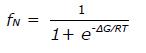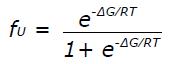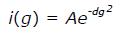Peer-review started: December 25, 2016
First decision: January 16, 2017
Revised: February 8, 2017
Accepted: February 28, 2017
Article in press: March 2, 2017
Published online: May 12, 2017
Processing time: 143 Days and 1.5 Hours
To further characterize the structure and nucleic acid binding properties of the 195 amino acid small delta antigen, S-HDAg, a study was made of a truncated form of S-HDAg, comprising amino acids 61-195 (∆60HDAg), thus lacking the domain considered necessary for dimerization and higher order multimerization.
Circular dichroism, and nuclear magnetic resonance experiments were used to assess the structure of ∆60HDAg. Nucleic acid binding properties were investigated by gel retardation assays.
Results showed that the truncated ∆60HDAg protein is intrinsically disordered but compact, whereas the RNA binding domain, comprising residues 94-146, adopts a dynamic helical conformation. We also found that ∆60HDAg fails to multimerize but still contains nucleic acid binding activity, indicating that multimerization is not essential for nucleic acid binding. Moreover, in agreement with what has been previously reported for full-length protein, no apparent specificity was found for the truncated protein regarding nucleic acid binding.
Taken together these results allowed concluding that ∆60HDAg is intrinsically disordered but compact; ∆60HDAg is not a multimer but is still capable of nucleic acid binding albeit without apparent specificity.
Core tip: The characterization of a truncated form of S-HDAg lacking amino acids 1-60, ∆60HDAg is reported. Structure of ∆60HDAg was assessed by circular dichroism and nuclear magnetic resonance and its nucleic acid binding properties were investigated using gel retardation assays. This study demonstrates for the first time that ∆60HDAg is intrinsically disordered and a monomer. Furthermore, ∆60HDAg can bind a wide variety of nucleic acids without apparent specificity.
- Citation: Alves C, Cheng H, Tavanez JP, Casaca A, Gudima S, Roder H, Cunha C. Structural and nucleic acid binding properties of hepatitis delta virus small antigen. World J Virol 2017; 6(2): 26-35
- URL: https://www.wjgnet.com/2220-3249/full/v6/i2/26.htm
- DOI: https://dx.doi.org/10.5501/wjv.v6.i2.26
Hepatitis delta virus (HDV) is the human pathogen with the smallest genome known to date. It is a defective virus that requires the presence of hepatitis B virus (HBV) to propagate infection, since the envelope is provided by the HBV surface antigens. HDV co-infection with HBV or super-infection of hepatitis B patients increases the severity of acute and chronic liver disease[1]. The HDV genome consists of a 1.7 kb single-stranded closed circular RNA of negative polarity. Replication of HDV genome results in the production of an intermediate complementary strand in which a single open reading frame coding for the delta antigen (HDAg) has been identified. During the replication cycle, site-specific editing of the antigenomic RNA by a host adenosine deaminase (ADAR 1) results in the expression of a second form of the HDAg. The mRNA for the original small delta antigen (S-HDAg), corresponding to a 195 amino acids long protein, has its stop codon changed to a tryptophan codon, giving rise to a 19 amino acid extension. This results in the translation of the large delta antigen (L-HDAg) with 214 amino acids. None of the HDAg forms have any known enzymatic activity and, although both antigens share most of their sequence and therefore functional domains, they display some different functions in the HDV replication cycle. S-HDAg is essential for HDV RNA accumulation while L-HDAg acts as a trans-dominant inhibitor of replication and is essential for virus assembly[1].
S-HDAg is considered an “intrinsically disordered protein” (IDP) as revealed by a meta-predictor as well as circular dichroism (CD) measurements[2]. In fact, S-HDAg has several characteristics commonly attributed to IDPs: They rarely display enzymatic activity and are commonly involved in nucleic acid binding and/or interactions with other proteins[3]. S-HDAg is a nucleic acid-binding protein, with multiple host protein partners identified over the years by different approaches[4-7]. S-HDAg is also a basic protein with a predicted net charge of +12 at neutral pH[8]. This is common for IDPs and may play an important role in the ability of the protein to bind negatively charged nucleic acids. Post-translational modification (PTM) sites are also a common feature to IDPs. S-HDAg is modified by phosphorylation, methylation, acetylation, and sumoylation, and distinct functions have been attributed to the different modified forms of the antigen[9-13]. The evidence showing that S-HDAg is an IDP can explain the difficulty in solving its 3-D structure. Despite several attempts it has not yet been possible to obtain crystals of the full-length antigen. However, crystals were readily obtained for a truncated form spanning amino acids 12 to 60 corresponding to a more ordered region of the protein[2,14]. This segment is involved in S-HDAg dimerization and was designated a coiled-coil domain (CCD) as dimers form an anti-parallel coiled-coil structure[14].
In the present study, we address the role of the C-terminal regions of S-HDAg in determining its conformational properties, multimerization and nucleic acid binding. A truncated form of S-HDAg lacking the first 60 amino acids (∆60HDAg) was generated and purified. We characterized the structural conformation of purified ∆60HDAg using CD and nuclear magnetic resonance (NMR). Additionally, we also investigated the multimerization and nucleic acid binding properties of ∆60HDAg.
Plasmid pR5δV5 was used to express full-length S-HDAg in Escherichia coli (E. coli) as previously described[15]. This plasmid was also used as template to amplify by PCR the region encoding amino acids 61-195 (forward primer: 5’-TTTCAATTGCCAAAGATAAAGATGGCG-3’, reverse primer: 5’-TTTCTCGAGTTACGGAAAGCC-3’). The amplified sequence was cloned into the EcoRI-XhoI cloning site of pGEX-6P-2 (GE Healthcare). The resulting plasmid, designated pGEX-6P-2-∆60S-HDAg, allowed further expression of the fusion protein GST-∆60HDAg.
Expression and purification of full-length S-HDAg was performed as described[6]. Expression of GST-∆60HDAg was performed in BL21 (DE3) codon plus competent cells (Novagen) and protein purification was performed as follows. Cells expressing GST-∆60HDAg were resuspended in PBS supplemented with protease inhibitors (cOmplete, Roche). Cell lysis was achieved by four freeze-thaw cycles after the addition of 0.1 mg/mL lysozyme. Lysates were treated with DNase 1 (Roche), sonicated, and centrifuged at 14000 × g for 10 min. Supernatants were analyzed by SDS-PAGE followed by western blot to detect the presence of recombinant protein. Total protein extracts were then used to purify recombinant GST-tagged protein as previously reported[6]. The GST tag was removed by PreScission protease (GE Healthcare) digestion following manufacturer’s instructions. Finally, purified ∆60HDAg was concentrated using the protein concentrating solution and dialysis cassettes (Pierce) using the protocol suggested by the manufacturer.
Plasmid pDL542 containing a T7 promoter, was used to express full-length antigenomic HDV RNA[16]. The plasmid was transcribed in vitro using a T7 RiboMax transcription system (Promega) following protocols provided by the manufacturer.
Protein samples were analyzed by electrophoresis in 12% SDS-PAGE gels and detected by Coomassie blue staining. In cross-linking experiments, protein samples were treated with 0.01% or 0.1% glutaraldehyde for 10 min at room temperature. Glutaraldehyde was inactivated by the addition of 100 mmol/L ammonium acetate and samples were analyzed by SDS-PAGE.
Regarding protein-nucleic acid interactions in vitro, protein samples were diluted in a standard binding buffer containing 150 mmol/L NaCl and 10 mmol/L Tris-HCl (pH 7.5) unless stated otherwise. Nucleic acids were then added, the mix was incubated for 10 min at room temperature and resolved by electrophoretic mobility shift assays. For the study of DNA-protein interactions we used the PCR product obtained in the cloning of the truncated protein. For the study of RNA-protein interactions we used full-length antigenomic HDV RNA. Mobility shift assays were performed in non-denaturing 1 × TBE 1.5% agarose gels stained with ethidium bromide.
The far-UV CD spectrum of approximately 10 μmol/L protein solution (in 20 mmol/L potassium phosphate buffer, pH 6.3) was acquired at 25 °C on an Aviv 62A spectropolarimeter (Aviv, Lakewood, NJ), using a 1 mm quartz cuvette. The CD spectrum was the average of five scans recorded in the far-UV region (195-250 nm) with a band pass of 2 nm. The temperature dependence was studied at a protein concentration of approximately 15 μmol/L by following the change in ellipticity at 225 nm upon increasing the temperature from 5 °C to 85 °C in 2 °C intervals, using a 2 nm bandwidth and a 2 mm quartz cuvette. Data average and temperature equilibrate times were 1 s and 12 s, respectively. The CD thermal scans were analyzed by nonlinear least squares analysis based on the Gibbs-Helmholtz equation. The fitting model used in this case assumes two-state transition with a temperature-independent heat enthalpy, ∆Cp = 0. The enthalpy of unfolding, ∆H, and the midpoint temperature of unfolding, Tm, were derived from:
∆G(Tm) = ∆H(Tm)(1-T/Tm) - ∆Cp[Tm - T + Tln(T/Tm)] (1)
where T is the sample temperature, Tm is the midpoint temperature of unfolding. ∆H is the enthalpy of unfolding at Tm, and ∆Cp is the heat capacity change.
Thermal unfolding curves monitored via the ellipticity at 225 nm, ∈255, were calculated as follows:
∈225 = fN∈N + fU∈U (2)
where fN and fU are the fractional populations of the native and unfolded state, respectively, and ∈N and ∈U are the corresponding ellipticity values. The temperature dependence of ∈N and ∈U is given by ∈N
∈N = ∈ + NsT (3)
∈U = ∈ui+ NUT (4)
where ∈Ni and ∈ui designate the initial ellipticity of the native and unfolded states, and Ns and Us are slopes in the pre- and post-transition regions. For a two-state equilibrium fN and fU depend on ∆G as follows:
Math 7
(5)
Math 8
(6)
For NMR sample preparation, the DNA sequence encoding residues 61-195 of S-HDAg (∆60HDAg) was commercially synthesized (LifeTechnology, GeneArt). The truncated gene was subcloned into the expression vector pET49b (Novagen) between BamHI and Hind III sites. Uniformly 15N-labeled fusion protein was prepared by growing previously transformed E. coli cells in M9 medium supplemented with 1 g/L of 15N NH4Cl. After removing GST by digestion with HRV 3C protease, the purified protein contained extra residues with the sequence GPGYNDP at its N-terminus. The protein (approximately 1 mmol/L) was dissolved in 50 mmol/L sodium phosphate and 50 mmol/L sodium chloride buffer, pH 6.3.
All NMR data were collected on a Bruker Avance II 600 MHz spectrometer equipped with a TCI cryoprobe. The spectra were recorded at 25 °C, processed using the Felix 2007 NMR software (http://www.felixnmr.com) and analyzed with the Sparky NMR assignment and integration software (http://www.cgl.ucsf.edu/home/sparky/).
NMR diffusion measurements were carried out with an NMR sample (300 μL and approximately 1 mmol/L containing 3 μL of 1% 4,4-dimethyl-4-silapentane-1-sulfonic acid, DSS, used as an internal standard) by using 1D 1H pulse gradient stimulated echo longitudinal encode-decode (PG-SLED) experiment with saturation of the water signal during relaxation delay[17]. A spin echo delay of 5 ms and a STE delay of 135 ms were used. The data were analyzed using TopSpin 3.2 (Bruker, MA, United States). Fitting the signal integrations from amide and aromatic ring protons (approximately 6.4-9.4 ppm) and DSS(i) as a function of gradient strength (g) to
Math 9
allowed to extraction the decay rates (d), which are proportional to the diffusion coefficients D. The protein hydrodynamic radius (Rhprot) was calculated according to the equation:
Rhprot = dref(Rhref)/dprot (7)
where dref and Rhref are the decay rate and hydrodynamic radius of a reference compound (DSS in the present case), and dprot is the measured decay rate (diffusion constant) of the protein. The hydrodynamic radii of native or denatured proteins are estimated from the number of residues, N, according to[18]:
Rh,i = ANiα (8)
with A = 4.75 and 2.21, and α = 0.29 and 0.57 for a compactly folded and a completely denatured protein, respectively. The effective hydrodynamic radius (Rh) of DSS was calculated as 3.38 Å from Rh: 18.4 Å of cytochrome c from the previous NMR PFG diffusion measurements (see Results).
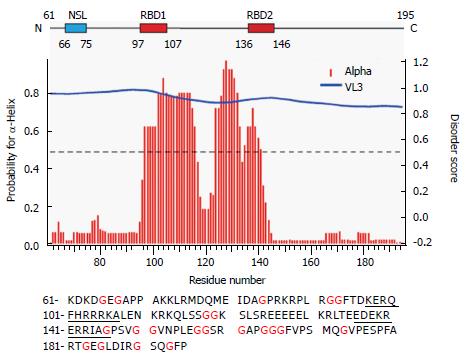
We have previously reported that full-length S-HDAg is able to form multimers in vitro, similar to the behavior of S-HDAg present in viral particles[2]. Furthermore, using PONDR-Fit, a Meta predictor of protein order that combines six neural network programs we found that, with the exception of the amino acid fragment 12-60, S-HDAg is intrinsically disordered. To address the possibility that the disordered regions of the antigen contribute to S-HDAg oligomerization, we focused here on a construct that lacks the previously characterized dimerization domain, ∆60HDAg (Figure 1).
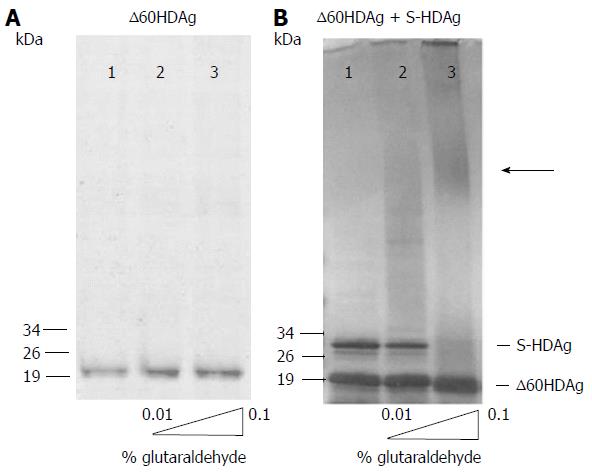
To determine whether the truncated antigen has a tendency to oligomerize, ∆60HDAg was analyzed by SDS-PAGE with and without prior glutaraldehyde crosslinking. The non-crosslinked protein displayed a well-defined single band and no changes in mobility were observed when the recombinant protein was pre-incubated with increasing amounts of glutaraldehyde (Figure 2A), indicating that ∆60HDAg is monomeric in solution. Although the truncated form of S-HDAg has an estimated weight of 15 kDa, in SDS-PAGE analysis the monomer appears to be a approximately 19 kDa protein. The difference in molecular weight relates with the reported observation that SDS-PAGE over estimates the size of full-length S-HDAg[19,20].
To investigate whether the truncated protein can form oligomers in the presence of full-length HDAg, we combined both forms at a 1:1 molar ratio and analyzed the mixture by SDS-PAGE, with and without prior crosslinking. In the absence of crosslinking (Figure 2B, lane 1) both proteins are readily detected, one with an apparent molecular weight of approximately 19 kDa and another with an apparent molecular weight of approximately 28 kDa, corresponding to the truncated and the full-length antigens, respectively. When the protein mixture was crosslinked, levels of monomeric full-length S-HDAg were reduced, forming oligomers with higher molecular weight as increasing concentrations of glutaraldehyde were present in the mixture (Figure 2B, lanes 2-3). In contrast, levels of monomeric truncated ∆60HDAg remained unchanged, with no oligomerization observed. This result shows that the intermolecular interaction occurs through the first 60 residues of S-HDAg, which includes the CCD, essential for the oligomerization of the full-length protein.
It is thought that IDPs may confer evolutionary advantages by allowing more flexible and diverse interactions with other proteins and nucleic acids. Using PONDR-Fit we have previously shown that, with the exception of portions of the N-terminal CCD, S-HDAg is a largely disordered protein[2]. The structured CCD domain is involved in the formation of dimers and higher order multimers that are believed to play important roles in the HDV replication cycle. Indeed, by using program VL3, the truncated version ∆60HDAg is also predicted to be disordered. As shown in Figure 1 (middle panel), disorder prediction of the truncated protein showed a high disorder score of approximately 0.8[21].
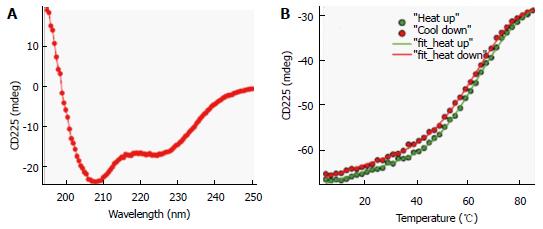
To investigate conformational preferences of the C-terminal S-HDAg for residues 61-195 (Δ60HDAg), we evaluated its secondary structure using CD spectroscopy. This technique allows determination of the average secondary structure content of a protein in solution[22]. Figure 3A shows the spectrum of the truncated recombinant protein exhibiting a strong negative peak around 208 nm and a weaker negative peak around 222 nm, very similar to that of the full-length protein[2]. A strong negative CD peak at 222 nm is characteristic of α-helical conformation and can be used to estimate the helix content. However, the lack of reliable protein concentration (there are no tryptophan and tyrosine residues in this peptide) made this estimate inaccurate. Nevertheless, the CD spectrum in Figure 3A clearly indicates that the truncated ∆60HDAg is at least partially helical rather than being fully disordered, as previously predicted.
To determine the thermostability of the helical conformation, we studied the thermal unfolding of ∆60HDAg by monitoring the CD signal at 225 nm as a function of temperature. As shown in Figure 3B, the negative CD peak at 225 nm became less pronounced with increasing temperature (10 °C-85 °C), indicating that the protein undergoes thermal melting transition. The signal measured upon cooling of the sample followed a very similar trend with only slightly more positive ellipticity at 225 nm, indicating that the transition is almost completely reversible (> 98%). In contrast to α-helical model peptides, which generally undergo a gradual helix-coil transition upon heating, the melting curve observed for ∆60HDAg has a clear sigmoidal character consistent with a cooperative (two-state) thermal unfolding transition. Indeed, we were able to quantitatively fit the data using the Gibbs-Helmholtz equation, which parametrizes a two-state thermal unfolding transition in terms of melting temperature, Tm, enthalpy change at Tm, ∆H, and heat capacity change, ∆Cp (see Materials and Methods). The value obtained for ∆H, 26 kcal/mol, is almost half of that obtained for the small globular protein erythropoietin, which has a similar amino acid length and is known to undergo cooperative (two-state) unfolding transition.
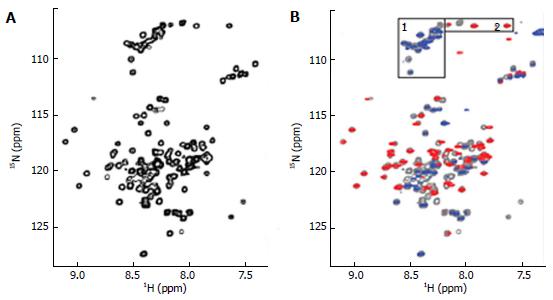
The ∆60HDAg was also analysed by NMR. A 1H-15N heteronuclear single quantum coherence (HSQC) spectrum of purified ∆60HDAg is shown in Figure 4A.
This spectrum served as a “fingerprint” of the protein as it contains a unique cross peak for the backbone NH of each non-proline residue. A peak count revealed that 124 out of a total of 131 non-proline residues (142 residues minus 11 proline residues) gave rise to resolved cross peaks in the HSQC spectrum. However, in contrast to the spectrum of a totally disordered protein, these cross-peaks are distributed over a relatively broad range (7.2-9.2 ppm in 1H and 106-128 ppm in 15N) being consistent with those of a typical globular protein with a dominated helical conformation. Taken together, the results from both CD and NMR suggest that ∆60HDAg is not totally disordered and adopts a non-random, dynamic, ensemble of interconverting conformations.
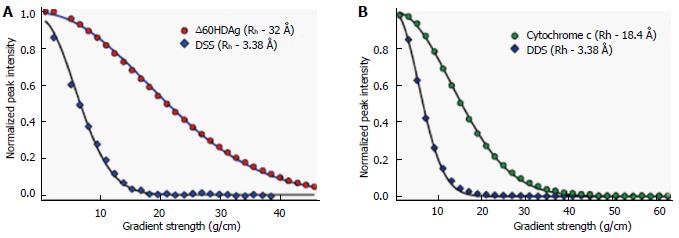
Furthermore, we used a pulsed-field gradient (PFG) diffusion NMR approach to determine the overall dimension of the polypeptide chain. These experiments yield translational diffusion coefficients and, indirectly, the hydrodynamic radius (Rh) of a protein. Figure 5A shows the intensity of a resolved methyl resonance of ∆60HDAg measured in a series of 1D NMR spectra as a function of gradient strength. The chemical shift marker DSS was used as an internal reference.
The effective hydrodynamic radius of DSS was measured in a separate control experiment relative to that of cytochrome c for which an Rh of 17.8 Å has been reported[18]. A similar Rh, 18.4 Å, was measured by dynamic light scattering (data not shown). The Rh we obtained for ∆60HDAg, 32.0 Å, is larger than that expected for a globular (folded) 142-residue protein (20.0 Å), but significantly smaller than the Rh of 37.2 Å expected for a fully unfolded protein of this size.
To identify any (partially) ordered regions within ∆60HDAg, we recorded a steady-state 1H-15N (heteronuclear) Nuclear Overhauser Effect (NOE) spectrum. Heteronuclear NOEs typically are strongly positive for a folded protein and weak or negative for a fully disordered protein[23]. If the protein is partially structured, however, the residues in disordered regions such as flexible loops or chain termini, undergo faster motion than the overall tumbling of the molecules (picosecond to nanosecond time scale). Thus, we expected the NOE peaks for residues in disordered regions to be less intense or of the opposite sign compared to those in structured regions of the protein. The heteronuclear NOE spectrum of ∆60HDAg (Figure 4B) shows 53 positive peaks with chemical shifts distributed over the range from 7.7 to 9.2 ppm in the amide proton dimension, which we assigned to relatively ordered regions of the protein (red contours). Of the remaining 71 residues with resolved peaks in the control HSQC spectrum (gray contours), 50 had negative 1H-15N NOEs (blue contours) and 21 were undetectable. These residues undergo local motion on a time scale that is fast relative to the rotational correlation time that characterizes overall tumbling of the molecule. The NH chemical shifts of these peaks fall within a relative narrow range (7.8-8.6 ppm), and can thus be assigned to residues in relatively disordered regions of the protein. The sequence of ∆60HDAg contains 23 glycines, which are expected to exhibit cross peaks near 8.33 ppm in the proton dimension and 109.1 ppm in the nitrogen dimension of the 1H-15N HSQC spectrum. Interestingly, in the heteronuclear NOE spectrum of ∆60HDAg (Figure 4B), the majority of glycine residues (approximately 20) gave rise to weak or negative peaks (gray or blue contours in box 1) while only three glycine cross-peaks were positive (red contours in box 2). This result indicates that most glycine residues are in disordered regions of the molecule while only three glycines are located in relatively ordered regions. The sequence of ∆60HDAg is especially rich in glycine residues in segments outside the RNA binding domain (RBD, residues 97-146, Figure 1 bottom). The RBD comprises two potential RNA binding motifs, residues 97-107 (KERQDHRRRKA) and 136-146 (EDERRERRIAG). Only 3 out of approximately 53 residues in the RBD are glycines. When Agadir (http://agadir.crg.es/agadir.jsp), an empirical algorithm based on the helix-forming tendencies of peptides, was applied to ∆60HDAg it provided a helix content of 62.2% at pH 7.0 and 5 °C for the sequence containing residues 94-146. The predicted helix content was highly temperature dependent, dropping to 44.7% at 26 °C. After removing the first three residues from the N-terminus, so that the sequence only contained the two RNA binding motifs and the linker (97-146), the estimated helix content dropped to 48.84% at pH 7.0 and 5 °C, and to 34.19% at 25 °C, indicating that the first three residues, FTD, are important for stabilizing the helical conformation.
We also used NetSurf1.1 (http://www.cbs.dtu.dk/services/NetSurfP/) to predict the protein secondary structure. The result showed that the RBD has a high propensity for α-helix formation while random coil structure predominated in all other regions (Figure 1 middle). Furthermore, NetSurf1.1 calculation shows that the RBD contains two helical segments, residues 96-117 and 124-142, respectively, with 30%-90% probability for α-helical structure. The two helical segments are linked by a six-residue, GGKSLS (positions 118-123), loop with low (< 30%) helix propensity.
In a previous work we have shown that full-length S-HDAg can bind a variety of nucleic acids as a multimer[2]. We reported no binding specificity in in vitro conditions, as the full-length S-HDAg was able to bind HDV RNA, non-HDV RNA and DNA from different sources. In the present study we analyzed whether ∆60HDAg is still capable of binding nucleic acids despite of the absence of the N-terminal CCD domain, which is responsible for antigen oligomerization.
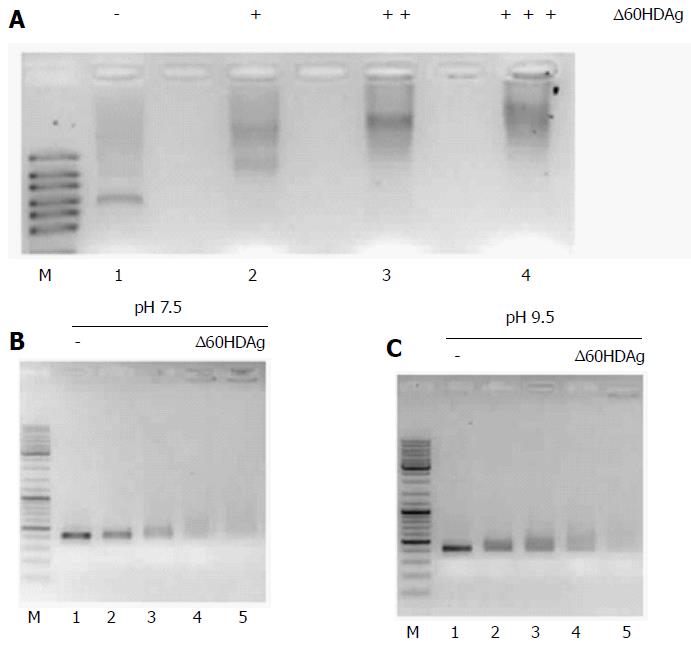
We first assessed binding of ∆60HDAg to RNA. In vitro synthetized HDV RNA was incubated with increasing amounts of recombinant ∆60HDAg and samples were resolved in gel retardation assays. Change in mobility of the complexes was readily observed even with the lowest concentration of ∆60HDAg used and responsive to increasing amounts of recombinant truncated protein (Figure 6A), indicating that ∆60HDAg retains the capacity of binding to RNA.
Then, we analyzed binding of ∆60HDAg to DNA. A 400-nucleotide PCR product was incubated with increasing amounts of recombinant ∆60HDAg and samples were resolved in gel retardation assays. Since ∆60HDAg has an estimated pI of 9.84 based on its amino acid composition, we performed this assay at pH 7.5 but also at pH 9.5, closer to ∆60HDAg pI. Thus, it is expected that at higher pH the net charge of S-HDAg is closer to neutral than at pH 7.5. Increasing amounts of ∆60HDAg led to a marked decrease in the amount of free dsDNA molecules in both conditions (Figure 6B and C), indicative of ∆60HDAg-dsDNA complexes formation. We conclude that, similarly to what was observed with RNA, ∆60HDAg has also the capacity to bind dsDNA molecules and that binding is not dependent on the charge of the antigen.
S-HDAg is essential for HDV genomic RNA accumulation in infected cells and plays a crucial role in the replication cycle of the virus. This 195 amino acid protein is highly promiscuous as it not only binds to HDV RNA to form viral ribonucleoproteins, but also interacts with a myriad of host factors[4-6]. S-HDAg is predicted to be an IDP, being consistent with many of its known characteristics, such as high net positive charge, ability to bind several partners and lack of enzymatic activity[2]. The high level of intrinsic disorder found in S-HDAg can explain the difficulties encountered to determine its structure. So far, only a peptide spanning the first 60 amino acids has been crystallized and its structure determined[14]. Here, we focused on a truncated version of S-HDAg, consisting of amino acids 61 through 195, lacking the CCD involved in S-HDAg multimerization. Surprisingly, our structural studies show that rather than being fully disordered, ∆60HDAg adopts a relatively compact ensemble of interconverting conformations with a partially ordered RBD. Our results show that it is possible to discriminate between ordered and disordered regions of the protein by using CD, NMR, and secondary structure prediction, without the need for laborious sequence-specific NMR resonance assignments. We show that the RNA binding domain of S-HDAg adopts a dynamic helical conformation.
The N-terminal CCD motif of S-HDAg is involved in oligomerization of the full-length protein. Consistently, cross-linking experiments with ∆60HDAg show that the truncated protein is unable to form homomultimers in vitro. We also show that ∆60HDAg does not interfere with multimerization of full-length S-HDAg and that multimerization of ∆60HDAg is not enhanced by the presence of full-length S-HDAg. Results from cross-linking, and specially NMR PFG diffusion measurements, show that overall dimensions are larger than those expected for a globular protein but smaller than those of fully unfolded (random coil) protein of this size. Thus, ∆60HDAg is a monomer under the experimental conditions used.
Similar to what has been previously shown for full-length S-HDAg, nucleic acid binding results show that ∆60HDAg can bind not only RNA but also dsDNA. Incubation of ∆60HDAg with in vitro transcribed RNA and dsDNA and further analysis of complex formation followed by electrophoretic mobility shift assays shows that this truncated protein, although failing to oligomerize, still displays nucleic acid binding activity. In vitro nucleic acid binding seems to be largely unspecific as it binds RNA and DNA molecules used in this study and RNA and DNA from other sources with unrelated sequences (data not shown). Noteworthy, when analyzing nucleic acid interactions with full-length S-HDAg, there was a clear shift from the unbound to the fully-bound state without any intermediate positions[2], which most likely reflects binding of S-HDAg multimers covering the whole sequence.
Earlier reported findings, using a different approach, suggested that S-HDAg specifically binds HDV RNAs with rod-like folding[24]. Moreover, S-HDAg RNA-binding specificity has also been recently reported when a C-terminal deletion mutant of the antigen was used and with requirements that the RNA must have a minimum of approximately 300 nt of rod-like folding for binding to occur[25]. Interestingly, this report also cited that in studies with full length S-HDAg no specificity for nucleic acid binding was found. Thus, one could suggest that some level of intrinsic disorder in the C-terminus may compromise specific binding to HDV RNAs. Furthermore, more recently it was shown that binding of HDAg to HDV RNA is not sequence specific but rather depends on secondary structure features, internal loops and bulges of the nucleic acid[26].
To get a deeper insight into the non-specific nucleic acid binding ability of the protein, structural information is also required. We used CD and NMR to characterize measurable structural features in ∆60HDAg. Disordered protein regions with a measured propensity for helical secondary structure have been found to act as preformed molecular recognition elements. Although ∆60HDAg is predicted to be extensively disordered, the relative large chemical shift dispersion in both proton and nitrogen in NMR HSQC spectrum indicates that this is not the case. Moreover, the CD spectrum further showed that ∆60HDAg has a measurable helical content. In conjunction with the sequence analysis, the heteronuclear NOE NMR experiment showed that the RBD domain contained a dynamic helical conformation, which is consistent with secondary structure prediction of a helix-turn-helix RNA binding motif. In addition, our study shows that qualitative NMR analysis such as 1H-15N HSQC, heteronuclear NOE, and PFG diffusion measurements, without need for laborious sequence-specific resonance assignments, can provide insight into structural and dynamic properties, even for disordered proteins. Such sequence-specific resonance assignments are usually a challenge as a consequence of peak overlap and live-width broadening due to conformational exchange observed in the spectra.
Accumulated data indicate that disordered regions in proteins are a common feature and may give rise to important properties as plasticity and reversibility in regulatory intermolecular interactions with their targets, such as proteins and nucleic acids. The mediation of IDR to binding can be achieved through interaction with binding domains and stabilizing their dynamic local structure upon interaction with their targets. Such regulatory functions are frequently modulated by post-translational modification in IDRs. These PTMs, namely phosphorylation, methylation, acetylation, and sumoylation, were reported for S-HDAg in different motifs, including in disordered regions of the protein[9-13]. Furthermore, modifications of S-HDAg are known to mediate the subcellular localization of S-HDAg thus facilitating its interaction with a broad number of cellular targets including the enzymatic machinery involved in the different steps of virus replication. Some of these modifications, namely phosphorylation, were reported to play important roles in the virus replication cycle, including in the accumulation of virus RNAs[11,12]. In addition, the plasticity of S-HDAg may form the basis of interaction with a vast number of cellular partners, inhibiting, redirecting or accelerating host metabolic functions contributing to promote more acute and adverse forms of the liver disease caused by HDV.
In conclusion, the information obtained in this study provides structural basis for future understanding of the non-selective nucleic acid binding property of S-HDAg.
The present in vitro studies show that a truncated form of the Delta antigen no longer multimerizes but still binds nucleic acids, although without specificity for HDV rodlike RNA. The lack of specificity may partially be due to an electrostatic interaction between the positively charged protein and the negatively charged nucleic acids, and mediated by the disordered regions. However, the antigen is extensively phosphorylated in vivo, which likely reduces its high net charge, limiting the ability to be involved in non-specific electrostatic interactions[9].
These results pave the way for more detailed future studies of structural properties of S-HDAg and its interactions with nucleic acids and other cellular partners.
Hepatitis delta virus small antigen (S-HDAg) is predicted to be an intrinsically disordered protein that interacts with multiple cellular targets and plays a crucial role in the virus replication cycle.
Intrinsically disordered proteins exhibit high plasticity, and like S-HDAg, rarely display enzymatic activity and are often involved in nucleic acid binding. It is well established that S-HDAg is necessary for accumulation of virus RNAs in infected cells, but its structure and precise role in hepatitis delta virus (HDV) replication cycle are still largely unknown.
The authors made use of circular dichroism and nuclear magnetic resonance NMR, as well as gel retardation assays to study a truncated form of S-HDAg, lacking the first 60 amino acids, that contain the dimerization and higher order multimerization domain (Δ60HDAg). The authors concluded that Δ60HDAg is intrinsically disordered but compact and is not a multimer under the experimental conditions used in this study. Moreover, Δ60HDAg is still capable of nucleic acid binding although without apparent specificity.
This study provides a structural basis for future understanding of the non-selective nucleic acid binding property of S-HDAg. Furthermore, it opens the way for more in-depth future investigations of structural properties of S-HDAg and its interactions with nucleic acids and other cellular partners.
IDP: Intrinsically disordered proteins lack an ordered conformation. They may range from fully unstructured to partially structured including some well characterized domains like random coils.
The manuscript by Alves et al describes the characteristics of the C-terminal region of S-HDAg using a truncated form of this protein. The methods used in this paper are straightforward.
Manuscript source: Invited manuscript
Specialty type: Virology
Country of origin: Portugal
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B, B, B, B
Grade C (Good): 0
Grade D (Fair): D
Grade E (Poor): 0
P- Reviewer: Chung YH, Chen CJ, Rodriguez-Frias F, Rossi LMG, Taylor JM S- Editor: Ji FF L- Editor: A E- Editor: Wu HL
| 1. | Rizzetto M. Hepatitis D: thirty years after. J Hepatol. 2009;50:1043-1050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 152] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 2. | Alves C, Cheng H, Roder H, Taylor J. Intrinsic disorder and oligomerization of the hepatitis delta virus antigen. Virology. 2010;407:333-340. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 3. | Uversky VN. Intrinsically disordered proteins from A to Z. Int J Biochem Cell Biol. 2011;43:1090-1103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 313] [Cited by in RCA: 337] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 4. | Chang MF, Baker SC, Soe LH, Kamahora T, Keck JG, Makino S, Govindarajan S, Lai MM. Human hepatitis delta antigen is a nuclear phosphoprotein with RNA-binding activity. J Virol. 1988;62:2403-2410. [PubMed] |
| 5. | Greco-Stewart V, Pelchat M. Interaction of host cellular proteins with components of the hepatitis delta virus. Viruses. 2010;2:189-212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 6. | Casaca A, Fardilha M, da Cruz e Silva E, Cunha C. The heterogeneous ribonuclear protein C interacts with the hepatitis delta virus small antigen. Virol J. 2011;8:358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 7. | Cao D, Haussecker D, Huang Y, Kay MA. Combined proteomic-RNAi screen for host factors involved in human hepatitis delta virus replication. RNA. 2009;15:1971-1979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 8. | Kuo MY, Goldberg J, Coates L, Mason W, Gerin J, Taylor J. Molecular cloning of hepatitis delta virus RNA from an infected woodchuck liver: sequence, structure, and applications. J Virol. 1988;62:1855-1861. [PubMed] |
| 9. | Mu JJ, Wu HL, Chiang BL, Chang RP, Chen DS, Chen PJ. Characterization of the phosphorylated forms and the phosphorylated residues of hepatitis delta virus delta antigens. J Virol. 1999;73:10540-10545. [PubMed] |
| 10. | Chen YS, Huang WH, Hong SY, Tsay YG, Chen PJ. ERK1/2-mediated phosphorylation of small hepatitis delta antigen at serine 177 enhances hepatitis delta virus antigenomic RNA replication. J Virol. 2008;82:9345-9358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 11. | Tseng CH, Jeng KS, Lai MM. Transcription of subgenomic mRNA of hepatitis delta virus requires a modified hepatitis delta antigen that is distinct from antigenomic RNA synthesis. J Virol. 2008;82:9409-9416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 12. | Mu JJ, Tsay YG, Juan LJ, Fu TF, Huang WH, Chen DS, Chen PJ. The small delta antigen of hepatitis delta virus is an acetylated protein and acetylation of lysine 72 may influence its cellular localization and viral RNA synthesis. Virology. 2004;319:60-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 51] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 13. | Li YJ, Stallcup MR, Lai MM. Hepatitis delta virus antigen is methylated at arginine residues, and methylation regulates subcellular localization and RNA replication. J Virol. 2004;78:13325-13334. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 70] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 14. | Zuccola HJ, Rozzelle JE, Lemon SM, Erickson BW, Hogle JM. Structural basis of the oligomerization of hepatitis delta antigen. Structure. 1998;6:821-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 73] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 15. | Dingle K, Bichko V, Zuccola H, Hogle J, Taylor J. Initiation of hepatitis delta virus genome replication. J Virol. 1998;72:4783-4788. [PubMed] |
| 16. | Lazinski DW, Taylor JM. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J Virol. 1994;68:2879-2888. [PubMed] |
| 17. | Wu DH, Johnson CS. Diffusion-Ordered 2d Nmr in the Fringe-Field of a Superconducting Magnet. J Magn Reson, Ser A. 1995;116:270-272. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 18. | Wilkins DK, Grimshaw SB, Receveur V, Dobson CM, Jones JA, Smith LJ. Hydrodynamic radii of native and denatured proteins measured by pulse field gradient NMR techniques. Biochemistry. 1999;38:16424-16431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 795] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 19. | Bergmann KF, Pohl C, Gerin JL. Characterization of proteins of hepatitis delta virus. Prog Clin Biol Res. 1987;234:105-110. [PubMed] |
| 20. | Gerlich WH, Heermann KH, Ponzetto A, Crivelli O, Bonino F. Proteins of hepatitis delta virus. Prog Clin Biol Res. 1987;234:97-103. [PubMed] |
| 21. | Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53 Suppl 6:566-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 337] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 22. | Johnson WC. Secondary structure of proteins through circular dichroism spectroscopy. Annu Rev Biophys Biophys Chem. 1988;17:145-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 458] [Cited by in RCA: 415] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 23. | Akke M, Carr PA, Palmer AG. Heteronuclear-correlation NMR spectroscopy with simultaneous isotope filtration, quadrature detection, and sensitivity enhancement using z rotations. J Magn Reson B. 1994;104:298-302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Chao M, Hsieh SY, Taylor J. The antigen of hepatitis delta virus: examination of in vitro RNA-binding specificity. J Virol. 1991;65:4057-4062. [PubMed] |
| 25. | Defenbaugh DA, Johnson M, Chen R, Zheng YY, Casey JL. Hepatitis delta antigen requires a minimum length of the hepatitis delta virus unbranched rod RNA structure for binding. J Virol. 2009;83:4548-4556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 26. | Griffin BL, Chasovskikh S, Dritschilo A, Casey JL. Hepatitis delta antigen requires a flexible quasi-double-stranded RNA structure to bind and condense hepatitis delta virus RNA in a ribonucleoprotein complex. J Virol. 2014;88:7402-7411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 2.3] [Reference Citation Analysis (0)] |