Peer-review started: June 27, 2016
First decision: September 5, 2016
Revised: October 9, 2016
Accepted: November 27, 2016
Article in press: November 29, 2016
Published online: February 12, 2017
Processing time: 224 Days and 20.6 Hours
To test whether a simple animal, Caenorhabditis elegans (C. elegans), can be used as an alternative model to study the interaction between hepatitis B virus antigens (HBsAg) and host factors.
Three plasmids that were able to express the large, middle and small forms of HBsAgs (LHBsAg, MHBsAg, and SHBsAg, respectively) driven by a ubiquitous promoter (fib-1) and three that were able to express SHBsAg driven by different tissue-specific promoters were constructed and microinjected into worms. The brood size, egg-laying rate, and gonad development of transgenic worms were analyzed using microscopy. Levels of mRNA related to endoplasmic reticulum stress, enpl-1, hsp-4, pdi-3 and xbp-1, were determined using reverse transcription polymerase reaction (RT-PCRs) in three lines of transgenic worms and dithiothreitol (DTT)-treated wild-type worms.
Severe defects in egg-laying, decreases in brood size, and gonad retardation were observed in transgenic worms expressing SHBsAg whereas moderate defects were observed in transgenic worms expressing LHBsAg and MHBsAg. RT-PCR analysis revealed that enpl-1, hsp-4 and pdi-3 transcripts were significantly elevated in worms expressing LHBsAg and MHBsAg and in wild-type worms pretreated with DTT. By contrast, only pdi-3 was increased in worms expressing SHBsAg. To further determine which tissue expressing SHBsAg could induce gonad retardation, we substituted the fib-1 promoter with three tissue-specific promoters (myo-2 for the pharynx, est-1 for the intestines and mec-7 for the neurons) and generated corresponding transgenic animals. Moderate defective phenotypes were observed in worms expressing SHBsAg in the pharynx and intestines but not in worms expressing SHBsAg in the neurons, suggesting that the secreted SHBsAg may trigger a cross-talk signal between the digestive track and the gonad resulting in defective phenotypes.
Ectopic expression of three forms of HBsAg that causes recognizable phenotypes in transgenic worms suggests that C. elegans can be used as an alternative model for studying virus-host interactions because the resulting phenotype is easily detected through microscopy.
Core tip: In the past, mouse and cell culture models have been used for studying the effects of hepatitis B virus antigens (HBsAg) on hosts. Both models have advantages and disadvantages in terms of economic and time concerns. In this study, we provide an alternative animal model, Caenorhabditis elegans (C. elegans), to demonstrate that SHBsAg can induce observable phenotypes which has never been reported in mouse and cell culture models. We suggest that C. elegans can serve as a new plateform for studying various viral pathogenesis.
- Citation: Chen YY, Lee LW, Hong WN, Lo SJ. Expression of hepatitis B virus surface antigens induces defective gonad phenotypes in Caenorhabditis elegans. World J Virol 2017; 6(1): 17-25
- URL: https://www.wjgnet.com/2220-3249/full/v6/i1/17.htm
- DOI: https://dx.doi.org/10.5501/wjv.v6.i1.17
Human hepatitis B virus (HBV), a member of the family Hepadnaviride, is a partially double-stranded DNA virus. The genome of HBV contains approximately 3200 nucleotides that encodes four open reading frames, namely surface (S), core (C), polymerase (P) and X in an overlapping but frame-shifted manner[1-3]. Infection with HBV inducers a broad range of clinical outcomes, from asymptomatic hepatitis to fulminant hepatitis. Chronic hepatitis B carriers are highly associated with the development of liver cirrhosis and hepatocellular carcinoma[4,5]. Molecular biology analyses from tumor samples have revealed that HBV DNA integration could activate genes associated with the cell cycles, leading to abnormal cell proliferation[6,7]. Pathogenesis and etiology studies have found that X and truncated preS proteins play oncogenic roles[8-10].
Woodchucks were the first animal model for studying liver carcinogenesis caused by natural woodchuck hepatitis virus (WHV) infection[11,12]. A high incidence of liver tumor formation occurs in newborn woodchucks infected with WHV. Molecular dissection has revealed that c-myc oncogene level are highly elevated in liver tumors[13,14]. Later, the transgenic mouse model was applied to express an individual viral protein, such as the large hepatitis B surface antigen (LHBsAg) and X protein, which are driven by the albumin promoter for specific expression in the liver, to study the mechanisms of liver carcinogenesis induced by viral proteins[15,16]. In combining molecular biology analyses of the HBV X (HBx) gene in transfected cells, numerous studies have elucidated that the X protein is multifunctional and induces transactivation activity, signal transduction and cell death[9,17,18]. Recently, Geng et al[19] employed Caenorhabditis elegans (C. elegans), a soil nematode, to express HBx under a heat shock control and found that HBx induced cell apoptosis and necrosis through the interaction of HBx and CED-9, a human homolog of Bcl-2.
C. elegans was first used as a model organism for studying development and the nervous system because the species is transparent throughout its life span and in its adult form, possesses approximately 300 neurons out of 1000 somatic cells[20,21]. Because of its short-life cycle, simplicity, numerous available mutated forms, and ease of handling for knocking-down specific gene, C. elegans is now a model for studying various biological topics, such as aging, human diseases, host-pathogen interaction and viral pathogenesis[22-25]. Because of a high percentage of genes in numerous cellular pathways is conserved across nematodes to vertebrates, a study of PEG-mediated Poxviridae infection in C. elegans revealed that the core genes of apoptosis (ced-3 and ced-4) control vaccinia virus replication in worms[26,27]. Therefore, C. elegans could serve as a new platform for virologists to study virus-host interaction and pathogenesis in addition to the currently used cell culture and mammalian models. In this study, we expressed three forms of HBsAg in C. elegans to determine different degrees of defects in gonad development.
Pfib-1::gfp::icr::SHBsAg: A 1.5 kb fragment excised from the Pfib-1::gfp::LD plasmid[28] by cutting with HindIII and AgeI and was isolated and then inserted into the HindIII and AgeI sites of pPD95.75 to generate Pfib-1::gfp. Pfib-1::gfp was then cut with EcoRI and ligated with a 0.8 kb of icr::SHBsAg fragment which was isolated from Pfib-1::LD::icr::SHBsAg to generate a 6.8 kb of Pfib-1::gfp::icr::SHBsAg. When microinjection of Pfib-1::gfp::icr::SHBsAg into N2 strain, worms expressed green fluorescence proteins (GFP) and HBV small surface antigens (SHBsAg).
Pfib-1::gfp::icr::linker: A linker was designed to contain EcoRI, NotI , BglII, SalI , NsiI and SacI cutting sites. This linker was ligated to Pfib-1::gfp and generated plasmid Pfib-1::gfp::linker. Pfib-1::gfp::linker was cut with EcoRI and NotI and ligated with an icr fragment which was isolated from Pfib-1::gfp::icr::SHBsAg to generate a 6.2 kb of Pfib-1::gfp::icr::linker plasmid.
Pfib-1::gfp::icr::MHBsAg: A 680 bp of MHBsAg DNA fragment was amplified from pMH3/3097[29] using primers HBVs(M)-NotI-F and HBVs-SalI-R. This fragment was ligated with Pfib-1::gfp::icr::linker to generate a 7.0 kb of Pfib-1::gfp::icr::MHBsAg. Transgenic worm carrying Pfib-1::gfp::icr::MHBsAg expressed GFP and MHBsAg.
Pfib-1::gfp::icr::LHBsAg: A 1.1 kb of LHBsAg DNA fragment was amplified from pMH3/3097[29] using primers HBVs(L)-NotI-F and HBVs-SalI-R. This fragment was ligated with Pfib-1::gfp::icr::linker to generate a 7.3 kb of Pfib-1::gfp::icr::LHBsAg. When microinjection of Pfib-1::gfp::icr::LHBsAg into N2 strain, worms expressed GFP and LHBsAg.
Pmyo-2::gfp::icr::SHBsAg: The fragment of fib-1 promoter was cut from Pfib-1GFP-icr-SHBsAg by digestion with HindIII and AgeI, and replaced with the myo-2 promoter which was isolated from Pmyo-2::gfp::icr::DsRed::LD[28] to create a 6.6 kb of Pmyo-2::gfp::icr::SHBsAg. Transgenic worms carrying this plasmid expressed both GFP and SHBsAg in pharynx.
Pmec-7::gfp::icr::SHBsAg: The plasmid was generated by substitution of the fib-1 promoter of Pfib-1::gfp::icr::SHBsAg with the mec-7 promoter which was isolated from Pmec-7::gfp::icr::DsRed::LD[28] to create a 6.3 kb of Pmec-7::GFP::icr::SHBsAg. Transgenic worms carrying this plasmid expressed GFP and SHBsAg in neurons.
Pges-1::gfp::icr::SHBsAg: The plasmid was generated by substitution of the fib-1 promoter of Pfib-1::gfp::icr::SHBsAg with the ges-1 promoter which was isolated from Pges-1::gfp::icr::DsRed::LD[28] to create a 6.6 kb of Pges-1::gfp::icr::SHBsAg. Transgenic worms carrying this plasmid expressed GFP and SHBsAg in intestinal cells.
For plasmid constructions and RT-PCR analyses, the following paired primers were used: linker-F: 5’-aat tcaaaaagcggccgcagatctgtcgacatgcatgagctc-3’; linker-R: 5’-gtttttcgccggcgtctagacagctgtacg tactcgagttaa-3’; HBVs(L)+NotI-F: 5’-gggaacaagag cggccgcatggggcag-3’; HBVs(M)+NotI-F: 5’-acactc atcgcggccgcatgcagtgg-3’; HBVs+SalI-R: 5’-gttttgtgtcgacttaaatgtataccc-3’; eft-2-F: 5’-ggtggtcaaatcatcccaac-3’; eft-2-R: 5’-tcctcgaaaacgtgtcctct-3’; endoplasmin-F: 5’-t gaaaa cctccaacagcaca-3’; endoplasmin-R: 5’-gcagtttccttg agccagtc-3’; hsp-4-F: 5’-ttttcgaggttcttgccact-3’; hsp-4-R: 5’-tctccggtattttcgacacc-3’; PDI-F: 5’-gccgtttcca aca aagaaga-3’; PDI-R: 5’-cccttgagc ccatcagtaga-3’; xbp-1-F: 5’-cgtcgtcta cgaagaagaagtcgtc-3’; xbp-1-R: 5’-gatg ata gttagatacatatccacactg-3’.
N2 (wild-type) worm was obtained from the Caenorhabditis Genetics Center (CGC, University of Minnesota) and cultured on Nematode Growth Medium (NGM) following standard methods[20]. Images of transgenic worms were acquired using Leica DM2500 equipped with CoolSNAP K4 (photometrics) and processed with a MetaMorph (version 6.1).
Plasmid DNA was prepared by using QIAprep spin miniprep kit and the concentration was adjusted at 100 ng/μL in injection buffer (20 mmol/L potassium phosphate, pH 7.5, 3 mmol/L potassium citrate, pH 7.5, 2% polyethylene glycol, M.W. 6000). The injection mixture also contained pRF-4 which was included as a screening marker. Worm was placed onto 2% agarose pads and injected by capillary needle loaded with DNA mixture using a FemtoJet system (Eppendorf AG, Hamburg, Germany). The glass capillaries were purchased from World Precision Instruments (Kwik-Fil™, borosilicate 16 glass capillaries, item number 1B100F-6, United States) and pull by Flaming/Brown micropipette puller (MODEL P-97, Sutter Instrument Co., United States).
Worms were first synchronized and placed one worm per a single plate. The offspring in each plate were counted every day.
For visualization of GFP expression in transgenic worms, an upright fluorescence microscope (Leica DM2500) was used. For visualization of gonad structure and development a differential interference contrast (DIC) microscope was used and images were captured using a cool CCD (CoolSNAP K4).
The total RNA was extracted from transgenic worms expressed both GFP and HBsAgs with TRIzol reagent. The reverse transcription reaction was first carried out with 4 μg of RNA, 2 μL of dNTP (10 mmol/L), 2 μL of Oligo-dT (10 mmol/L), and added DEPC H2O to 12 μL. After incubated at 68 °C for 5 min, the mixture was then added 4 μL of 5 × first-strand buffer (invitrogen), 2 μL of DTT (0.1 mmol/L, invitrogen), 1 μL of RNase inhibitor (invitrogen) and 1 μL of Reverse Transcriptase (invitrogen) and incubated at 42 °C for 50 min, and then 70 °C for 15 min. The primers used in PCR analyses were listed as above.
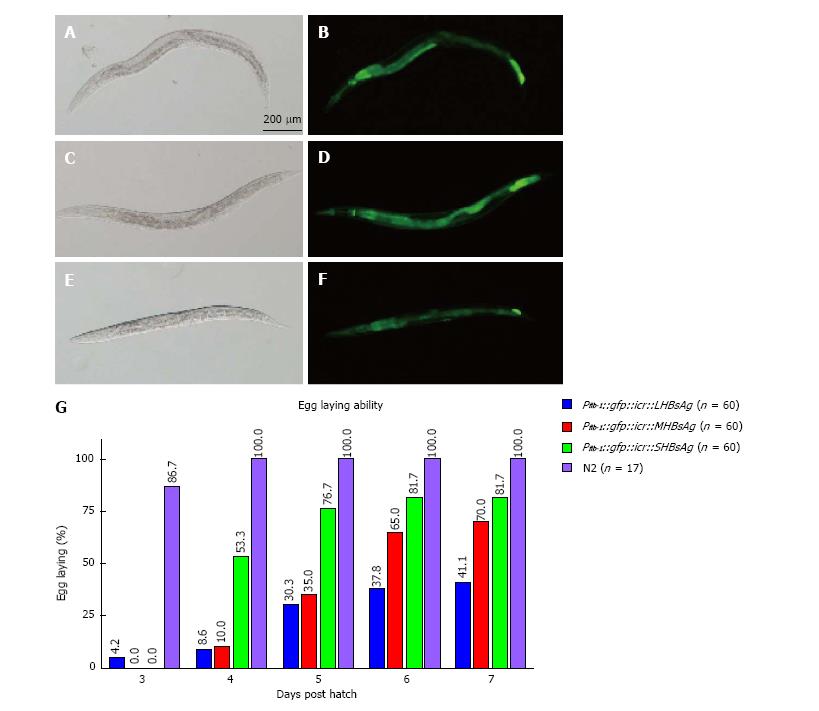
To determine whether C. elegans can be a new platform for studying virus-host interaction we ectopically expressed three lengths of HBsAg (SHBsAg, MHBsAg, and LHBsAg) in worms under the control of the ubiquitous promoter fibrillarin (fib-1). Three HBsAg gene sequences were individually placed in a bicistronic vector behind a reporter gene, green fluorescence protein (GFP), which was used as a selection marker[30]. Transgenic worms were selected for the expression of GFP (Figure 1A-F) and maintained to characterize phenotypes. After synchronization, transgenic worms were singled out and placed on single plates, and the numbers of eggs produced by individual worms were counted every day. The results showed different egg-laying averages in a total of 60 transgenic animals in three groups expressing SHBsAg, MHBsAg, and LHBsAg (Figure 1G). On the fourth day after hatching, the wild-type worms (N2) displayed an egg-laying rate of 100% whereas those worms expressing SHBsAg, MHBsAg, and LHBsAg demonstrated laying-egg rates of approximately 8.6%, 10%, and 53.3%, respectively. Although the egg-laying rates of the three lines of transgenic worms increased in the following days, the maximum egg-laying rate was 41.1% for worms expressing SHBsAg, 70% for worms expressing MHBsAg and 81.7% for worms expressing LHBsAg at 7 d post-hatching (Figure 1B). The reduced rate of egg-laying in the three lines of HBsAg-expressing worms was unlikely to have caused by the ectopic expression of GFP because worms carrying a plasmid with the sole function of expressing GFP throughout the body exhibited egg-laying capability of 100% (Table 1).
| Construct | Strain | Ecotopic proteins | Protein expression site | Egg-laying ability (%) | Brood size |
| N2 | 100 (n = 17) | 290 ± 15 (n = 17) | |||
| Pfib-1::gfp::icr | N2 | GFP | Whole worm | 100 (n = 10) | 268 ± 29 (n = 18) |
| Pfib-1::gfp::icr::SHBsAg | N2 | GFP, SHBsAg | Whole worm | 9 (n = 60) | 66 ± 15 (n = 14) |
| Pfib-1::gfp::icr::MHBsAg | N2 | GFP, MHBsAg | Whole worm | 10 (n = 60) | 175 ± 50 (n = 15) |
| Pfib-1::gfp::icr::LHBsAg | N2 | GFP, LHBsAg | Whole worm | 54 (n = 60) | 239 ± 14 (n = 15) |
| Pmyo-2::gfp::icr::SHBsAg | N2 | GFP, SHBsAg | Pharynx | 83 (n = 60) | 163 ± 20 (n = 26) |
| Pges-1::gfp::icr::SHBsAg | N2 | GFP, SHBsAg | Intestine | 97 (n = 60) | 203 ± 50 (n = 32) |
| Pmec-7::gfp::icr::SHBsAg | N2 | GFP, SHBsAg | Neuron | 100 (n = 60) | 270 ± 42 (n = 26) |
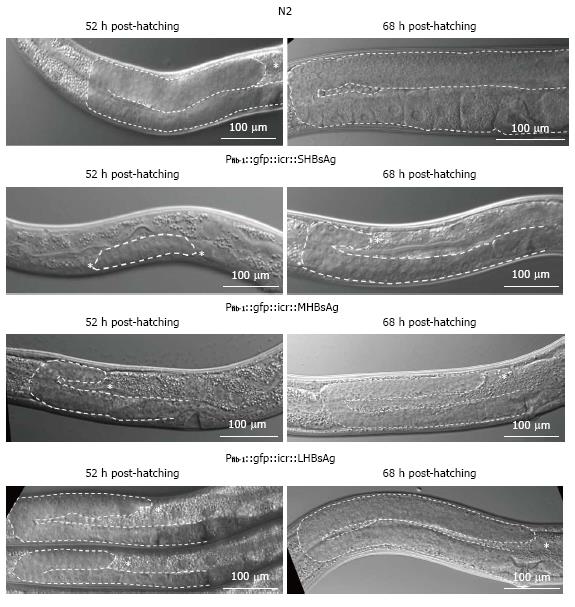
To understand why the expression of various lengths of HBsAg in transgenic worms caused a reduction in egg-laying capability, we examined the gonad development of the three types of transgenic worms under a DIC microscope. As shown in Figure 2 (upper two rows), at 52 h after hatching, wild-type worms had nearly completed the gonad development; by contrast, worms expressing SHBsAg exhibited a dramatic retardation of gonad development in larval stage 3 (L3). The process of oogenesis was observed in N2 worms 68 h post hatching whereas the gonads of worms expressing SHBsAg were only just beginning to turn as mid-stage of larva 4 as 72 h post-hatching. Oogenesis was observed in some worms expressing SHBsAg until 96 h post-hatching (data not shown). Worms expressing MHBsAg and LHBsAg at 52 h and 68 h post-hatching showed a retardation of gonad development that was less severe than that observed in worms expressing SHBsAg at a similar stage (Figure 2, lower two rows). The severity of gonad retardation clearly reflected the reduced percentage of egg-laying (9%, 10%, and 54%, respectively) and average brood size (66, 175, and 239, respectively) in the three lines of transgenic worms, as shown in Table 1.
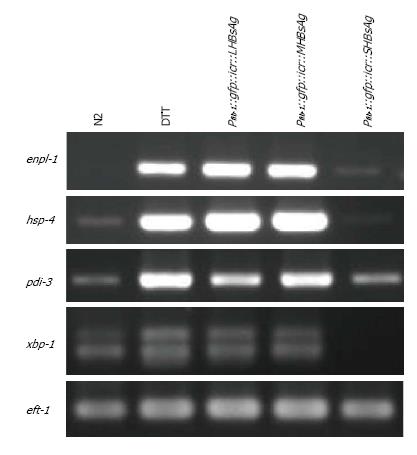
A previous study reported that endoplasmic reticulum (ER) stress could cause retardation of gonad development[31]. To determine whether the defective phenotypes of the three lines of transgenic worms resulted from ER stress, we performed RT-PCR analysis of ER stress markers. Total RNA from the three lines of transgenic worms and N2 worms with or without DTT treatment was isolated and analyzed for the expression levels of enpl-1, hsp-4, pdi-3 and xbp-1 through RT-PCR. The results of the gel-electrophoresis of RT-PCR products indicated that the levels of enpl-1, hsp-4 and pdi-3 were substantially elevated in worms expressing LHBsAg and MHBsAg, being similar to those N2 worms pretreated with the ER stress-inducer DTT whereas the xbp-1 level had increased only slightly (Figure 3). Only a slight increase in the level of pdi-3 was observed in worms expressing SHBsAg compared with that of wild-type worms, and no obvious elevation of other ER stress-related transcripts was detected. We concluded that the defective phenotypes caused by the expression of LHBsAg and MHBsAg were likely attributable to ER stress signals. By contrast, the defect induced by the expression of SHBsAg might have been caused by other unknown pathways.
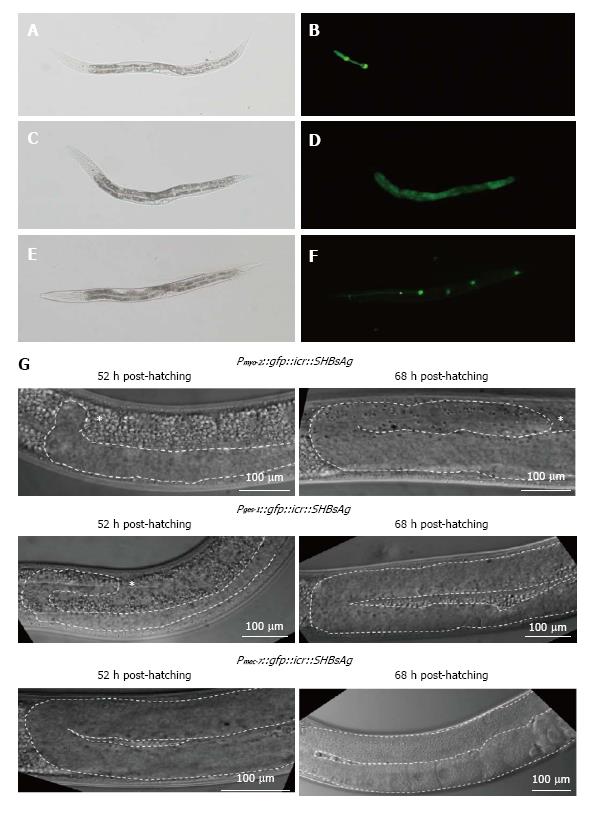
To determine which tissues expressing SHBsAg were responsible for gonad retardation, we substituted the fib-1 promoter with myo-2, ges-1, and mec-7, to express SHBsAg in the pharynx, intestines, and neurons, respectively. Transgenic worms expressing GFP in the pharynx, intestines, and neurons were selected (Figure 4A) and analyzed for egg-laying capability and gonad development. As shown in Table 1, 100% and 97% egg-laying capability were found in worms expressing SHBsAg in the neurons and intestines, respectively, compared with 83% egg-laying capability in worms expressing SHBsAg through myo-2. When examining the gonad development of the three lines of transgenic worms using DIC, we found no obvious retardation in worms expressing SHBsAg in neuron at 52 h and 68 h post-hatching and a moderate level of gonad retardation in worms expressing SHBsAg in the pharynx and intestines (Figure 4B). This suggested that cross-talk between the digestive and reproductive system may be triggered by secreted SHBsAg, causing defects in a small portion of the population.
In this study, we demonstrated that transgenic worms expressing three forms of HBsAgs throughout the body exhibited lower rates of egg-laying, reduced brood sizes and retardation of gonad development to various degrees. Unexpectedly, worms expressing SHBsAg displayed the most severe defects (Table 1). No study has yet reported that the expression of SHBsAg can induce detectable phenotypes in cultured cells or animals; however, ER-stress and tumor formation have been observed in cells and animals expressing LHBsAg and MHBsAg[16,32]. Consistent with previous studies, worms expressing LHBsAg and MHBsAg were found to possess higher levels of enpl-1, hsp-4, pdi-3 and xbp-1 transcripts as did N2 worms pretreated with DTT (Figure 3). Because the gonad is the organ most sensitive to environmental changes[33], we suggest that ER-stress signal occurring autonomously or non-autonomously in the gonad can lead to gonad retardation, a reduced rate of egg-laying, and a smaller brood sizes in transgenic worms expressing LHBsAg and MHBsAg.
The unexpected results of the most severe phenotypes induced by the expression of SHBsAg might be explained by the different nature of the three forms of HBsAg. In general, SHBsAg can form subviral particles of approximately 22 nm and be constantly secreted outside of cells whereas MHBsAg is less efficiently secreted and LHBsAg is usually retained in the ER[29,32]. This hypothesis is supported by the results shown in Figure 3, namely that four ER-stress related transcripts (enpl-1, hsp-4, pdi-3 and xbp-1) were substantially elevated in worms expressing LHBsAg and MHBsAg but only one transcript (pdi-3) displayed a slight elevation in worms expressing SHBsAg. The secretion of SHBsAg might either trigger signals inhibiting gonad development or titrate out secretion factors that are required for gonad development, although ER-stress signals might also play a minor role (Figure 4). Nevertheless, the underlying mechanism that leads to the most severe phenotypes in worms expressing SHBsAg remains unknown and will be elucidated by performing rescue and genetic cross experiments in the future.
C. elegans has been used for studying viral pathogenesis and virus-host interaction for more than a decade[27,34]. In comparison with the number of publications using C. elegans study viral, bacterial and fungal pathogenesis, relatively few papers have focused on viral pathogenesis and virus-host interaction in the past 10 years. The bottleneck could be due to the difficulty of creating transgenic worms expressing viral antigens. Currently, two methods for delivering ectopic genes into C. elegans are microinjection and gene bombardment, neither of which are easily achievable in general biology laboratories. To use C. elegans as a platform for studying virus-host interaction, virologists must collaborate with worm scientists. Alternatively, virologists could engineer the three viruses (Orsay, Santeui, and Le Blanc virus[35,36]), that naturally infect C. elegans to become versatile vectors for the easy deliver of different viral genes into worms through infection.
We would like to thank Wallace Academic Editing for editing the manuscript, the Caenorhabditis Genetics Center for providing worm strains, and Kuei-Ching Hsiung for preparing the figures. This work is supported by grants from the Chang Gung Memorial Hospital (CMRPD1C0812, CMRPD1C0813 and BMRP742) to to Lo SJ.
The purpose of this research was to develop a model for human hepatitis B viral antigen interaction with host factors in the standard model animal Caenorhabditis elegans (C. elegans).
The hepatitis B viral surface proteins (large, small and middle sized) (LHBsAg, MHBsAg, SHBsAg) are essential in viral assembly and infection. Here the three viral surface proteins were synthesized in C. elegans after microinjection of bacterial plasmids containing the genes for the proteins expressed under the control of a ubiquitous animal promoter. Severe reduction in egg laying and brood size as well as gonad retardation occurred with expression of the small hepatitis B virus (HBV) surface antigen in the worm. Smaller effects were found with the middle and larger sized surface antigens.
The specific effects of human HBV surface antigen expression of the C. elegans worm demonstrates the worm as a useful model for understanding viral infection and its effects on animal tissues.
Simple and rapid animal cell and model animal systems are essential for understanding of the infection process and disruptions caused by important human disease agents such as HBV. Here a new and useful model is established and its characteristics presented.
HBV is the familiar abbreviation for human hepatitis B virus, although with HAV and HCV, a major cause of liver damage and morbidity. LHBsAg, MHBsAg and SHBsAg are the abbreviations for the large, middle-sized and small versions of the viral surface antigen, the proteins involved in the initial phase of virus surface attachment and infection.
The paper is well written.
Manuscript source: Invited manuscript
Specialty type: Virology
Country of origin: Taiwan
Peer-review report classification
Grade A (Excellent): A
Grade B (Very good): B, B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Ghiringhelli PD, Liu ZW, Striker R S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Tiollais P, Pourcel C, Dejean A. The hepatitis B virus. Nature. 1985;317:489-495. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 934] [Cited by in RCA: 928] [Article Influence: 23.2] [Reference Citation Analysis (0)] |
| 2. | Ganem D, Varmus HE. The molecular biology of the hepatitis B viruses. Annu Rev Biochem. 1987;56:651-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 805] [Cited by in RCA: 827] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 3. | Locarnini S, Littlejohn M, Aziz MN, Yuen L. Possible origins and evolution of the hepatitis B virus (HBV). Semin Cancer Biol. 2013;23:561-575. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 4. | Brechot C, Kremsdorf D, Soussan P, Pineau P, Dejean A, Paterlini-Brechot P, Tiollais P. Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC): molecular mechanisms and novel paradigms. Pathol Biol (Paris). 2010;58:278-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 5. | Beasley RP, Hwang LY, Lin CC, Chien CS. Hepatocellular carcinoma and hepatitis B virus. A prospective study of 22 707 men in Taiwan. Lancet. 1981;2:1129-1133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 6. | Wang J, Chenivesse X, Henglein B, Bréchot C. Hepatitis B virus integration in a cyclin A gene in a hepatocellular carcinoma. Nature. 1990;343:555-557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 482] [Cited by in RCA: 489] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 7. | Wang J, Zindy F, Chenivesse X, Lamas E, Henglein B, Bréchot C. Modification of cyclin A expression by hepatitis B virus DNA integration in a hepatocellular carcinoma. Oncogene. 1992;7:1653-1656. [PubMed] |
| 8. | Wang HC, Chang WT, Chang WW, Wu HC, Huang W, Lei HY, Lai MD, Fausto N, Su IJ. Hepatitis B virus pre-S2 mutant upregulates cyclin A expression and induces nodular proliferation of hepatocytes. Hepatology. 2005;41:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 106] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 9. | Bouchard MJ, Schneider RJ. The enigmatic X gene of hepatitis B virus. J Virol. 2004;78:12725-12734. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 362] [Cited by in RCA: 386] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 10. | Yen TT, Yang A, Chiu WT, Li TN, Wang LH, Wu YH, Wang HC, Chen L, Wang WC, Huang W. Hepatitis B virus PreS2-mutant large surface antigen activates store-operated calcium entry and promotes chromosome instability. Oncotarget. 2016;7:23346-23360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Tennant BC, Toshkov IA, Peek SF, Jacob JR, Menne S, Hornbuckle WE, Schinazi RD, Korba BE, Cote PJ, Gerin JL. Hepatocellular carcinoma in the woodchuck model of hepatitis B virus infection. Gastroenterology. 2004;127:S283-S293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 105] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 12. | Gerin JL, Cote PJ, Korba BE, Tennant BC. Hepadnavirus-induced liver cancer in woodchucks. Cancer Detect Prev. 1989;14:227-229. [PubMed] |
| 13. | Transy C, Fourel G, Robinson WS, Tiollais P, Marion PL, Buendia MA. Frequent amplification of c-myc in ground squirrel liver tumors associated with past or ongoing infection with a hepadnavirus. Proc Natl Acad Sci USA. 1992;89:3874-3878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 46] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 14. | Hsu T, Möröy T, Etiemble J, Louise A, Trépo C, Tiollais P, Buendia MA. Activation of c-myc by woodchuck hepatitis virus insertion in hepatocellular carcinoma. Cell. 1988;55:627-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 135] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Wu BK, Li CC, Chen HJ, Chang JL, Jeng KS, Chou CK, Hsu MT, Tsai TF. Blocking of G1/S transition and cell death in the regenerating liver of Hepatitis B virus X protein transgenic mice. Biochem Biophys Res Commun. 2006;340:916-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 16. | Chisari FV, Klopchin K, Moriyama T, Pasquinelli C, Dunsford HA, Sell S, Pinkert CA, Brinster RL, Palmiter RD. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145-1156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 518] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 17. | Kim A, Kwon OS, Kim SO, He L, Bae EY, Lee MS, Jeong SJ, Shim JH, Yoon DY, Kim CH. Caspase-3 activation as a key factor for HBx-transformed cell death. Cell Prolif. 2008;41:755-774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 18. | Zhang XD, Wang Y, Ye LH. Hepatitis B virus X protein accelerates the development of hepatoma. Cancer Biol Med. 2014;11:182-190. [PubMed] |
| 19. | Geng X, Harry BL, Zhou Q, Skeen-Gaar RR, Ge X, Lee ES, Mitani S, Xue D. Hepatitis B virus X protein targets the Bcl-2 protein CED-9 to induce intracellular Ca2+ increase and cell death in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2012;109:18465-18470. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 73] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 21. | Ankeny RA. The natural history of Caenorhabditis elegans research. Nat Rev Genet. 2001;2:474-479. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 95] [Cited by in RCA: 74] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Diogo J, Bratanich A. The nematode Caenorhabditis elegans as a model to study viruses. Arch Virol. 2014;159:2843-2851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 23. | Kurz CL, Ewbank JJ. Caenorhabditis elegans: an emerging genetic model for the study of innate immunity. Nat Rev Genet. 2003;4:380-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 143] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 24. | Culetto E, Sattelle DB. A role for Caenorhabditis elegans in understanding the function and interactions of human disease genes. Hum Mol Genet. 2000;9:869-877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 188] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 25. | Olsen A, Vantipalli MC, Lithgow GJ. Using Caenorhabditis elegans as a model for aging and age-related diseases. Ann N Y Acad Sci. 2006;1067:120-128. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 76] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 26. | Zarski JP, Kuhns M, Berck L, Degos F, Schalm SW, Tiollais P, Bréchot C. Comparison of a quantitative standardized HBV-DNA assay and a classical spot hybridization test in chronic active hepatitis B patients undergoing antiviral therapy. Res Virol. 1989;140:283-291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 27. | Liu WH, Lin YL, Wang JP, Liou W, Hou RF, Wu YC, Liao CL. Restriction of vaccinia virus replication by a ced-3 and ced-4-dependent pathway in Caenorhabditis elegans. Proc Natl Acad Sci USA. 2006;103:4174-4179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 34] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 28. | Lee LW, Chang TY, Lo HW, Lo SJ. Hepatitis D antigens cause growth retardation and brood-size reduction in C. elegans. Front Biosci (Elite Ed). 2011;3:380-390. [PubMed] |
| 29. | Sheu SY, Lo SJ. Biogenesis of the hepatitis B viral middle (M) surface protein in a human hepatoma cell line: demonstration of an alternative secretion pathway. J Gen Virol. 1994;75:3031-3039. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 30. | Lee LW, Lo HW, Lo SJ. Vectors for co-expression of two genes in Caenorhabditis elegans. Gene. 2010;455:16-21. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 31. | Safra M, Ben-Hamo S, Kenyon C, Henis-Korenblit S. The ire-1 ER stress-response pathway is required for normal secretory-protein metabolism in C. elegans. J Cell Sci. 2013;126:4136-4146. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 32. | Hung JH, Su IJ, Lei HY, Wang HC, Lin WC, Chang WT, Huang W, Chang WC, Chang YS, Chen CC. Endoplasmic reticulum stress stimulates the expression of cyclooxygenase-2 through activation of NF-kappaB and pp38 mitogen-activated protein kinase. J Biol Chem. 2004;279:46384-46392. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 221] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 33. | Hubbard EJ, Greenstein D. The Caenorhabditis elegans gonad: a test tube for cell and developmental biology. Dev Dyn. 2000;218:2-22. [PubMed] |
| 34. | Lu R, Maduro M, Li F, Li HW, Broitman-Maduro G, Li WX, Ding SW. Animal virus replication and RNAi-mediated antiviral silencing in Caenorhabditis elegans. Nature. 2005;436:1040-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 334] [Cited by in RCA: 333] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 35. | Jiang H, Franz CJ, Wang D. Engineering recombinant Orsay virus directly in the metazoan host Caenorhabditis elegans. J Virol. 2014;88:11774-11781. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 36. | Franz CJ, Renshaw H, Frezal L, Jiang Y, Félix MA, Wang D. Orsay, Santeuil and Le Blanc viruses primarily infect intestinal cells in Caenorhabditis nematodes. Virology. 2014;448:255-264. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |