Copyright
©The Author(s) 2015.
World J Transl Med. Apr 12, 2015; 4(1): 11-24
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
Published online Apr 12, 2015. doi: 10.5528/wjtm.v4.i1.11
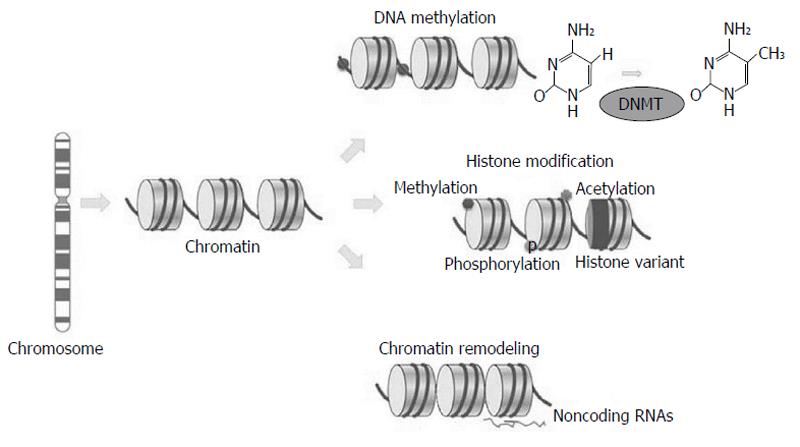
Figure 1 Epigenetic mechanisms.
Variations in chromatin structure without DNA sequence modifications by (1) DNA methylation; (2) histone modifications methylation, phosphorylation and acetylation; (3) histone variant composition (dark); and (4) chromatin remodeling (sparse or dense nucleosome occupancy), and noncoding RNAs. DNMT: DNA methyltransferases. Adapted by Choi et al[1], 2013.
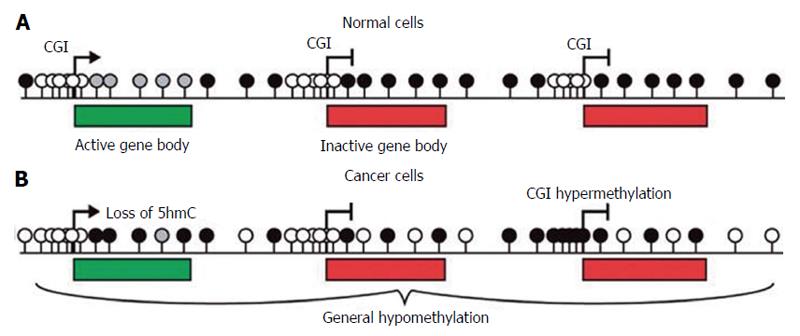
Figure 2 The methylation scenario of normal and cancer cells.
A: In the mammalian genome the amount of CpGs is low and most these sites are methylated (black lollipops). CGIs are usually located in gene promoters and are generally unmethylated (white lollipops), irrespective of gene expression status. The bodies of active genes are enriched in hydroxymethylated CpGs (grey lollipops); B: In cancer cells both DNA methylation and hydroxymethylation are reduced even if some CpG island have been found to be aberrantly hypermethylated. Adapted by Sproul et al[4], 2013. 5hmC: 5 hydroxymethylcytosine.
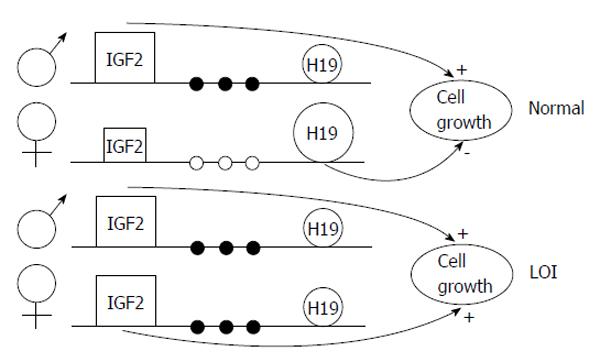
Figure 3 Model of loss of imprinting of insuline-like growth factor 2, H19 and methylation of the H19 promoter in Wilms’ tumor.
In normal cells, the paternal IGF2 and maternal H19 genes are expressed (shown large). Several sites upstream of H19 are methylated on the paternal allele (filled circles) and unmethylated on the maternal allele (open circles). In tumors with LOI, the maternal chromosome reverses to a paternal epigenotype, with a paternal pattern of methylation of the H19 promoter, IGF2 turned on, and H19 turned off, causing increased cell growth. LOI of H19 on the maternal chromosome, when it occurs, could occur independently or could be influenced by events in the paternal chromosome. Adapted by Steenman et al[9], 1994. LOI: Loss of imprinting; IGF2: Insuline-like growth factor 2.
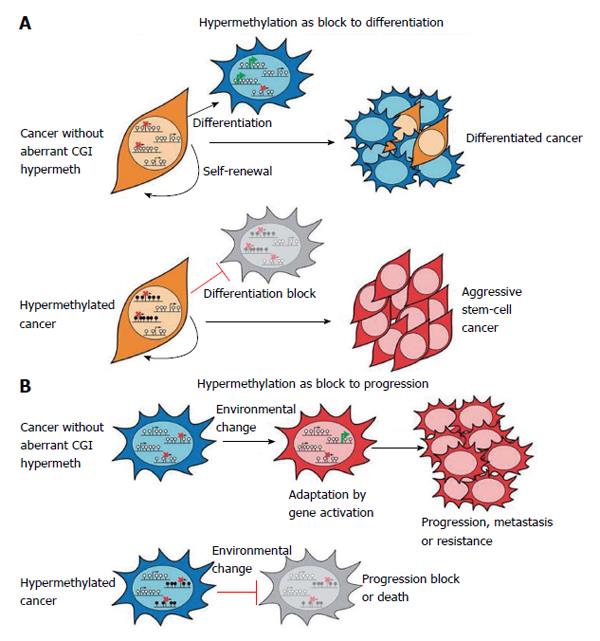
Figure 4 Hypermethylation consequences in cancer.
A: Key genes required for normal cellular differentiation become hypermethylated in cancer, resulting in a block to their activation and to normal differentiation processes. Thus cancer cells express a more aggressive, stem-cell like phenotype; B: Hypermethylation of repressed CpG island promoters might prevent the activation of genes facilitating survival in changing conditions such during metastatisation. Thus, widespread hypermethylation might restrict the potential for epigenetic adaptation and result in block to progression. Adapted by Sproul et al[4], 2013.
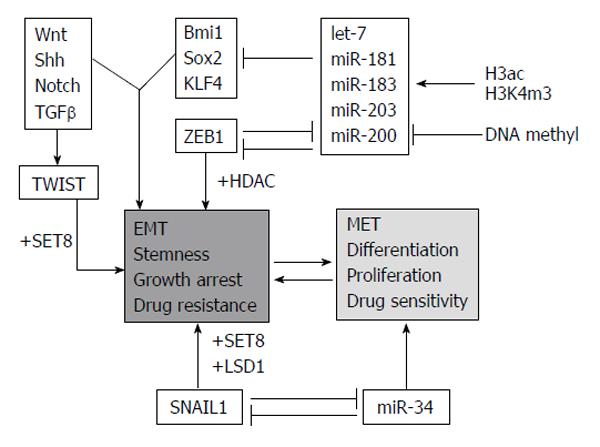
Figure 5 Molecular regulators regulating epithelial vs mesenchymal cell phenotypes and reverse process.
In gray/italicised factors/processes involved in epigenetic control are highlighted. ac: Acetylation; EMT: Epithelial-mesenchymal transition; MET: Mesenchymal-epithelial transition; miR: miRNA; m3: Trimethylation; Shh: Sonic hedgehog; TGFβ: Transforming growth factor beta; KLF4: Kruppel-like transcription factor4; HDAC: Histone deacetylase inhibitor; LSD1: Lysine-specific demethylase 1. Adapted by Kiesslich et al[11], 2013.
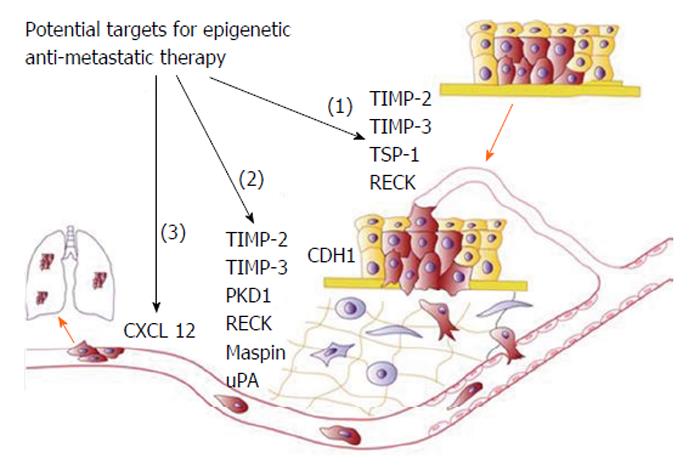
Figure 6 Schematic representation of progressive steps from initial tumour formation to establishment of metastasis include (1) tumour growth, angiogenesis and localised invasion; (2) intravasation and survival; and (3) extravasation and formation of distant tumours.
For each step some genes promoting these processes and regulated by DNA methylation are indicated. Adapted by Cock-Rada et al[8], 2013. TIMP-2: Tissue inhibitor of metalloproteinase 2.
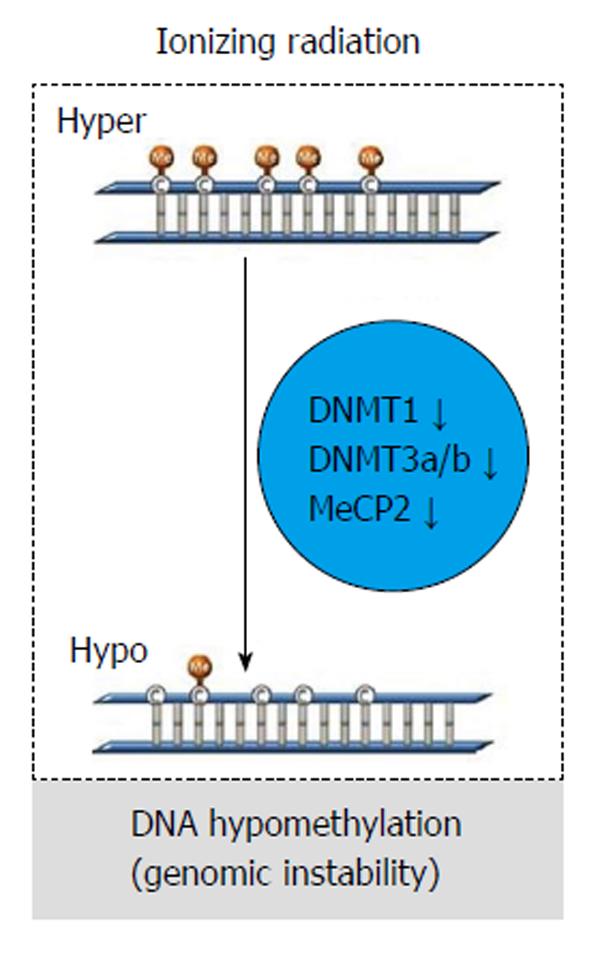
Figure 7 The figure represents the general change of global DNA methylation after radiation exposure in cancer cells.
Radiation might induce a decrease in DNA methyltransferases, including DNA methyltransferase 1 (DNMT1), DNMT3a, DNMT3b, and methyl CpG binding protein 2 leading to global DNA hypomethylation and genomic instability. Adapted by Kim et al[6], 2013.
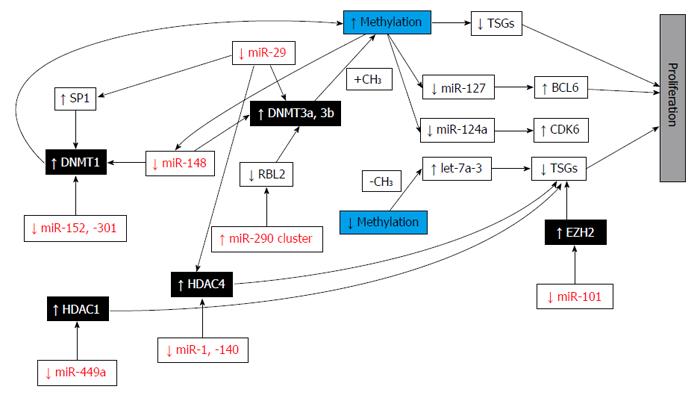
Figure 8 Epi-miRNA functions in cancer cells.
Epi-miRNAs (in red) directly target epigenetic effectors (black boxes) and indirectly affect the expression of epigenetically regulated miRNAs and protein coding genes (white boxes), contributing to carcinogenesis. TSGs: Tumor suppressor genes; DNMT: DNA methyltransferase; HDAC: Histone deacetylase; EZH2: Enhancer of zeste homolog 2; BCL6: B-cell CLL/lymphoma 6; CDK6: Cyclin-dependent kinase 6; SP1: Sp1 transcription factor; RBL2: Retinoblastoma-like 2 (p130); CH3: Methyl group. Adapted by Fabbri et al[25], 2013.
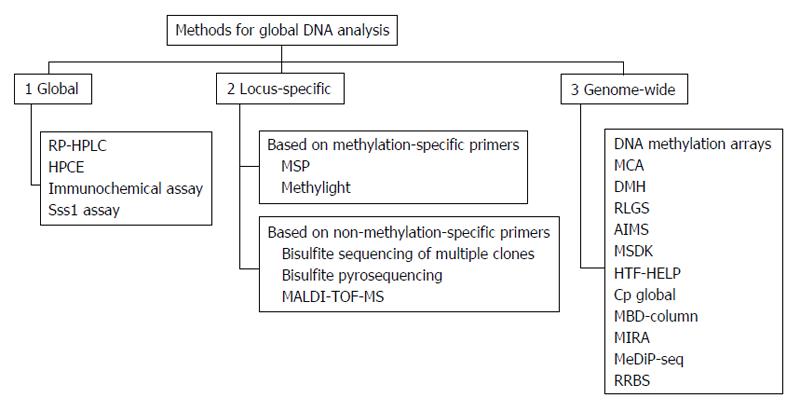
Figure 9 Methods for genomic DNA methylation analysis, classified as global, locus-specific and genome-wide.
In the case of locus-specific approaches, techniques were divided depending on the use of methylation-specific primers or not. RP-HPLC: Reverse-Phase high-performance liquid chromatography; HPCE: High performance capillary electrophoresis; Sss1 assay: Methyl group acceptance assay; MSP: Methylation-specific polymerase chain reaction (PCR); MALDI-TOF-MS: Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry; MCA: Methylation CpG island amplification; DMH: Differential methylation hybridization; RLGS: Restriction-landmark genomic scanning; AIMS: Amplification of inter-methylated sites; MSDK: Methylation-specific digital karyotyping; HTF-HELP: HPAII tiny fragment enrichment by ligation-mediated PCR; MBD-column: Methylated DNA binding column; MIRA: Methylated CpG island recovery assay; MeDIP-seq: Methyl-DNA immunoprecipitation and sequencing; RRBS: Reduced Representation Bisulfite Sequencing. Adapted by Toraño et al[33], 2011.
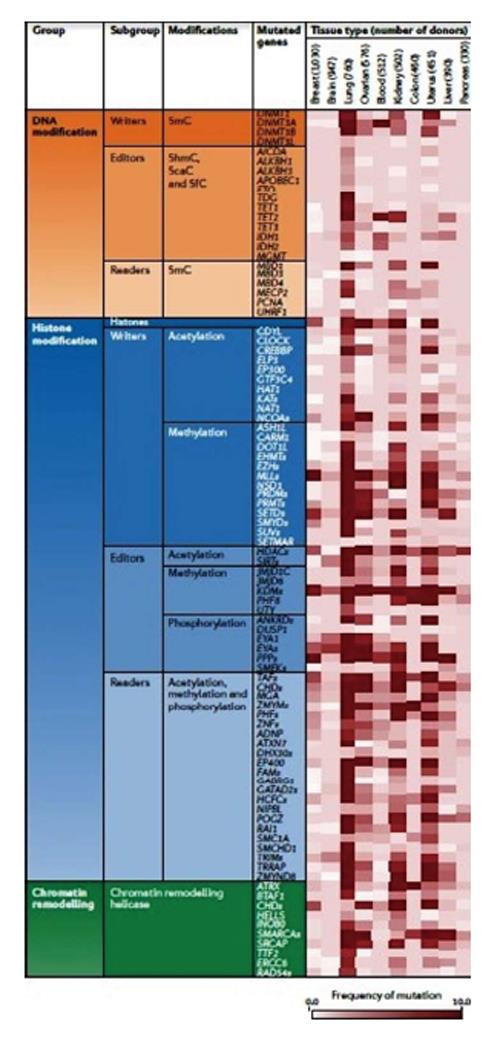
Figure 10 Some of the most known mutated genes classified in groups based on DNA modification, histone modification and chromatin remodelling enzymes.
The number of analysed tumour tissues is given. Several epigenetic enzymes present high frequencies of mutations in distinct tumor types. These data are not adjusted for chromosomal instability or mutator phenotypes, hence the frequencies reflect a combination of probable driver mutations in epigenetic regulators and the background mutation rate for the tumour type. 5caC: 5 carboxylcytosine; 5fC: 5 formylcytosine; 5hmC: 5 hydroxymethylcytosine; 5mC: 5 methylcytosine. Adapted by Plass et al[42], 2012.
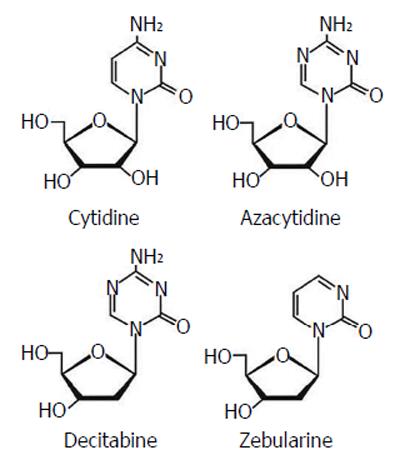
Figure 11 DNA methyltranferase inhibitors, analog to nucleoside.
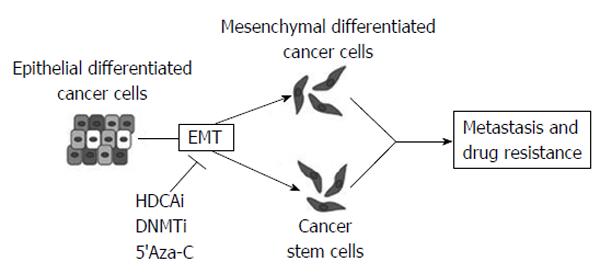
Figure 12 Different classes of epigenetic drugs could inhibit epithelial-mesenchymal transition which plays a crucial role in tumor progression generating both mesenchymal differentiated and stem cancer cells.
HDACi: Histone deacetylase inhibitor; DNMTi: DNA methyltransferase inhibitor; 5’Aza-C: 5-azacytidine are given as examples of demethylating agent. Adapetd by Kiesslich et al[11], 2013.
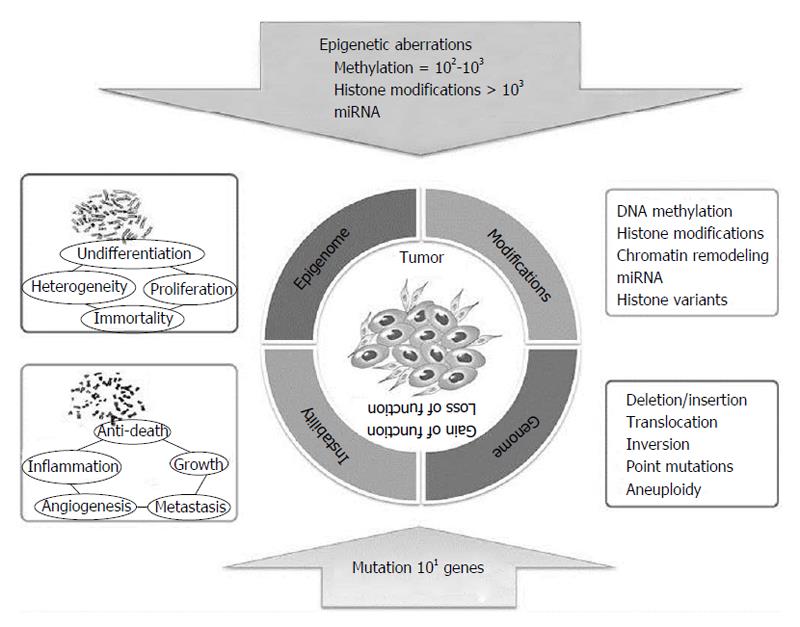
Figure 13 Epigenetics and genetics cooperates in cancerogenesis.
Adapted by Choi et al[1], 2013.
- Citation: Lattanzio L, Lo Nigro C. Epigenetics and DNA methylation in cancer. World J Transl Med 2015; 4(1): 11-24
- URL: https://www.wjgnet.com/2220-6132/full/v4/i1/11.htm
- DOI: https://dx.doi.org/10.5528/wjtm.v4.i1.11