Copyright
©The Author(s) 2016.
World J Transl Med. Apr 12, 2016; 5(1): 1-13
Published online Apr 12, 2016. doi: 10.5528/wjtm.v5.i1.1
Published online Apr 12, 2016. doi: 10.5528/wjtm.v5.i1.1
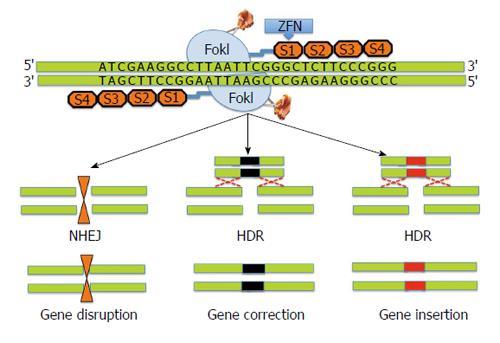
Figure 1 Schematic diagram showing structure and design of a typical zinc finger nuclease.
Zinc finger nucleases (ZFNs) use a modular array of 3-6 ZFNs (4 shown) specifically designed to bind to the target DNA together with the FokI cleavage domain. The FokI cleavage domains can be engineered to function as heterodimers or homodimers to achieve desired cleavage specificity. ZFNs typically recognize 24-36 bp unique sequence within the genome to achieve target specificity. ZFN mediated cleavage of the target leads to double strand breaks, which in turn induces either non-homologous end joining pathway (NHEJ) or homology directed repair (HDR) processes. NHEJ leads to gene disruption due to small insertions or deletions (indels) while HDR leads to gene correction.
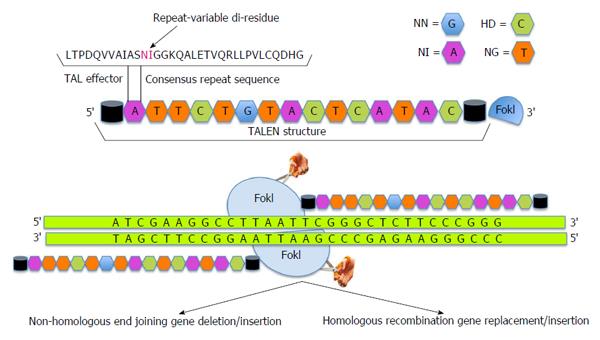
Figure 2 Transcription activator-like effector nucleases.
In transcription activator-like effector nucleases (TALENs) the nuclease effector domains of FokI are fused to TALE DNA binding domains. Since FokI is active only as a dimer, pair of TALENs are constructed to position FokI nuclease domains adjacent to genomic target sites. Like zinc finger nucleases, dimerization of TALENs leads to double strand breaks that is repaired by either error prone non-homologous end joining pathway thereby leading to frameshift mutations (deletions, insertions or frameshift) if exons are targeted or homology directed repair which can be utilized to introduce non-random mutations, targeted deletion or addition of large fragments.
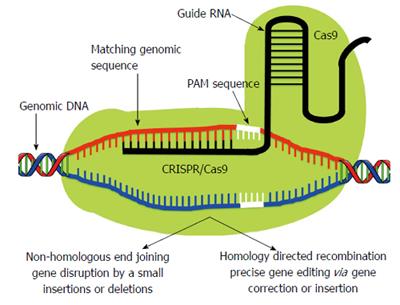
Figure 3 Clustered Regularly Interspaced Short Palindromic Repeat/Clustered Regularly Interspaced Short Palindromic Repeat Associated Systems.
In contrast to Like zinc finger nucleases and transcription activator-like effector nucleases, Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR)-associated protein (Cas9) monomer possess innate nuclease activity which catalyzes double strand breaks leading to random knockout phenotypes via non-homologous end joining pathway. Therefore Cas9 requires a single guide RNA (sgRNA) to recognize its target site. The sgRNA is composed of two separately expressed RNAs including a CRISPR RNA (crRNA) and a trans-activating crRNA (tracrRNA), which are processed by endogenous bacterial machinery to yield the mature gRNA. The current CRISPR/Cas9 system employs a single chimeric sgRNA, which is a fusion of crRNA and tracrRNA. Currently used sgRNA typically contains a 17-20 nucleotide long variable region, which is complementary to the genomic target sequence. A short region immediately 3’ to the target sequence known as protospacer adjacent motif has NGG sequence which is a major specificity determinant of Cas9. PAM: Protospacer-adjacent motif.
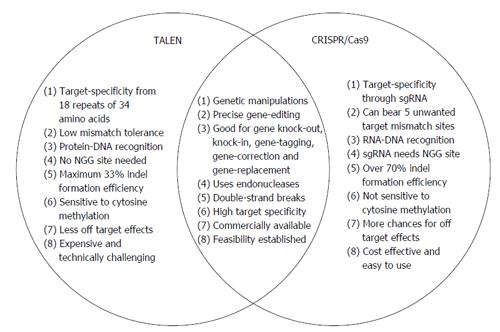
Figure 4 Venn diagram of transcription activator-like effector nucleases and Clustered Regularly Interspaced Short Palindromic Repeat.
The schematic Venn diagram shows potential differences and similarities between transcription activator-like effector nucleases (TALENs) and Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) systems. The gold standard to decipher the gene function is to selectively knockout or disrupt the gene expression and analyze the resulting phenotypes. Both TALENs and CRISPR are promising and powerful gene editing tools that allow complete loss-of-function reverse genetics approaches to study gene function. sgRNA: Single guide RNA; Cas9: Clustered Regularly Interspaced Short Palindromic Repeats associated protein 9.
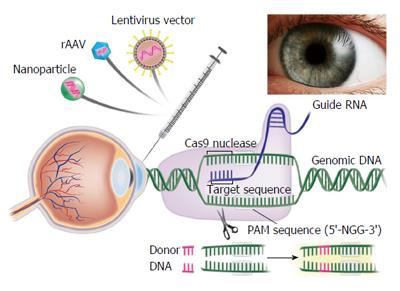
Figure 5 Application of Clustered Regularly Interspaced Short Palindromic Repeat/Clustered Regularly Interspaced Short Palindromic Repeat Associated Systems to develop novel therapies for corneal diseases.
Corneal Delivery of Clustered Regularly Interspaced Short Palindromic Repeat/Clustered Regularly Interspaced Short Palindromic Repeat Associated System using recombinant adeno-associated virus, integrase deficient lentiviral vectors and nanovectors can be used to potentially target multiple corneal diseases especially Fuchs’ endothelial corneal dystrophy to develop novel disease models as well as innovative personalized gene and stem cell therapies. PAM: Protospacer adjacent motif. Cas9: Clustered Regularly Interspaced Short Palindromic Repeats associated protein 9.
- Citation: Raikwar SP, Raikwar AS, Chaurasia SS, Mohan RR. Gene editing for corneal disease management. World J Transl Med 2016; 5(1): 1-13
- URL: https://www.wjgnet.com/2220-6132/full/v5/i1/1.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v5.i1.1