE) consortium profiled a variety of cell lines for 12 histone modifications and variants including H3K4me3 and acetylation of histone 3 at lysine 9 (H3K9ac) to disclose regulatory regions in the human genome[28].
The resulting data allow us to extend the knowledge on tissue-specific and ubiquitous miRNA promoters. Analysis of 329 miRNA promoters revealed that 300 overlapped with or were close to a DNase I- hypersensitive site. All these analyses might permit us to estimate the number of tissue-specific miRNA promoters as suggested for miR-21[21].
For example, miR-9-1 has been associated with a CpG island 200 bp upstream and has been found to be hypermethylated in breast cancer, melanoma, and head and neck cancer. Also, miR-200 family members have been found to be near to a CpG island[29]. CpG island methylation correlated with down-regulated miRNA expression in breast and prostate cancer cell lines[30]. Moreover, a correlation between loss of miRNA expression and acquisition of mesenchymal features have been observed in tumour progression[21].
The ENCODE consortium is about to publish the genomewide DNA methylation data, completing analysis of epigenetic regulation of all gene classes including miRNAs in cell lines. In addition, cancer methylomes are analyzed and will be made publicly available by the ICGC which, for example, provided the methylomes of patients with chronic lymphatic leukemia. In conclusion, integrating data sets from different sources will enable scientists to estimate the global influence of DNA methylation on the regulation of miRNA and their aberrant behaviour in cancer[21].
Furthermore, there has been demonstrated a possible role of miRNAs in IR-induced response in vitro and in vivo. Indeed, in murine models IR cause sex- and tissue-specific alterations in miRNA expression[6].
miRNAs are likely epigenetically regulated but it is already well known that they can also affect expression of epigenetically regulated genes by targeting key enzymes responsible for epigenetic reactions. This group of miRNAs is called epi-miRNAs (Figure 8)[25].
Recently, many in vivo models of carcinogenesis have been developed in order to investigate epigenetic mechanisms and cancer progression. These models are usually derived from transgenic manipulation or toxicant exposure, inducing a tissue-specific cancer. As a consequence, these models could be useful in characterizing molecular pathways of carcinogenesis and elucidating the contribution of epigenetic and genetic alterations transforming carcinoma in situ to metastatic disease[7].
Recently, these mouse models have been used to study the efficacy of epigenetic-modifying drugs (i.e., 5-azacytidine, decitabine and zebularine), as well as to determine their toxicity, by treating xenograft mice and evaluating tumour size or metastasis formation. However, these results cannot be directly translated into the clinic, since because tumour biology and response to drugs in mice may be substantially different from patients[8]. In oncology, the greatest challenge is the integration of human and animal results from translational research. This integration may shed light on how or when epigenetic dysregulation could occur in tumor and how environmental and dietary hits may influence the tumour phenotype. Additionally, these studies will provide information on susceptibity to therapy that target epigenetics (DNA demethylating agents, histone deacetylases inhibitors, or a other promising epigenetic therapies currently in trials). Thus, investigation of genetic and epigenetic profiles in cancer patients is a crucial step in the improvement of any personalized cancer therapy[7].
A biomarker is an indicator of normal biological processes, pathogenic processes, or pharmacologic response to therapeutic intervention. Biomarkers have many valuable applications in disease detection and monitoring, even if the validation and qualification of biomarkers for use with patients is time-consuming. Currently, many validated biomarkers should be used for personalized therapy. In cancer, several biomarkers have been used to reflect the extent of tumor growth and metastasis or as tools for screening and monitoring of disease. For example, our group identified TFPI2 gene as a novel biomarker of metastatic melanoma, demonstrating that its methylation correlates with metastatic state of the disease. Moreover, we observed that circulating, methylated TFPI2 DNA was undetectable in sera from healthy individuals but detectable in sera from patients with primary and metastatic melanomas. In particular, the presence of methylated TFPI2 DNA in serum was strongly associated with metastatic disease, thus defining TFPI2 a sensitive and specific biomarker of metastatic melanoma[25].
New biomarkers, useful in clinical oncology and based on DNA methylation, are coming from epigenomic analyses. As a consequence of the large set of alterations in methylation discovered in different tumours, a myriad of DNA methylation-based biomarkers of several human neoplasia have been reported, principally involving hypermethylation of tumor suppressor CGIs[5].
DNA methylation is an epigenetic event that usually occurs in specific genes or in viral genome regions that are quite promising as independent diagnostic and prognostic markers. Several of these markers could be in common with two or more cancers, while others appear to be tumor-specific, providing an opportunity to determine the origin of metastases of uncertain origin. Moreover, information derived by new biomarkers could help in distinguishing similarities or differences between diseases. However, there is a growing need for evaluation and selection of the most appropriate biomarker sets, standardisation of the methods for assessment of each type of alteration, and clinical validation[31]. This could hamper and delay implementation of useful epigenetic biomarkers.
We suggest that the readers refer to a detailed review concerning the discovery and validation of clinically relevant DNA methylation biomarkers in cervix and prostate cancers[32].
One of the open questions in the epigenetic field is which method of analysis of DNA methylation should be the standard in order to show evidence of clinical utility.
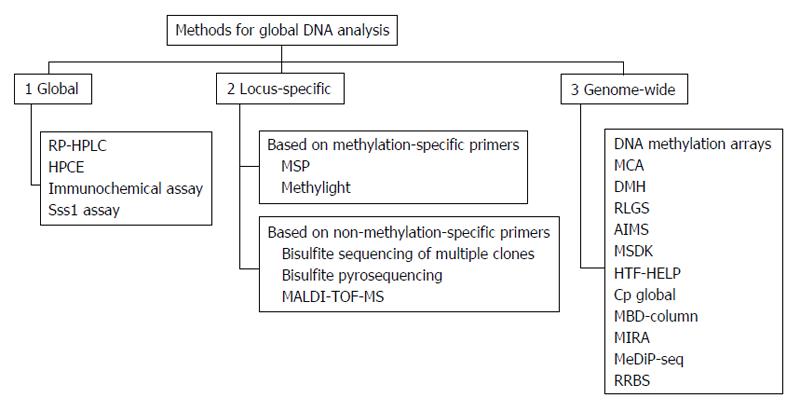
Healthy cells show a specific DNA methylation pattern; however, alterations in this pattern, such as hypomethylation or hypermethylation, can lead to diseases, including cancer. Methylation status is currently used to classify and characterize cancers and could be of clinical significance at three levels: detection, prognosis, and prediction of treatment responses. In recent years, different methods have been developed to identify aberrant methylation signatures and may be used to identify specific biomarkers useful for tumor subtypes classification. All these technologies have been commonly classified as: (1) global approaches for detection of gross DNA methylation; (2) locus-specific methods for analysis of specific methylated CpG regions; and (3) genome-wide approaches developed to identify methylation hot-spots in the whole genome sequence (Figure 9)[33].
Two of the most used analyses in DNA methylation are methylation-specific polymerase chain reaction (PCR) (MSP) and bisulphite sequencing PCR. These methods needed an initial bisulphite reaction converting unmethylated cytosines to “uracil” bases read as thymidines (T) after amplification by PCR. This allows to not modify methylated cytosines (“C”) in 5mCpG dinucleotides that remain “C”. Thus, a hypothetical bisulphite-converted sequence of 5’-AATCmCGTACTmCGCCTG-3’ would be read as 5’-AATTCGTATTCGTTTG-3’, where the Ts in italics derive from unmethylated Cs, whereas methylated CpG remains CpG (here underlined). After bisulphite transformation, DNA could be analysed to specifically distinguish between methylated and unmethylated cytosines.
In MSP, two distinct set of primers containing at least two CpG dinucleotides within the primer sequences are used: U primers detect unmethylated CpGs while M primers detect methylated CpGs. In a methylated sample, only M primers produce a PCR band but not U primers; vice versa in samples not methylated. Requiring a simple PCR machine, MSP is adequate in the analysis of large numbers of clinical samples and has been successfully used in tumour methylation studies. The main limit of MSP is that the result obtained is purely qualitative. For quantification of methylation levels, bisulphite sequencing PCR has been developed. In this method, after PCR, amplified DNA need to be appropriately cloned into a vector and then 5-10 clones independently sequenced in order to read all CpG sites included in the amplified sequence, giving a global representation of the cellular methylation status[34].
More recently, many methods focusing on specific single-CpG have been developed, such as combined bisulfite restriction analysis (COBRA)[35], MethyLight[36], and bisulfite pyrosequencing[37]. In COBRA bisulfite conversion and PCR amplification are maintained; then, PCR product are digested by restriction enzyme in a methylation-dependent manner. Digestion proceeds in the recognition sequence only if the CpG site is protected from bisulfite conversion by methylation. Thus, the presence of restriction products indicates methylation in the PCR amplicon. MethyLight is a bisulfite-dependent, fluorescence-based, quantitative real-time PCR method for DNA methylation. This technique includes specific priming combined with methylation-specific fluorescent probing, allowing one to sensitively detect very low frequencies of hypermethylated alleles.
Another method based on chemical modification of genomic DNA with sodium bisulfite is pyrosequencing. This technique allows quantification of methylation at individual CpG dinucleotides into an amplified DNA fragment. Different from MSP, primers for amplification are designed from regions which contain no CpG dinucleotides and differences between methylated and unmethylated sequences are seen only after pyrosequencing. The main advantage of this method is that it allows high-resolution analysis of methylation and detection of small changes in methylation at each CpG, and also in samples containing large amounts of normal DNA.
All the above-mentioned methods are sensitive, specific, and relatively inexpensive, but none allows one to analyse the whole genome, which includes about 28 million CpGs. For a global analysis, recent microarray-based methods have been designed, including direct hybridization[38], methylated DNA immunoprecipitation (MeDIP)[39] and HELP assay (HpaII tiny fragment enrichment by ligation-mediated PCR)[40]. Direct hybridization to CpG island arrays is able to detect DNA methylation in several CpG sites. It is based on the use of methylation-specific oligonucleotides arrayed on glass slides, detecting all possible methylation in target genes. MeDIP is a genome-wide method based on an antibody that recognises 5-methylcytosine in methylated DNA sequences. This technique is used for either array-based hybridization (MeDIP-chip) or high-throughput sequencing (MeDIP-seq). However MeDIP presents a significant limitation: restricted resolution typical of array-based technology. The HELP assay is comparative isoschizomer profiling of DNA methylation. DNA is digested by HpaII in parallel with MspI (resistant to DNA methylation), and then the HpaII and MspI products are either amplified by ligation-mediated PCR and hybridized using separate fluorochromes to a customized array, or directly sequenced[41].
All these methods based on next generation sequencing technology which produces a huge amount of information on methylomes. Generally, genome-wide technologies are very useful for genome-wide DNA methylation analysis but they are relatively expensive and cannot be currently introduced in routinely clinical studies (Table 4). However, methylation profiling could be a useful tool to better understand the biological mechanism at the basis of tumorigenesis and provide insight into prevention strategies to reduce the burden of cancer.
| Methylation arrays | Ultra-deep sequencing | |
| CpG coverage | + | +++ |
| Sensitivity | +++ | ++/+ (antibody-based) |
| Time consuming | ++ | ++ |
| Data analysis | +++ | + |
| High-throughput | +++ | + |
| Price | ++ | +/++ (price decreasing) |
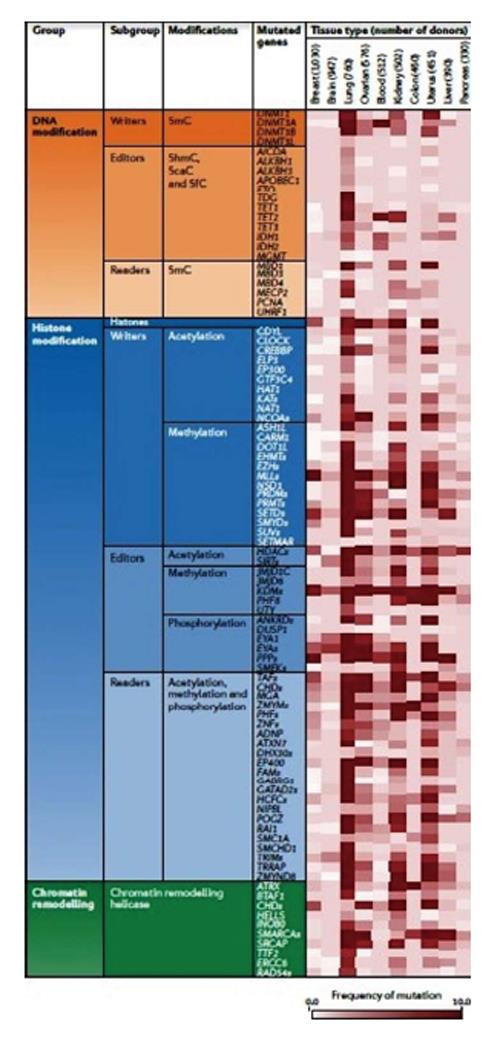
Thanks to advances in sequencing technologies thousands of cancer genomes and methylomes have been re-sequenced and new coding-gene mutations, genetic rearrangements, DNA copy-number alterations and alterations in either regulatory sequences or epigenetic patterns have been discovered. In addition, abnormalities in epigenetic enzymes and pathways, including DNA methylation or demethylation, histone modification, and chromatin remodelling processes, have been highlighted (Figure 10). For example, novel gene mutations have been uncovered in different tumours (IDH1 or DNMT3A in acute myeloid leukaemia, mitochondrial succinate dehydrogenase in paragangliomas or gastrointestinal stromal tumours, AT-rich interactive domain 1A-ARID1A in NSCLC; CREB-binding protein-CREBBP, E1A-binding protein p300 and MLL in small-cell lung cancer; H3F3A in paediatric glioblastoma; and MLL2 and SWI/SNF-related, matrix-associated, actin-dependent regulator of chromatin (SMARCA4) in medulloblastoma[42].
A major challenge for researchers will be to investigate the role of these mutations in tumorigenesis.
Beside the discovery of new mutations, genome-wide association studies have identified various single nucleotide polymorphisms (SNPs) correlated with increased risk of cancer. Interestingly, those SNPs are preferentially located in functional enhancers in ES cells and might confer cancer susceptibility by altering the cellular chromatin setting. In fact, a correlation between genetic variations and in gene expression changes have been demonstrated to involve chromatin accessibility of transcription factor (TF) binding sites, such us SNPs (CpG SNPs) that create or delete CpGs and influence the binding of specific TFs. Further and deeper studies on this association could reveal the functional link among epigenetic, genetic variation and phenotype[43].
The major clinical impact of the rising knowledge of epigenetic mechanisms is the possibility of defining epigenetic cancer therapy which inhibit methylation events in order to increase therapeutic efficacy. Recently, many epigenetic-modifying drugs have been introduced in combination with standard chemotherapy treatments in cancer patients. Nonetheless, those drugs may lack specificity, since they modulate global expression more than being gene-specific. However, different from other drugs, they could be able to restore TSG expression or loss-of-function phenotypes; thus, combined therapy could be a good therapeutic strategy.
Unfortunately, it is evident that epigenetic biology is complex; indeed, there are quite a number of scientific and pragmatic challenges, many of which are summarized in Table 5[44].
| Category | Issues |
| Target selection | Few activating mutations, translocations or syntethic lethal relationships known limited high-quality antibodies to epigenetic proteins and histone marks (e.g., confirm target expression linkage of target to mark) Biology driving cancer phenotype unknown or poorly understood Post-translation modification of histone vs non-histone substrates by "epigenetic" targets unclear |
| Chemistry | Existing chemical librairies may not have adeguate diversity to provide goog strating points Few crystal structures solved; are structrues relevant if not reflecting complete complex? |
| Assay development | Few reference compunds to establish assy signal window, sensitivity, reproducibility Are binding or enzyme configured to properly reflect physiological context? Production of actibe enzymes is difficult, may require multimeric complex and specific sunstrate (nucleosome, histone, non-histone) Limited high-quality antibodies to epigenetic proteins and histone marks (quantify mark or target gene product) |
| In vivo biology | Histone marks and target genes slow to change, require longer-duration studies to assess engagement (PD biomarker) May necessitate higer compund requirement to conduct studies, earlier optimation of PK properties than traditional paradigm May require novel models for tumors with mutation or traslocations |
| Toxicology | Acute and/or chronic liabilities of specific isofrom targed epigenetic therapies currently unknown Knockout animal data limited; inducibile knockouts, dominant negatives preferred but more scarce and technically challening |
| Clinical | Identify and implement appropriate patient selection markers, more challenging if not activating mutation (overexpression, gene profile?) Identify and implement suitable PD marker (posttranslational modification or mark, target gene, surrogate tissue or tumor?) Epigenetic changes at metastatic sites can differ from primary tumor, which should be targed clinically? |
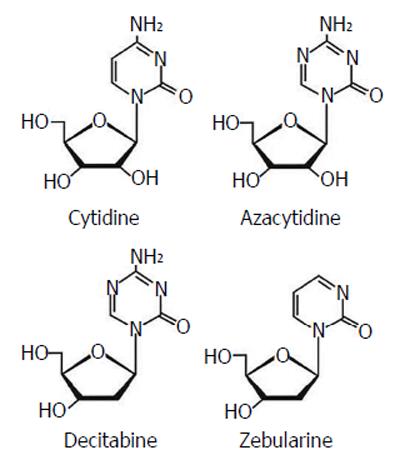
Currently, the most common epigenetic drugs are the DNMT inhibitors azacytidine and decitabine, although their clinical efficacy is limited by toxicity and chemical instability. Zebularine [1-(β-D-ribofuranosyl)-1,2-dihydropyrimidin-2-one] is a DNMT inhibitor characterized by more stability and less toxicity with a inhibitory effect on cytidine deaminase (Figure 11).
All these drugs are based on the rationale that, unlike genetic mutations, epigenetic alterations is potentially reversible, thus being an attractive target for cancer therapy.
Since hypermethylation of tumor suppressor genes and overexpression of DNMTs are crucial events for tumor progression, the possibility of de-methylating DNA sequences seems a good strategy for cancer therapy. DNMT inhibitors, in fact, can allow re-expression of aberrantly silenced genes and restore their normal function. Azacytidine (Vidaza; Celgene) and decitabine (5 aza 2’ deoxycytidine) (Dacogen; SuperGen) have been approved by the Food and Drug Administration (FDA) for current management of acute myeloid leukemia and myelodyplastic syndrome. Azacytidine has also been approved by the FDA and the European Medicines Agency for use against chronic myelomonocytic leukemia. More recently these two drugs have also been introduced in clinical trials in patients with solid tumors.
Zebularine is a novel member of the nucleoside DNMT inhibitor family, not yet used routinely in clinical practice. Although much in vitro data show good results, especially in terms of less toxicity compared to azacytidine or decitabine, zebularine use for future clinical trials is needed[45].
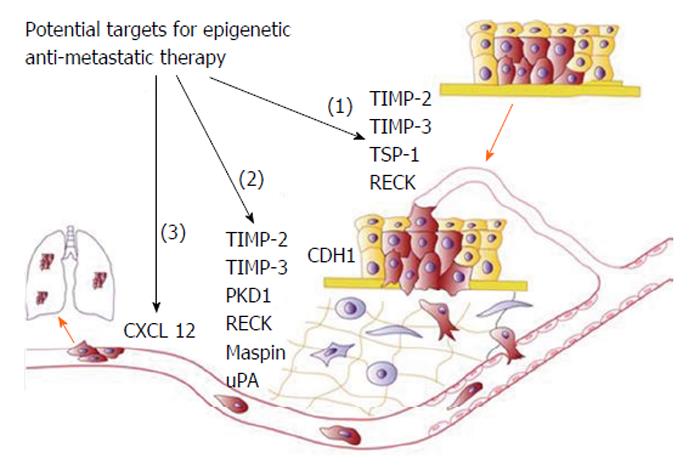
Moreover, recent studies have also demonstrated a possible use of epigenetic-modifying drugs in targeting invasion, metastasis and drug resistance, all involving EMT (Figure 12)[11].
It is already well known that cancer is a heterogeneous disease and an integrated genome, epigenome, and transcriptome analysis seems necessary to help clinicians to find good therapeutic strategies to treat this complex disease.
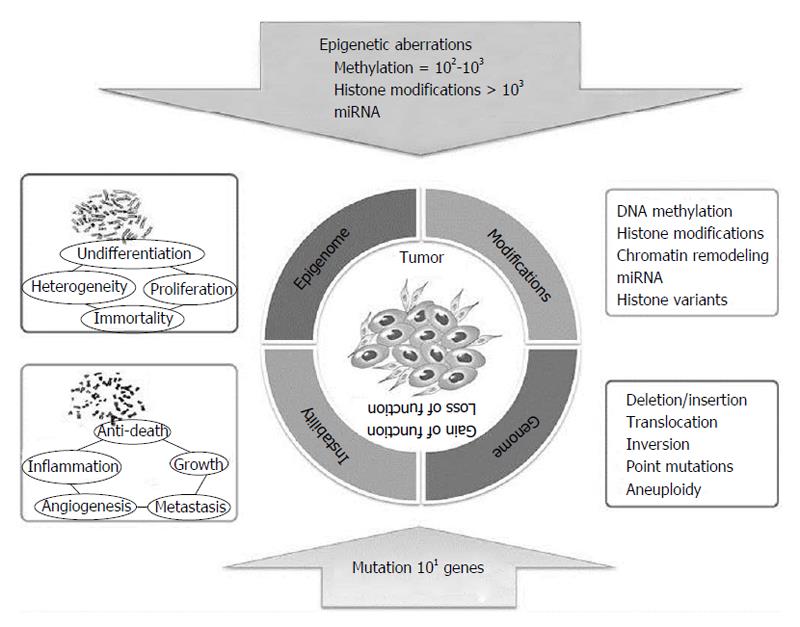
Only in recent years have researchers begun to integrate data deriving from both genetic and epigenetic alteration analyses, including mutations, CNVs, structural changes, epigenetic profiles, and expression changes in both coding and non-coding RNAs. Thanks to these studies, it is now well accepted that epigenetic abnormalities can play a crucial role in tumor initiation and development. In addition, it is now recognized that tumor cells present epigenetic silencing at higher frequency than mutations. Indeed, in tumour cells, hyper- or hypomethylation, histone modifications, and miRNA expression dysregulation are present in thousands of genes, while mutations affect only tens of genes, although all of them determine gene inactivation (Figure 13).
Epigenetic events can be useful biomarkers for detecting disease and predicting therapeutic efficacy. The epigenome undergoes all the above described during tumor initiation driving tumor cell heterogeneity, and consequently progression. Notably, those patterns are stable but reversible depending on the cellular environment, while mutations remain irreversibly locked into the cancer genome. For this reason, epigenetic events could be “drugable” targets for reversing epimutational effects and associated phenotypes.
Basing on the hypothesis that epigenetic agents may enhance sensitivity to conventional drugs (e.g., platinum or taxane chemotherapy), many efforts have been made for using epigenetic agents to re-sensitize tumours recurrent or refractory to first-line treatment.
Advances in genome-wide methylation analyses and the combination of new epigenetic drugs with conventional therapies could offer new hope for cancer patients, providing in the near future more effective patient-tailored treatments.
P- Reviewer: Garfield D, Gervois P, Wang M S- Editor: Song XX L- Editor: A E- Editor: Liu SQ
| 1. | Choi JD, Lee JS. Interplay between Epigenetics and Genetics in Cancer. Genomics Inform. 2013;11:164-173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 2. | Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1920] [Cited by in RCA: 2274] [Article Influence: 174.9] [Reference Citation Analysis (0)] |
| 3. | Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143-153. [PubMed] |
| 4. | Sproul D, Meehan RR. Genomic insights into cancer-associated aberrant CpG island hypermethylation. Brief Funct Genomics. 2013;12:174-190. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 92] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 5. | Sandoval J, Esteller M. Cancer epigenomics: beyond genomics. Curr Opin Genet Dev. 2012;22:50-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 330] [Article Influence: 25.4] [Reference Citation Analysis (0)] |
| 6. | Kim JG, Park MT, Heo K, Yang KM, Yi JM. Epigenetics meets radiation biology as a new approach in cancer treatment. Int J Mol Sci. 2013;14:15059-15073. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 7. | Virani S, Colacino JA, Kim JH, Rozek LS. Cancer epigenetics: a brief review. ILAR J. 2012;53:359-369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 61] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 8. | Cock-Rada A, Weitzman JB. The methylation landscape of tumour metastasis. Biol Cell. 2013;105:73-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 9. | Steenman MJ, Rainier S, Dobry CJ, Grundy P, Horon IL, Feinberg AP. Loss of imprinting of IGF2 is linked to reduced expression and abnormal methylation of H19 in Wilms’ tumour. Nat Genet. 1994;7:433-439. [PubMed] |
| 10. | Maeda M, Johnson KR, Wheelock MJ. Cadherin switching: essential for behavioral but not morphological changes during an epithelium-to-mesenchyme transition. J Cell Sci. 2005;118:873-887. [PubMed] |
| 11. | Kiesslich T, Pichler M, Neureiter D. Epigenetic control of epithelial-mesenchymal-transition in human cancer. Mol Clin Oncol. 2013;1:3-11. [PubMed] |
| 12. | Lee EJ, Lee BB, Han J, Cho EY, Shim YM, Park J, Kim DH. CpG island hypermethylation of E-cadherin (CDH1) and integrin alpha4 is associated with recurrence of early stage esophageal squamous cell carcinoma. Int J Cancer. 2008;123:2073-2079. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 13. | Sarkar S, Goldgar S, Byler S, Rosenthal S, Heerboth S. Demethylation and re-expression of epigenetically silenced tumor suppressor genes: sensitization of cancer cells by combination therapy. Epigenomics. 2013;5:87-94. [PubMed] |
| 14. | Seo DW, Li H, Guedez L, Wingfield PT, Diaz T, Salloum R, Wei BY, Stetler-Stevenson WG. TIMP-2 mediated inhibition of angiogenesis: an MMP-independent mechanism. Cell. 2003;114:171-180. [PubMed] |
| 15. | Galm O, Suzuki H, Akiyama Y, Esteller M, Brock MV, Osieka R, Baylin SB, Herman JG. Inactivation of the tissue inhibitor of metalloproteinases-2 gene by promoter hypermethylation in lymphoid malignancies. Oncogene. 2005;24:4799-4805. [PubMed] |
| 16. | Pulukuri SM, Patibandla S, Patel J, Estes N, Rao JS. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene. 2007;26:5229-5237. [PubMed] |
| 17. | Gu P, Xing X, Tänzer M, Röcken C, Weichert W, Ivanauskas A, Pross M, Peitz U, Malfertheiner P, Schmid RM. Frequent loss of TIMP-3 expression in progression of esophageal and gastric adenocarcinomas. Neoplasia. 2008;10:563-572. [PubMed] |
| 18. | Cho HJ, Kim SY, Kim KH, Kang WK, Kim JI, Oh ST, Kim JS, An CH. The combination effect of sodium butyrate and 5-Aza-2’-deoxycytidine on radiosensitivity in RKO colorectal cancer and MCF-7 breast cancer cell lines. World J Surg Oncol. 2009;7:49. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Kuhmann C, Weichenhan D, Rehli M, Plass C, Schmezer P, Popanda O. DNA methylation changes in cells regrowing after fractioned ionizing radiation. Radiother Oncol. 2011;101:116-121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Chaudhry MA, Omaruddin RA. Differential DNA methylation alterations in radiation-sensitive and -resistant cells. DNA Cell Biol. 2012;31:908-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 21. | Baer C, Claus R, Plass C. Genome-wide epigenetic regulation of miRNAs in cancer. Cancer Res. 2013;73:473-477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 759] [Reference Citation Analysis (0)] |
| 22. | Lujambio A, Esteller M. How epigenetics can explain human metastasis: a new role for microRNAs. Cell Cycle. 2009;8:377-382. [PubMed] |
| 23. | Zhang H, Li Y, Lai M. The microRNA network and tumor metastasis. Oncogene. 2010;29:937-948. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 155] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 24. | Kasinski AL, Slack FJ. Epigenetics and genetics. MicroRNAs en route to the clinic: progress in validating and targeting microRNAs for cancer therapy. Nat Rev Cancer. 2011;11:849-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 730] [Cited by in RCA: 794] [Article Influence: 56.7] [Reference Citation Analysis (0)] |
| 25. | Fabbri M, Calore F, Paone A, Galli R, Calin GA. Epigenetic regulation of miRNAs in cancer. Adv Exp Med Biol. 2013;754:137-148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 115] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 26. | Mendell JT. miRiad roles for the miR-17-92 cluster in development and disease. Cell. 2008;133:217-222. [PubMed] |
| 27. | Costinean S, Zanesi N, Pekarsky Y, Tili E, Volinia S, Heerema N, Croce CM. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci USA. 2006;103:7024-7029. [PubMed] |
| 28. | ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57-74. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14554] [Cited by in RCA: 12949] [Article Influence: 996.1] [Reference Citation Analysis (0)] |
| 29. | Kunej T, Godnic I, Ferdin J, Horvat S, Dovc P, Calin GA. Epigenetic regulation of microRNAs in cancer: an integrated review of literature. Mutat Res. 2011;717:77-84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 151] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 30. | Vrba L, Jensen TJ, Garbe JC, Heimark RL, Cress AE, Dickinson S, Stampfer MR, Futscher BW. Role for DNA methylation in the regulation of miR-200c and miR-141 expression in normal and cancer cells. PLoS One. 2010;5:e8697. [RCA] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 234] [Cited by in RCA: 236] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 31. | Hatzimichael E, Syed N, Lo Nigro C, Rao B, Crook T. How detection of epigenetic alterations of blood-borne DNA could improve melanoma diagnosis. Expert Rev Mol Diagn. 2014;14:639-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 32. | Lorincz AT. Cancer diagnostic classifiers based on quantitative DNA methylation. Expert Rev Mol Diagn. 2014;14:293-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 33. | Toraño EG, Petrus S, Fernandez AF, Fraga MF. Global DNA hypomethylation in cancer: review of validated methods and clinical significance. Clin Chem Lab Med. 2012;50:1733-1742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 34. | Choo KB. Epigenetics in disease and cancer. Malays J Pathol. 2011;33:61-70. [PubMed] |
| 35. | Xiong Z, Laird PW. COBRA: a sensitive and quantitative DNA methylation assay. Nucleic Acids Res. 1997;25:2532-2534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 913] [Cited by in RCA: 884] [Article Influence: 31.6] [Reference Citation Analysis (0)] |
| 36. | Eads CA, Danenberg KD, Kawakami K, Saltz LB, Blake C, Shibata D, Danenberg PV, Laird PW. MethyLight: a high-throughput assay to measure DNA methylation. Nucleic Acids Res. 2000;28:E32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1017] [Cited by in RCA: 1057] [Article Influence: 42.3] [Reference Citation Analysis (0)] |
| 37. | Dupont JM, Tost J, Jammes H, Gut IG. De novo quantitative bisulfite sequencing using the pyrosequencing technology. Anal Biochem. 2004;333:119-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 189] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 38. | Gitan RS, Shi H, Chen CM, Yan PS, Huang TH. Methylation-specific oligonucleotide microarray: a new potential for high-throughput methylation analysis. Genome Res. 2002;12:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 189] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 39. | Weber M, Davies JJ, Wittig D, Oakeley EJ, Haase M, Lam WL, Schübeler D. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 2005;37:853-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1315] [Cited by in RCA: 1291] [Article Influence: 64.6] [Reference Citation Analysis (0)] |
| 40. | Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C. Comparative isoschizomer profiling of cytosine methylation: the HELP assay. Genome Res. 2006;16:1046-1055. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 320] [Cited by in RCA: 297] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 41. | Jang H, Shin H. Current trends in the development and application of molecular technologies for cancer epigenetics. World J Gastroenterol. 2013;19:1030-1039. [PubMed] |
| 42. | Plass C, Pfister SM, Lindroth AM, Bogatyrova O, Claus R, Lichter P. Mutations in regulators of the epigenome and their connections to global chromatin patterns in cancer. Nat Rev Genet. 2013;14:765-780. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 330] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 43. | You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 794] [Cited by in RCA: 802] [Article Influence: 61.7] [Reference Citation Analysis (0)] |
| 44. | Campbell RM, Tummino PJ. Cancer epigenetics drug discovery and development: the challenge of hitting the mark. J Clin Invest. 2014;124:64-69. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 45. | Gnyszka A, Jastrzebski Z, Flis S. DNA methyltransferase inhibitors and their emerging role in epigenetic therapy of cancer. Anticancer Res. 2013;33:2989-2996. [PubMed] |