Published online Aug 12, 2013. doi: 10.5528/wjtm.v2.i2.27
Revised: May 13, 2013
Accepted: July 4, 2013
Published online: August 12, 2013
AIM: To evaluate GenoType methicillin-resistant Staphylococcus aureus (MRSA) Direct assay and cultivation for the identification of MRSA by using mecA polymerase chain reaction (PCR) as the “gold standard” assay.
METHODS: In total of 61 nasal specimens from patients at the intensive care unit were studied by GenoType MRSA Direct test, conventional culture method and automated bacterial identification system. The results of GenoType MRSA Direct assay were compared to conventional culture method the identification of MRSA and mecA gene PCR as the “gold standard” method. The sensitivity, specificity, positive predictive value and negative predictive value were calculated.
RESULTS: In total, 61 specimens were studied. Fifty-four specimens (88.5%) were negative by all three methods. Six swabs (9.8%) were found positive by GenoType MRSA Direct test, conventional culture method and automated bacterial identification system. The presence of mecA in these strains was confirmed by PCR. One swab sample was negative for culture methods but MRSA and mecA gene were detected by GenoType MRSA Direct test and mecA PCR respectively. GenoType MRSA Direct test had a sensitivity of 100% (6/6) and a specificity of 100% (55/55), with a positive predictive value of 100% and a negative predictive value of 98%. Culture method of MRSA had a sensitivity of 83.3% (5/6) and a specificity of 98.2% (55/56).
CONCLUSION: It was found that the GenoType MRSA Direct assay, which is a rapid and accurate test, is of the same sensitivity and specificity with mecA PCR. The GenoType MRSA Direct assay can be a better tool for rapid and accurate detection of MRSA in diagnostic laboratories.
Core tip: For the identification of methicillin-resistant Staphylococcus aureus (MRSA), GenoType MRSA Direct assay and cultivation were evaluated by using mecA polymerase chain reaction (PCR) as the “gold standard” assay. Fifty-four specimens (88.5%) were negative by all three methods. Six swabs (9.8%) were found positive by GenoType MRSA Direct test, conventional culture method and automated bacterial identification system. The presence of mecA in these strains was confirmed by PCR. One swab sample was negative for culture methods but MRSA and mecA gene were detected by GenoType MRSA Direct test and mecA PCR respectively. It was found that the GenoType MRSA Direct assay, which is a rapid and accurate test, is of the same sensitivity and specificity with mecA PCR.
-
Citation: Gamze Sener A, Kirdar S, Afsar I, Demirci M. Evaluation of three methods for detection of methicillin-resistant
Staphylococcus aureus . World J Transl Med 2013; 2(2): 27-31 - URL: https://www.wjgnet.com/2220-6132/full/v2/i2/27.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v2.i2.27
Methicillin-resistant Staphylococcus aureus (MRSA) has become increasingly prevalent worldwide. Since its discovery during the 1960s, MRSA has emerged as a common cause of nosocomial infection. Infections caused by MRSA is one of the major sources of morbidity and mortality nosocomial infections especially in the intensive care units (ICU). Prevention of nosocomial infections caused by MRSA in ICU has been recommended for several years[1,2]. The spread of MRSA can be controlled by effective preventive measures and to limit this spread, a rapid and sensitive test for detection of MRSA colonization is required. However, conventional tests for the identification of MRSA require at least 48 h to be completed[3]. In recent years, there has been a growing emphasis on the use of molecular methods to detect not only just infectious agents but also antimicrobial-resistance genes carried by microorganisms[4]. Several DNA based tests have been developed for the rapid detection of MRSA[3].
Methicillin resistance in Staphylococcus aureus (S. aureus) is mediated by the production of an altered penicillin-binding protein, PBP 2a[5]. The mec gene complex regulates the production of PBP 2a. The detection of the mecA gene or of PBP 2a provides much more accurate detection of methicillin resistance in S. aureus[5,6]. MecA gene detection tests based on polymerase chain reaction (PCR) are considered as the gold standard for methicillin resistance[5].
Screening of the patients with risk factors for MRSA carriage is important for a successful MRSA control policy. At our hospital, infection control precautions are taken immediately after a positive MRSA result becomes available from diagnostic and surveillance specimens. Since molecular methods are rapid, with turnaround times of 2 to 4 h, these tests are able to improve the utilization of infection control resources. We compared GenoType MRSA Direct assay and culture for identification of MRSA using PCR for mecA as the “gold standard” assay.
A total of 61 consecutive patients were screened for MRSA. Of the patients, 32 (52.4%) were female and 29 (47.5%) were male. Ethical approval was received from Ataturk Training and Research Hospital Ethics Committee.
Nasal specimens were obtained from patients at ICU. Two concurrent specimens were obtained from each site swabbed. First swab was used for culture and second swab was used for molecular assays. Swabs were transported at room temperature and processed within 1 to 3 h of collection.
The swabs were inoculated on blood agar plates directly on the day of receipt of the swab, incubated at 35 °C in O2, and read after 24 and 48 h. A colony suggestive of Staphylococcus was confirmed as S. aureus by using a tube coagulase and DNase test, while methicillin resistance was confirmed with cefoxitin susceptibility testing according to the Clinical and Laboratory Standards Institute method[7]. The Phoenix (Becton Dickinson, Sparks, MD, United States) was used for confirmation of strains. Control strains, MRSA strain (ATCC 43300), and methicillin-susceptible S. aureus (ATCC 25923) were used in all tests.
DNA extraction and amplification: The swabs were processed using the GenoType MRSA Direct (Hain Lifescience, Nehren, Germany) method. According to the manufacturers’ recommendations, the swabs were washed in 300 μL of lysis buffer before DNA extraction. Bacterial DNA was released by incubation of the lysis buffer for 10 min at 95 °C, followed by centrifugation for 5 min at 6000 g. Portions (5 μL) of the supernatant were used for amplification. In brief, 45 μL of primer nucleotide mix (provided with the kit), MgCl2 to a final concentration of 2.5 mmol/L and 1 U of HotStart Taq polymerase (Qiagen, Hilden, Germany) were added, followed by amplification on a PE 9700 thermocycler (Applied Biosystems, Weiterstadt, Germany) for 15 min at 95 °C, 35 cycles of 95 °C for 30 s, 55 °C for 40 s and 72 °C for 40 s, and a final extension at 70 °C for 8 min. Each run included a negative control sample to demonstrate the absence of contaminating DNA. The sensitivity of amplification and hybridisation was monitored using an internal control.
Hybridization protocol: Briefly, the assay uses a specific oligonucleotide probe, targeting the SCCmec chromosomal cassette of MRSA immobilized on membrane strips. During the detection process PCR amplicons hybridise with this probe. Hybridization and detection were performed in an automated washing and shaking device (Profiblot; Tecan, Maennedorf, Switzerland). PCR products (20 μL) were mixed for 5 min with 20 μL of denaturing reagent (provided with the kit) at room temperature in separate troughs of a plastic tray. After addition of 1 mL of pre-warmed hybridization buffer, the membrane strips in the kit were added to every trough. Hybridization was at 45 °C for 30 min, followed by two washing steps at 45 °C for 30 min with 1 mL of prewarmed stringent wash solution. Streptavidin-conjugated alkaline phosphatase and the appropriate substrate were added for colourimetric detection of hybridised amplicons. After final washing, the strips were air-dried and fixed on a data sheet. DNA isolation, amplification and hybridisation, were monitored using an internal control to improve the reliability of the test.
The following primers were used: M1 (TGGCTATCGTGTCACAATCG) and M2 (CTGGAACTTGTTGAGCAGAG) that amplify a 310-bp fragment of the mecA gene, which codes for the PBP 2a protein. The mixture for PCR consisted of 5 μL of PCR buffer 10 (final concentration: 50 mmol/L KCl, 0.01% gelatin, 10 mmol/L Tris-HCl; pH 8.3), 1.5 mmol/L MgCl2, 0.1 mmol/L dNTP (dATP, dCTP, dGTP and dTTP), 0.4 pmol/mL of each of the specific primers, 2 U of tag polymerase and 5 μL of the template to get a final reaction volume of 50 μL. All components were mixed in polystyrene tubes and subjected to amplification temperatures of the nucleic acid in a thermocycler. DNA from the methicillin resistant S. aureus reference strain (ATCC 43300) was used as positive control of the reaction and sterile bi-distilled water as negative control.
The mixture was initially denatured at 94 °C for 5 min and then underwent 35 cycles of: 30 s at 94 °C, 30 s at 52 °C and 30 s at 72 °C. The reaction was finished with 5 min at 72 °C. The amplification products were detected with agarose gel electrophoresis (2% in TBE 0.5X buffer) at 100 V for 35 min.
The results of GenoType MRSA Direct assay were compared to conventional culture method the identification of MRSA and mecA gene PCR as the “gold standard” method. The sensitivity, specificity, positive predictive value and negative predictive value were calculated.
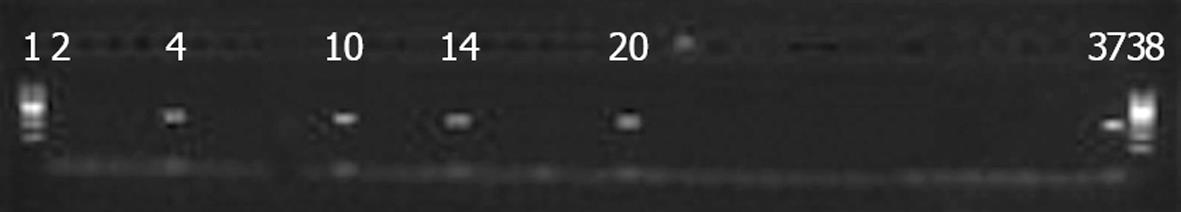
In total, 61 specimens were studied. Fifty-four specimens (88.5%) were negative by all three methods. Six swab samples (9.8%) were positive by conventional culture method and automated bacterial identification system (Table 1). The presence of mecA in these strains was confirmed by PCR and GenoType MRSA Direct test. Although one swab was negative by conventional culture method and automated bacterial identification system, it was positive for mecA gene detected by GenoType MRSA Direct test and mecA PCR respectively. The results of 61 specimens studied were shown in Table 1. GenoType MRSA Direct test had a sensitivity of 100% (6/6) and a specificity of 100% (55/55), with a positive predictive value of 100% and a negative predictive value of 98%. Culture method of MRSA had a sensitivity of 83.3% (5/6) and a specificity of 98.2% (55/56). Images of mecA PCR and GenoType MRSA Direct test are shown in Figures 1 and 2.
| n | Genotype MRSADirect test | Culture | MecA PCR |
| 54 | - | - | - |
| 6 | + | + | + |
| 1 | + | - | + |
| Total = 61 |
The prevalence of MRSA carriage on hospital admission is important in determining the effect of implementing any screening policy. Standard culture methods require at least 24 to 48 h for the recovery and identification of S. aureus and additional confirmatory tests[8] or susceptibility testing methods to determine methicillin resistance. Genotype MRSA direct test has been completed in approximately 4 h for detecting MRSA in the present study. Early and specific diagnosis of MRSA infections is significant in preventing their spread. Delays in detection of MRSA lead to the increased transmission of MRSA among patients, higher numbers of MRSA infections, and increased hospital costs[3]. A rapid and reliable test for the identification of MRSA would be desirable so that effective therapy could be initiated immediately. In recent years, there has been a growing emphasis on the use of molecular methods to detect not just infectious agents but also antimicrobial-resistance genes carried by microorganisms[4]. A study investigating the value of rapid diagnostic tests for MRSA when used for admission screening to a critical care area reported a reduction in the incidence of transmission of MRSA from 13.89/1000 patient days to 4/1000 patient days[9].
PCR tests are valuable for the rapid detection of MRSA carriers[10]. Conventional culture based detection methods for MRSA are time-consuming which leads to delayed isolation. Rapid molecular detection assays such as conventional PCR, real-time PCR and gene probe hybridization assays. Real-time PCR, has improved the sensitivity, and specificity, enables detection of resistance in a shorter time and lower risk of contamination than conventional PCR[11,12]. IDI-MRSA and the genotype MRSA tests can detect MRSA within a few hours directly from screening swabs with good sensitivity and specificity of 81% to 92% and 93% to 98%, respectively[10]. GenoType MRSA Direct test was evaluated to detect for the rapid detection of MRSA from nasal specimens in the present study. We evaluated the results of GenoType MRSA Direct test with the results obtained from conventional culture assay and PCR as the gold standard. We found a sensitivity of 100% and a specificity of 100%, with a positive predictive value of 100% and a negative predictive value of 98% with three tests. Our results were similar previous studies[13-15]. Warren et al[13] used a commercially available real-time PCR kit to detect MRSA directly from nasal swabs of 288 patients. They reported a sensitivity of 91.7%, and a specificity of 93.5%, with a positive predictive value of 82.5% and a negative predictive value of 97.1%. Similar results were reported by Huletsky et al[14,15] with the same system from 331 nasal swab specimen, with a sensitivity of 100%, specificity of 96.5%, with a positive predictive value of 89.4% and a negative predictive value of 100%.
Rising colonization rates lead to increased infection rates in the community and in hospitals. It has also been reported that rapid detection of carriage has an important role to play in such a “search-and-destroy” strategy[16,17]. van Hal et al[18] compared the relative sensitivities and specificities of the IDI-MRSA and GenoType MRSA Direct assays and three selective MRSA agars, MRSA ID, MRSASelect, and CHROMagar MRSA, with swabs from the three most commonly screened sites, i.e., the nose, groin and axilla. They informed that IDI-MRSA was the most sensitive method for the detection of MRSA with nasal swabs, with 90% sensitivity. GenoType MRSA Direct test had a sensitivity of 69%. However, Holfelder et al[3] found the sensitivity 94.5% of GenoType MRSA Direct test in their study. Swabs from 242 patients at risk for MRSA carriage were analysed by standard culture method and the PCR assay. They reported that the GenoType MRSA Direct assay provides a rapid, sensitive and specific method, in comparison with selective culture, for direct detection of MRSA in clinical swab specimens.
Tokue et al[6] tested mecA gene in 58 clinical isolates by the PCR and Southern blot analyses. Six PCR-positive strains were classified as methicillin susceptible by the conventional susceptibility test. They reported that the PCR assay appears to be more reliable than routine susceptibility testing. In the present study, one swab was negative for culture method but MRSA and mecA gene were detected by GenoType MRSA Direct assay and mecA PCR respectively. However, the broad use of MRSA PCR assays is hampered by high costs for PCR[19]. PCR tests are valuable for the rapid detection of MRSA, but high costs require the careful evaluation of their use. In patient populations with low MRSA endemicity, the broad use of PCR may not be cost-effective. But the rapid detection of MRSA carriers is important with low MRSA prevalence, since MRSA control is easiest when rates are still low, and maximal efforts should be made to maintain such epidemiology[10]. Metan et al[12] reported that the molecular assays would be appropriate for tertiary hospitals considering the upfront costs and requirement of expert laboratory staff.
Although conventional tests for identification of MRSA require at least 48 h, the GenoType MRSA Direct assay has rapid turnaround time of 4 h. This assay provides same day results and reduces the isolation time required for patients at risk of MRSA carriage. te Witt et al[20] emphasized that, nucleic acid-amplification techniques offer clear benefits over traditional culture-based assays, in particular, a reduced time to identification and an improved specificity and sensitivity. Luteijn et al[21] reported that it was a significantly higher sensitivity was found for the PCR in the recent article. Continual monitoring of clinical isolates is necessary to develop and maintain an effective strategy against S. aureus infection in the hospital setting[22].
As a conclusion, for the screening of MRSA in clinical swab specimens, it was found that the GenoType MRSA Direct assay, which is rapid and accurate test, is of the same sensitivity and specificity with mecA PCR in the present study. The GenoType MRSA Direct assay can be a better tool for the rapid and accurate detection of MRSA in diagnostic laboratories.
Thanks to Hain Lifescience-Turkey for providing GenoType MRSA Direct test.
Methicillin-resistant Staphylococcus aureus (MRSA) has become increasingly prevalent worldwide. The spread of MRSA can be controlled by effective preventive measures and to limit this spread, a rapid and sensitive test for detection of MRSA colonization is required. Screening of the patients with risk factors for MRSA carriage is important for a successful MRSA control policy. Since molecular methods are rapid, with turnaround times of 2 to 4 h, these tests are able to improve the utilization of infection control resources. Authors compared GenoType MRSA Direct assay and culture for identification MRSA using polymerase chain reaction (PCR) for mecA as the “gold standard” assay.
Infections caused by MRSA is one of the major sources of morbidity and mortality nosocomial infections especially in the intensive care units (ICU). Infection control precautions should taken immediately after a positive MRSA result becomes available from diagnostic and surveillance specimens. The article’s significance originates from its emphasis on the area of hospital infections.
In recent years, there has been a growing emphasis on the use of molecular methods to detect not just infectious agents but also antimicrobial-resistance genes carried by microorganisms. A study investigating the value of rapid diagnostic tests for MRSA when used for admission screening to a critical care area reported a reduction in the incidence of transmission of MRSA from 13.89/1000 patient days to 4/1000 patient days. Warren et al used a commercially available real-time PCR kit to detect MRSA directly from nasal swabs of 288 patients. They reported a sensitivity of 91.7%, and a specificity of 93.5%, with a positive predictive value of 82.5% and a negative predictive value of 97.1%. Similar results were reported by Huletsky et al. The present study was performed in a tertiary hospital. Metan et al reported that the molecular assays would be appropriate for tertiary hospitals considering the upfront costs and requirement of expert laboratory staff.
Besides their advantages like high sensitivity and specificity, molecular methods require experienced staff and laboured work. Yet, automatized molecular methods, being effective after in-house PCR, provides standardization along with an ease of use. As more advanced molecular methods are introduced, these methods will be preferred routinely.
This article is considered to be helpful for the clinicians about predicting MRSA infections among ICU patients. This consideration appears to be approved explicitly by the clinicians.
P- Reviewers Abraham WR, Taiwo SS S- Editor Gou SX L- Editor A E- Editor Zheng XM
| 1. | Lowy FD. Staphylococcus aureus infections. N Engl J Med. 1998;339:520-532. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4167] [Cited by in RCA: 4277] [Article Influence: 158.4] [Reference Citation Analysis (0)] |
| 2. | Lowy FD. Antimicrobial resistance: the example of Staphylococcus aureus. J Clin Invest. 2003;111:1265-1273. [PubMed] |
| 3. | Holfelder M, Eigner U, Turnwald AM, Witte W, Weizenegger M, Fahr A. Direct detection of methicillin-resistant Staphylococcus aureus in clinical specimens by a nucleic acid-based hybridisation assay. Clin Microbiol Infect. 2006;12:1163-1167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Tenover FC, Rasheed JK. Detection and characterization of antimicrobial resistance genes in bacteria. Manual of Clinical Microbiology. 8th ed. Washington, DC: American Society for Microbiology 2003; 1196-1212. |
| 5. | Chambers HF. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781-791. [PubMed] |
| 6. | Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and a conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:6-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 83] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 7. | Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing; twenty-first informational supplement. CLSI document M100-S21 (ISBN 1-56238-742-1). Wayne, PA: Clinical and Laboratory Standards Institute 2011; . |
| 8. | Louie L, Matsumura SO, Choi E, Louie M, Simor AE. Evaluation of three rapid methods for detection of methicillin resistance in Staphylococcus aureus. J Clin Microbiol. 2000;38:2170-2173. [PubMed] |
| 9. | Cunningham R, Jenks P, Northwood J, Wallis M, Ferguson S, Hunt S. Effect on MRSA transmission of rapid PCR testing of patients admitted to critical care. J Hosp Infect. 2007;65:24-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 114] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 10. | Bühlmann M, Bögli-Stuber K, Droz S, Mühlemann K. Rapid screening for carriage of methicillin-resistant Staphylococcus aureus by PCR and associated costs. J Clin Microbiol. 2008;46:2151-2154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 11. | Arbique J, Forward K, Haldane D, Davidson R. Comparison of the Velogene Rapid MRSA Identification Assay, Denka MRSA-Screen Assay, and BBL Crystal MRSA ID System for rapid identification of methicillin-resistant Staphylococcus aureus. Diagn Microbiol Infect Dis. 2001;40:5-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 12. | Metan G, Zarakolu P, Unal S. Rapid detection of antibacterial resistance in emerging Gram-positive cocci. J Hosp Infect. 2005;61:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 13. | Warren DK, Liao RS, Merz LR, Eveland M, Dunne WM. Detection of methicillin-resistant Staphylococcus aureus directly from nasal swab specimens by a real-time PCR assay. J Clin Microbiol. 2004;42:5578-5581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 160] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 14. | Huletsky A, Lebel P, Picard FJ, Bernier M, Gagnon M, Boucher N, Bergeron MG. Identification of methicillin-resistant Staphylococcus aureus carriage in less than 1 hour during a hospital surveillance program. Clin Infect Dis. 2005;40:976-981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 88] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 15. | Huletsky A, Giroux R, Rossbach V, Gagnon M, Vaillancourt M, Bernier M, Gagnon F, Truchon K, Bastien M, Picard FJ. New real-time PCR assay for rapid detection of methicillin-resistant Staphylococcus aureus directly from specimens containing a mixture of staphylococci. J Clin Microbiol. 2004;42:1875-1884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 288] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Kim T, Oh PI, Simor AE. The economic impact of methicillin-resistant Staphylococcus aureus in Canadian hospitals. Infect Control Hosp Epidemiol. 2001;22:99-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 131] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 17. | Bootsma MC, Diekmann O, Bonten MJ. Controlling methicillin-resistant Staphylococcus aureus: quantifying the effects of interventions and rapid diagnostic testing. Proc Natl Acad Sci USA. 2006;103:5620-5625. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 199] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 18. | van Hal SJ, Stark D, Lockwood B, Marriott D, Harkness J. Methicillin-resistant Staphylococcus aureus (MRSA) detection: comparison of two molecular methods (IDI-MRSA PCR assay and GenoType MRSA Direct PCR assay) with three selective MRSA agars (MRSA ID, MRSASelect, and CHROMagar MRSA) for use with infection-control swabs. J Clin Microbiol. 2007;45:2486-2490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 66] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 19. | Kunori T, Cookson B, Roberts JA, Stone S, Kibbler C. Cost-effectiveness of different MRSA screening methods. J Hosp Infect. 2002;51:189-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 20. | te Witt R, van Belkum A, van Leeuwen WB. Molecular diagnostics and genotyping of MRSA: an update. Expert Rev Mol Diagn. 2010;10:375-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 21. | Luteijn JM, Hubben GA, Pechlivanoglou P, Bonten MJ, Postma MJ. Diagnostic accuracy of culture-based and PCR-based detection tests for methicillin-resistant Staphylococcus aureus: a meta-analysis. Clin Microbiol Infect. 2011;17:146-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 60] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 22. | Song Y, Du X, Li T, Zhu Y, Li M. Phenotypic and molecular characterization of Staphylococcus aureus recovered from different clinical specimens of inpatients at a teaching hospital in Shanghai between 2005 and 2010. J Med Microbiol. 2013;62:274-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |