Peer-review started: September 7, 2015
First decision: October 8, 2015
Revised: October 28, 2015
Accepted: December 4, 2015
Article in press: December 8, 2015
Published online: February 12, 2016
Processing time: 156 Days and 8.9 Hours
Herpes simplex virus 1 (HSV-1) is a ubiquitous human pathogen that establishes latent infection in ganglia neurons. Its unique life cycle requires a balanced “conquer and compromise” strategy to deal with the host anti-viral defenses. One of HSV-1 α (immediate early) gene products, infected cell protein 0 (ICP0), is a multifunctional protein that interacts with and modulates a wide range of cellular defensive pathways. These pathways may locate in different cell compartments, which then migrate or exchange factors upon stimulation, for the purpose of a concerted and effective defense. ICP0 is able to simultaneously attack multiple host pathways by either degrading key restrictive factors or modifying repressive complexes. This is a viral protein that contains an E3 ubiquitin ligase, translocates among different cell compartments and interacts with major defensive complexes. The multiple functional domains of ICP0 can work independently and at the same time coordinate with each other. Dissecting the functional domains of ICP0 and delineating the coordination of these domains will help us understand HSV-1 pathogenicity as well as host defense mechanisms. This article focuses on describing individual ICP0 domains, their biochemical properties and their implication in HSV-1 infection. By putting individual domain functions back into the picture of host anti-viral defense network, this review seeks to elaborate the complex interactions between HSV-1 and its host.
Core tip: Due to the genomic limitation, viruses often use multifunctional proteins to ensure viral replication. Coordination of the multiple viral functions is critical for a successful viral infection. Infected cell protein 0 (ICP0) is notoriously multi-functional in terms of simultaneously targeting many host machineries located in different cellular compartments. Understanding the molecular basis of ICP0 multifunctionality is important for not only the elucidation of herpes simplex virus pathogenicity but also the delineation of host defense mechanisms.
- Citation: Gu H. Infected cell protein 0 functional domains and their coordination in herpes simplex virus replication. World J Virol 2016; 5(1): 1-13
- URL: https://www.wjgnet.com/2220-3249/full/v5/i1/1.htm
- DOI: https://dx.doi.org/10.5501/wjv.v5.i1.1
Herpes simplex virus 1 (HSV-1) is a ubiquitous virus that infects over 70% of the world adult population. It causes a wide range of clinical manifestations, including cold sores, genital ulceration, keratitis, and herpes encephalitis. Once infected, HSV-1 establishes a lifelong latency in human trigeminal ganglia. Its sporadic reactivation nourishes a wide spread of the virus. It is one of the most prevalent opportunistic pathogens that can cause severe diseases in newborns or immunocompromised patients. Infected cell protein 0 (ICP0), an α (immediate early, IE) gene product of HSV-1, is a key regulator that activates viral gene expression in both lytic and latent infections[1]. This multifunctional protein plays a critical role in viral counteractions against the host anti-viral defenses.
In early studies, viral proteins expressed in HSV-1 infection were classified into two groups: Virion proteins and infected cell proteins (ICPs)[2]. Both groups were numbered in the order of their descending molecular weight, with number “1” representing the largest protein on high resolution polyacrylamide gels[2]. ICP0 was named outside of the natural numbers for two reasons. First, the protein level of ICP0 was significantly lower than other ICPs. ICP0 was not detected in the initial efforts of numbering the ICPs[2]. It was only discovered after a cycloheximide treatment, which augmented mRNA accumulation and boosted a sudden protein production following the cycloheximide withdrawal[3]. The second reason why ICP0 was named differently was its anomalous mobility in denaturing polyacrylamide gel electrophoresis. The relative position of ICP0 vs other ICPs was not consistent on gels with different acrylamide concentrations, which made it impossible to give ICP0 a fixed position in the descending order of molecular weight.
Later on ICP0 was found to be extensively post-translationally modified[4-8] and to undergo quick turnover at early infection[9,10]. The complex biochemical properties of ICP0 likely contribute to the aforementioned low abundancy and abnormal mobility. Three decades of studies have showed that ICP0 is an important viral multifunctional protein to counteract against host anti-viral defenses. It is essential for low multiplicity infection in cultured cells and for latency reactivation in animal models. However, the complexity of how ICP0 carries out those biological functions is not well understood. Understanding the biochemical foundations of ICP0 at different infection phases will help to elucidate the molecular basis of ICP0 functionality. Individual functions of ICP0 as E3 ubiquitin ligase or chromatin remodeler have been discussed elsewhere[11-16]. This review will focus on dissecting ICP0 biochemical properties and seek to understand the profound coordination in the multiple functions of ICP0.
Initially, ICP0 was found to transactivate HSV-1 promoters when co-transfected in mammalian cells, similar to many other IE viral proteins such as ICP4 of HSV[17,18], T antigen of SV40[19], and E1A of adenovirus[20]. However, it was quickly realized that the mechanism of ICP0 transactivation was quite different from that of other viral gene activators. For example, ICP4 is essential for viral replication. Deletion of ICP4 led to abnormal viral expression and defective DNA replication[21,22]. In the case of ICP0, gene deletion did not affect viral expression or DNA replication at high multiplicity of infection (MOI) but it had great impact on the viral yield when MOI was lower than 0.1[23]. In experimental animals, deletion of ICP0 mildly reduced the efficiency of latency establishment but completely abolished the latency reactivation[24], whereas ICP4 or ICP27 deletion rendered the mutant virus neither able to replicate in the eyes nor to establish latent infection[24]. Moreover, many viral IE proteins contain a DNA binding domain and they work in mechanisms similar to cognate transcription activators such as GAL4, but ICP0 did not bind to the DNAs it activated[25,26]. Extensive functional analysis showed that ICP0 can transactivate a wide range of cellular promoters or promoters from other DNA or RNA viruses, with no requirement of a specific cis-sequence[27-29]. Therefore, ICP0 is defined as a promiscuous transactivator.
The unique functionality of ICP0 energized a great amount of interests in the virology field. In early 1990s, a series of mutagenesis analyses identified a cysteine-rich region required for the ICP0 transactivation activity[30-32], which was later determined as a C3HC4 zinc binding really interesting new gene (RING) finger motif[33-35]. Conserved RING finger sequences were found in a large family of E3 ubiquitin ligases[36,37]. Later on, ICP0 was also proven to be an E3 ubiquitin ligase[38,39]. The discovery that various ICP0 substrates imposed restrictions on viral expression in the absence of ICP0[40-45] eventually led to a conclusion that one major function of ICP0 is to target host defensive molecules for ubiquitin-mediated proteasomal degradation. By degrading the restrictive host factors, ICP0 alleviates host defense and promotes viral gene expression.
Starting in the late 1990s, several labs made the efforts to identify ICP0 interacting proteins. From pull-down assays, yeast-2-hybrid screenings and coimmuno-precipitations, a wide range of cellular proteins were found to interact with ICP0[46-49]. Therefore ICP0 carries out viral counteractions by degrading restrictive factors and modulating repressive complexes, and consequently ICP0 enhances viral expression and replication. To better understand the coordination of ICP0 functional domains in counteracting host defenses, this review summarizes the current knowledge of ICP0 domains and ICP0 binding partners, and discusses their implications in HSV-1 infection.
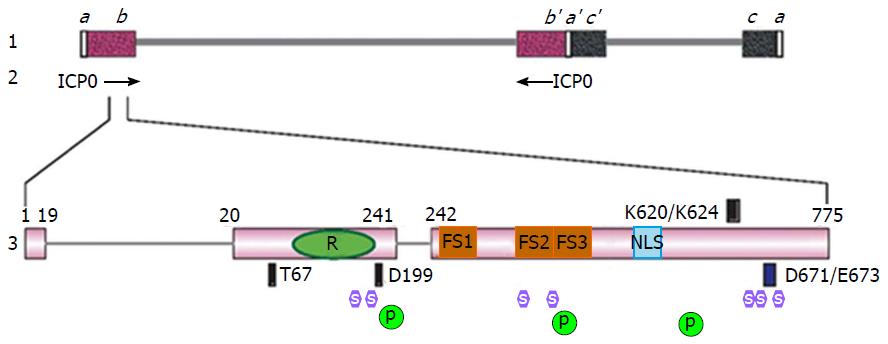
The gene that encodes for ICP0 protein, also called α0 gene, is located within the inverted sequences ab and b’a’ that flank the unique long (UL) region[50] (Figure 1). Therefore, the ICP0 gene is one of the few HSV-1 genes that are diploid in the genome. The ICP0 gene is also among the few HSV-1 genes that contain introns[51]. There are two introns of 765 and 136 nucleotides, respectively, intervening the three exons that encode for ICP0 amino acids 1-19, 20-241 and 242-775[51]. It is quite curious why the ICP0 gene would evolve to bear introns because these introns do not seem to have significant functions in viral replication and alternative splicing of ICP0 has not been observed in infected cells. In one report, the ICP0 cDNA virus had a slight delay of gene expression depending on the cell-type used[52], whereas in another report differences between wild type virus and ICP0 cDNA viruses were not observed[53]. In animal models, recombinant viruses containing ICP0 deleted of introns showed no obvious defects in latency establishment and reactivation[54].
There is an in-frame stop codon located inside intron 2, which predicts a truncated form of ICP0 (ICP0R) if alternative splicing occurs. Overexpression of ICP0R inhibited the transactivation activity of the co-transfected wild type ICP0[55,56], suggesting ICP0R can work as a dominant negative to repress ICP0 activity. Although a band at the size of ICP0R was detected at low level in some cell lines[57], it remains unclear whether this is a product from alternative splicing or a product of proteolytic cleavage of ICP0. The function of ICP0R in the infection context is unknown.
One important fact about the ICP0 gene is that the coding strand of ICP0 is anti-sense to the latency-associated transcript (LAT), the only transcript that is abundantly expressed in latently infected ganglia neurons[58,59]. The concept of LAT functioning as the anti-sense RNA to ICP0 mRNA has been explored and microRNAs identified in the LAT region have been shown to regulate ICP0 expression. Likely these actions fine-tune the basal level expression in latency maintenance and reactivation[60-62].
The three exons of ICP0 gene encodes for a 775-amino acid protein. It contains many functional domains and interacts with multiple binding partners (Table 1). The most important functional domain of ICP0 is the aforementioned C3HC4 zinc containing RING finger, which is located within exon 2 and spans through residues 116-156[63] (Figure 1). The promiscuous transactivator ability of ICP0 relies on a functional RING finger domain. Deletions or mutations of the consensus cysteine or histidine in the RING finger domain completely abolish the transactivation activity[63,64]. Recombinant viruses containing such deletions or mutations replicate at a rate similar to that of the ICP0-null virus[53,63,65]. This region is highly conserved among α-herpesviruses[34,63]. The structure of ICP0 RING finger has been solved by nuclear magnetic resonance (NMR)[35].
| Domain | Location | Function in HSV-1 replication | Section | Ref. |
| ICP0 cis-elements | ||||
| RING finger | aa 116-156 | E3 ubiquitin ligase, degrading PML, Sp100, etc. | RING finger domain and E3 ubiquitin ligase activity | [63-65] |
| Proline-rich region | aa 241-553 | Containing redundant ND10-fusion segments | Proline-rich region and ND10-fusion | [105] |
| NLS | aa 500-506 | Nuclear localization | Nuclear localization domain and ICP0 nuclear/cytoplasmic translocation | [90] |
| Dimerization domain | aa 617-711 | ICP0 self-dimerization, in vivo functions unclear | Dimerization | [115-117] |
| ND10-retention domain | aa 669-775 | Retaining ICP0 at ND10 | ND10-retention | [53] |
| SLSs | SUMO interaction motif and ICP0 substrate recognition | [113] | ||
| SLS-4 | aa 361-367 | Binding to SUMO-1/2/3, stimulating in vitro polyubiquitination | ||
| SLS-5, SLS-7 | aa 651-655, 681-685 | Binding to SUMO-1, cooperating with SLS-4 | ||
| ICP0 binding partners | ||||
| RNF8 | T67 | Degrading RNF8 to regulate DNA damage responses | RNF8 | [42,43] |
| Cyclin D3 | D199 | Involved in nuclear-to-cytoplasmic translocation of ICP0 | Cyclin D3 | [46,133-135] |
| BMAL1 | aa 20-241 | Activating viral transcription via BMAL1/CLOCK | BMAL1 | [48,140] |
| EF-1δ | aa 543-768 | Inhibiting translation in vitro, in vivo functions unclear | EF-1δ interaction | [96] |
| USP7 | K620/K624 | USP7 degradation, Cell-dependent ICP0 stabilization, | USP7 interaction | [47,85,88,123] |
| CoREST | D671/E673 | Dislodging HDAC from REST/CoREST/HDAC repressor | CoREST interaction | [49,124] |
| WDR11 | N/A | Regulating virion assembly and egress | WD repeat protein 11 | [143] |
The RING finger domain of ICP0, like many RING superfamily members[36,66,67], works as an E3 ubiquitin ligase. Mediated by the E2 conjugating enzyme UbcH5a[68,69], ICP0 uses this domain to ubiquitinate its substrate proteins and targets them for proteasomal degradation. The first two ICP0 substrates, promyelocytic leukemia (PML) and Sp100 (speckled 100 kDa) were identified by Chelbi-Alix and de Thé[40]. PML and Sp100 are the major organizer proteins for the dynamic nuclear bodies called nuclear domains 10 (ND10s) or PML nuclear bodies (for reviews, see references[70,71]). ND10s are nuclear structures that are composed of over 150 constituents[72]. They are involved in many cellular functions including gene regulation[73,74], cell cycle arrest[75], apoptosis[76], DNA repair[77] and anti-viral defense[78]. Degradation of PML and Sp100 by ICP0 leads to the dispersal of ND10 bodies[79]. In ICP0-null virus infection, depletion of PML and Sp100 was shown to compensate for the loss of ICP0 and to increase viral replication[41,80]. In PML-/- mouse embryotic fibroblasts (MEF), interferon (IFN) caused minimal effects on low multiplicity HSV-1 infection, whereas IFN treatment of PML+/+ MEF reduced viral growth at least 1000 folds[81], suggesting that PML can mediate the IFN inhibition on viral replication. Taken together, PML is an important factor in host defense pathways and ICP0 targets PML, and maybe also Sp100, to alleviate anti-viral repressions.
Additional ICP0 substrates identified up to date include DNA-dependent protein kinase K (DNAPK)[82], centromeric proteins C and A (CENP-C and CENP-A)[83,84], ubiquitin-specific protease 7 (USP7)[85], RNF8[43], the 111-kDa isoform of poly (ADP-Ribose) glycohydrolase[86], interferon inducible protein 16 (IFI16)[44], and tripartite motif (TRIM) protein TRIM27[87]. Among these substrates, siRNA knock-down of RNF8 or IFI16 promoted the replication of ICP0-null virus[43,45], suggesting the involvement of these two proteins in host anti-viral defenses. However, depletion of TRIM27 reduced the viral yield in the absence of ICP0[87], and overexpression of USP7 accelerated gene expression in wild type HSV-1 infection[88]. These results indicate that not all ICP0 substrates place simple direct repressions on viral gene expression. Some of the substrate proteins may be degraded to regulate a more complicated cell network in order to benefit the overall viral outcome, especially the balanced actions in latent infection.
The E3 ubiquitin ligase activity of ICP0 RING finger is highly regulated by multiple factors, including its subcellular location, its phosphorylation status, and its other functional domains. For example, a failure of ICP0 to completely merge with ND10 bodies blocked substrate access and abolished PML degradation[53], and two amino acid substitutions in the C-terminal CoREST binding site (D671A/E673A) also negatively affected PML degradation[89]. The regulatory mechanisms of ICP0 E3 are not completely understood. Some of the known regulations will be discussed more in detail as we describe other important ICP0 properties in this review.
ICP0 contains a nuclear localization signal (NLS) mapped to the short stretch of basic amino acids VRPRKRR located at residues 500-506[90] (Figure 1). This arginine-rich NLS is sufficient and necessary for the nuclear localization of transiently transfected ICP0[90]. However, in infected cells, ICP0 is not an exclusively nuclear protein. Its subcellular distribution is regulated by many other factors in addition to the NLS.
First of all, ICP0 undergoes localization changes during the infection process. Early in infection, newly synthesized ICP0 is immediately transported into the nucleus in the presence of the NLS. Once inside the nucleus, ICP0 is immediately localized to the dynamic nuclear structure ND10[91]. This leads to the aforementioned degradation of ND10 organizers, PML and Sp100[40], and the subsequent disruption of ND10 nuclear bodies[79]. The dynamic interaction between ICP0 and ND10 is critical for the efficient access of ICP0 to its substrates, PML and Sp100, and their subsequent degradation[53], which will be discussed in depth in section “Proline-rich region and ND10-fusion”.
After the dispersal of ND10 bodies, ICP0 diffuses throughout the nucleus. Once its nuclear functions are completed, ICP0 is translocated into the cytoplasm[92,93]. Many important ICP0 functions are carried out in the nucleus, where ICP0 degrades PML and interacts with REST/CoREST chromatin repressor (see section “CoREST interaction”) early in infection. Pre-transfection of irrelevant DNA before infection can prolong ICP0 nuclear localization and delay the cytoplasmic translocation, especially in cell lines that poorly express transgenes[93]. These results suggest that ICP0 is kept within the nucleus until its nuclear functions are completed[93].
It is not yet clear how the NLS containing ICP0 protein is translocated into the cytoplasm at late infection. Either the NLS is modified late in infection so that newly translated ICP0 cannot enter the nucleus, or a nuclear export signal (NES) is unmasked late in infection so that nuclear ICP0 is exported. So far, a functional NES has not been identified.
Multiple viral factors have been found to participate in regulating the nuclear-to-cytoplasmic translocation of ICP0. For example, deletion of ICP4 caused ICP0 to lose its nuclear localization. Even at early infection, ICP0 expressed in the ICP4-null virus infected cell was solely found in the cytoplasm[94]. On the other hand, deletion of ICP27 retained ICP0 within nucleus throughout the infection and overexpression of ICP27 facilitated ICP0 export into the cytoplasm[94]. Since ICP27 is highly expressed in ICP4-null virus infected cells, ICP27 is likely the factor promoting ICP0 export. Another viral protein, VP22, has also been reported to play a role in the ICP0 cytoplasmic translocation. Deletion or mutation in VP22 restricted a series of viral proteins, including ICP0, inside the nucleus[95]. Whether or not VP22 affects a general nuclear export pathway and therefore indirectly delays ICP0 translocation remains unclear.
Functions of cytoplasmic ICP0 are not understood either. Kawaguchi et al[96] reported an interaction between ICP0 and translation elongation factor 1δ (EF-1δ) (also see in section “EF1δ interaction”) and showed that ICP0 inhibited in vitro translation via this interaction. However, regulation of cellular translation by ICP0 is yet to be seen in vivo. Paladino et al[97] showed that ICP0 lacking NLS stayed in the cytoplasm and blocked IRF3 activation in infected cells. It remains unknown whether ICP0 directly interacts with IRF3 or secondary mediators are involved in this inhibition. Small amount of ICP0 has also been found in the tegument of purified virions[98,99]. Although the function of virion-associated ICP0 is not clear, it has been reported that ICP27 dependent cytoplasmic translocation of ICP0 is required for the incorporation of ICP0 into virions[100]. Delboy et al[101,102] also showed that an active ubiquitination was important for ICP0 to be incorporated into virions. Both RING finger mutation and proteasome inhibition precluded ICP0 from associating with virions. Since defective ubiquitination sequesters ICP0 within the ND10 bodies and prevents the cytoplasmic translocation of ICP0[89,92], Nicola’s results are consistent with the observation that cytoplasmic localization of ICP0 in late infection is a prerequisite for the incorporation of ICP0 into virions. Since up to 49 cellular proteins have also been found in purified virions[99], the selection mechanism of low copy tegument proteins and their biological significance are not clear.
In the center of ICP0 protein, there is a long stretch of proline-rich region spanning residues 241 to 553. Initial deletion mapping found that serial deletions from the carboxyl-end of this region resulted in a progressive loss of the ICP0 transactivator activity[55], indicating the importance of this region in ICP0 functions. Multiple repeats of the PxxP motif in this region can interact with the Src homology 3 (SH3) domain in Cbl-interacting protein 85 kDa (CIN85), and a few other Src kinase family members[103,104]. Recently, Zheng et al[105] demonstrated that the proline-rich sequences were important to direct the fusion of ICP0 with ND10 nuclear bodies. As discussed above, ICP0 is localized to ND10 at early infection. This colocalization process is composed of three sequential dynamic steps: ND10-adhesion, ND10-fusion and ND10-retention[53]. Among these steps, a successful ICP0-ND10 fusion is essential for the ICP0 E3 ligase to access and degrade its substrate PML[53]. The proline-rich region of ICP0 is critical for the ND10-fusion step[105]. Zheng et al[105] showed that three proline-rich segments located at residues 242-291, 343-391, and 393-441, termed ND10-FS1, ND10-FS2 and ND10-FS3, respectively (Figure 1), redundantly facilitated the ND10-fusion of ICP0. Deletion of one or two ND10-FSs did not substantially affect the fusion process. However when all three ND10-FSs were deleted, ICP0 was blocked from entering the ND10 bodies[105]. Since most of the cellular PML is located at ND10, the ICP0-ND10 fusion ensures a quick access of ICP0 to large amount of substrate and leads to an effective PML degradation. This likely increases the efficiency of ICP0 destroying the host restrictive factor PML and therefore enhances gene expression. The redundancy in proline-rich segments indicates the importance of ND10-fusion process in HSV-1 infection. Whether the redundant ND10-FSs synergistically improve the speed of ND10 fusion is a very important question waiting to be answered. It is also unknown whether ND10-FSs work via interacting with a SH3 domain or other proline-interacting motifs.
Small ubiquitin-like modifier (SUMO) is a unique type of post-translational modification found on a variety of proteins. Protein SUMOylation functions in almost every aspect of a cell’s life, including cell cycle, genome integrity, subcellular transport, and host immune defenses (for reviews, see references[15,106-108]). The SUMO moiety is recognized by hydrophobic sequences called the SUMO-interaction motif (SIM)[109,110]. RING-type E3 ubiquitin ligases that contain a SIM and specifically recognize SUMOylated substrates are classified as SUMO-targeted ubiquitin ligases (STUBL)[111,112]. Boutell et al[113] identified seven putative SIM-like sequences (SLSs) scattering throughout the ICP0 open reading frame (Figure 1). In yeast-2-hybrid assays, mutations in SLS-4 abolished the interaction between ICP0 and SUMO-2/3, whereas mutations in SLS-5 and SLS-7 did not affect such binding. SLS-4 was also found to be necessary for the in vitro ubiquitination of a SUMO-2 chain, indicating that ICP0 can work as a STUbL to preferentially recognize SUMOylated proteins for ubiquitination[113]. However, a recombinant virus containing mutant SLS-4 did not affect the degradation of endogenous PML in infected cells, while PML with all SUMOylation sites mutated were still degraded by ICP0[113], suggesting a more complex regulation on ICP0 substrate recognition in addition to the SUMO-SIM interaction. Moreover, although mutations in SLS-5 and SLS-7 did not interfere with the binding between ICP0 and SUMO-2/3, a recombinant virus carrying triple mutations in SLS-4/5/7 greatly demolished the ability of ICP0 to degrade PML[114]. This suggests there may be differences in the SLS affinities and multiple SLSs may work synergistically in PML degradation.
The C-terminus of ICP0, broadly defined for the region from downstream of NLS to the carboxyl-end, may be the most active but also the least understood region of ICP0. At least five major functions or interactions have been described in this region.
Dimerization: First, ICP0 is a protein known to aggregate and dimerize in vitro and in vivo[115-117]. In chromatography purification, ICP0 was fractionated at a much bigger molecular weight[117]. When wild type and mutant ICP0 were co-transfected into the same cell, the wild type ICP0 was able to correct the subcellular distribution of a mislocated mutant ICP0. The dimerization domain has been mapped to C-terminal residues 617-711[115]. The biological function of ICP0 dimerization is not yet clear.
ND10-retention: The second function of ICP0 C-terminus is related to the ND10 localization property of ICP0. Initial data showed that ICP0 lacking the C-terminus was evenly dispersed throughout the nucleus, compared to the full-length ICP0 that was colocalized to the ND10 bodies[117]. This led to an assumption that the C-terminus of ICP0 is responsible for ND10 localization[117,118]. However, recent results from Gu et al[53] showed that the C-terminus of ICP0 was not involved in the recruitment of ICP0 to ND10. In the absence of C-terminus, ICP0 did not aggregate at ND10 but had the ability to degrade PML. When a double mutant of both C-terminal truncation and RING finger mutation was introduced, ICP0 was found to localize at ND10. These results suggest that the C-truncated ICP0 undergoes adhesion and fusion steps to enter ND10, but it cycles in and out of ND10 in a more accelerated mode. Only when the inactive RING blocks the enzymatic reaction into a transition state, can the ICP0-ND10 colocalization be observed in a steady-state immunofluorescence staining. Therefore the C-terminus of ICP0 is responsible for the retention, but not the recruitment, of ICP0 to ND10.
USP7 interaction: The C-terminus of ICP0 also interacts with various proteins, such as USP7[47], CoREST[49] and EF-1δ[96], which are from proteasome pathway, chromatin repressor complex and translational machinery, respectively.
USP7 is the first ICP0 interacting protein identified via a GST pull-down/protein sequencing assay[47,119]. This is a deubiquitinase that regulates the ubiquitination status of many important cell check point proteins, such as p53[120], RE1-silencing transcription factor (REST)[121], and phosphatase and tensin homolog (PTEN)[122]. The minimum sequences required for the strong binding between the two are amino acids 615-633 of ICP0 and amino acids 535-889 of USP7[123]. The crystal structure of USP7 C-terminal ubiquitin-like domains bound with ICP0 peptide has been solved. Salt bridges between K620/K624 of ICP0 and D762/D764 of USP7 are critical for the interaction, while the peripheral residues form a binding pocket to support the strong ICP0-USP7 interaction[123]. Consistent structural data have also been obtained from NMR assays[124].
Initial in vitro ubiquitination assays showed that ICP0-USP7 interaction inhibited ICP0 autoubiquitination but promoted USP7 polyubiquitination[8,85]. Consistent with these observations, the ICP0-USP7 interaction was found essential for the degradation of USP7 by ICP0 in infected cells[85,88]. However, regarding to ICP0 autoubiquitination, different groups have reported contradictory results[85,88]. Boutell et al[85] used HSV-1 (strain 17+) and reported that wild type ICP0 stayed at a steady level after cycloheximide treatment, whereas an R623L/K624I mutant virus, of which ICP0 was incapable of binding to USP7 and was quickly degraded in the presence of cycloheximide. On the other hand, Roizman and colleagues demonstrated that wild type ICP0 of HSV-1 (strain F) underwent rapid degradation at early infection and was only stabilized late in infection[9,10]. Furthermore, they found that a K620I mutant virus that abolished ICP0-USP7 interaction had enhanced, not reduced, viral gene expression but showed defects in plaque formation[88]. Therefore, ICP0-USP7 interaction may have profound biological significances, depending upon the virus strains and cell lines. Since both ICP0 and USP7 have a wide range of different substrates that are involved in critical cellular pathways, the interaction between ICP0 and USP7 may be more important in fine-tuning the ubiquitin status of these check point proteins than simply regulating ICP0 self-stability. A complex balance of these proteins may in return affect ICP0 stability in a cell type dependent manner.
CoREST interaction: CoREST binding to ICP0 was discovered by co-immunoprecipitation[125]. CoREST is the corepressor partner for REST[126]. REST/CoREST are the key components of a chromatin regulatory complex that determines neural cell fate during development[127]. The CoREST binding of ICP0 is mapped to the amino acids D671/E673[89]. Gu et al[125] showed that ICP0-CoREST interaction depended on the presence of viral kinases US3 and UL13, and a prolonged infection resulted in less binding, suggesting that ICP0-CoREST interaction is a regulated transient process. This interaction was found essential for the dissociation of HDAC1 from REST/CoREST complex in HSV-1 infection[89,125]. A recombinant virus carrying a dominant negative CoREST incapable of HDAC1-binding showed a higher viral productivity in the absence of ICP0, which means the disruption of HDAC1-CoREST interaction is beneficial for viral replication[49]. Furthermore, on the molecular level, a recombinant virus containing D671A/E673A mutations had less acetylated histone H3 compared with the wild type virus or a mutant virus that kept the effective ICP0-CoREST interaction[128]. In contrast to these results, Everett showed that depletion of CoREST did not improve the yield of ICP0-null virus[129]. The seemingly contradictory results are reconciled from the fact that lysine-specific demethylase-1 (LSD1), another important component in the REST/CoREST complex, is required in HSV-1 replication[130]. Therefore the stoichiometry of REST/CoREST/LSD1/HDAC components[127] may play a role in determining the interaction to different viral proteins at different infection phases.
EF1δ interaction: Interaction between ICP0 and EF-1δ was identified through a yeast-2-hybrid screening[96]. The binding has been mapped to the C-terminal residues 543-768 and found to inhibit in vitro translation[96]. However, in vivo function of this interaction is not clear.
For all these different C-terminal functions it is not clear how these seemingly unrelated activities coordinate in this region. Are there different subsets of ICP0 distributed in distinct subcellular compartments? Or some of the components from different pathways converge at certain cellular hubs, such as ND10? Answers to these questions will be the key to understanding the complex functions of ICP0 in both lytic and latent infections.
Cyclin D3: Cyclin D3 is identified as an ICP0-interacting protein by a yeast-2-hybrid screening[46]. D-type cyclins form complexes with cyclin-dependent kinases to regulate G1 to S phase transition[131,132], which can be manipulated by many DNA viruses for the purpose of promoting DNA synthesis in infected cells[133]. ICP0 interacts with Cyclin D3 through its amino acid D199 located in exon 2, downstream to the RING finger domain (Figure 1). The D199-Cyclin D3 interaction is important in the nuclear-to-cytoplasmic translocation of ICP0. Mutation in cyclin D3 binding site or treatment by CDK4 inhibitor during the infection prevented ICP0 from translocating to the cytoplasm[134,135], whereas insertion of cyclin D3 gene into the HSV-1 genome to overexpress cyclin D3 led to an accelerated cytoplasmic translocation[135,136]. The regulation of the cell cycle during HSV-1 infection is a profound event involving multiple factors. For example, HSV-1 ICP22 and UL13 are found to participate in G2/M transition[137], and CDK inhibitor roscovitine inhibits HSV-1 gene transcription without affecting PML degradation[138,139]. Moreover, the D199 dependent nuclear-to-cytoplasmic translocation of ICP0 is a process that depends on viral DNA replication and the expression of a late protein(s)[92]. Therefore different cell cycle regulatory pathways are interwoven with ICP0 phosphorylation, translocation and possibly other infection events. The concerted efforts from both viral and cellular sides determine the ultimate productivity of an HSV-1 infection.
Brain and muscle ARNT-like protein 1: Brain and muscle ARNT-like protein 1 (BMAL1) interacting with ICP0 is also identified by a yeast-2-hybrid screening[48]. The interaction site to BMAL1 is located in the exon 2 of ICP0[48]. BMAL1 and circadian locomotor output cycles kaput (CLOCK), a histone acetyltransferase, forms a heterodimer to regulate mammalian circadian oscillation[140]. During HSV-1 infection, CLOCK is stabilized and recruited to ND10, which acts as a transcription activator to stimulate viral transcription and replication[141].
RNF8: The identification of ICP0-RNF8 interaction was based on the observation that RNF8 was degraded by ICP0 in HSV-1 infection[42,43]. RNF8 is an RING type E3 ubiquitin ligase that plays a key role in histone ubiquitination and chromatin remodeling upon DNA double-stranded break (DBS) damage[142,143]. ICP0-RNF8 binding is mapped to the phosphorylated amino acid T67 of ICP0, and amino acid R42 of RNF8[43]. A recombinant virus carrying the T67A mutation did not degrade RNF8 but had no problems in degrading DNAPK or USP7, which means ICP0-RNF8 interaction is likely important for a specific RNF8 substrate recognition[43]. Interestingly, knock-down of RNF8 only mildly delayed ICP27 gene transcription and had no effects on viral DNA replication, suggesting that the involvement of ICP0-RNF8 interaction in responding to DBS DNA damage is, again, a complex action.
WD repeat protein 11: WD repeat protein 11 (WDR11) is a newly reported ICP0 interacting protein identified by co-immunoprecipitation[144]. Taylor et al[144] showed that the trans-Golgi network localized WDR11 pulled down several viral proteins including gB, VP16 and VP5 in additional to ICP0, suggesting its possible role in virion assembly and egress.
ICP0 protein contains 775 amino acids, but the apparent molecular weight of ICP0 is about 110 kDa[3], suggesting the presence of post-translational modifications for ICP0. First of all, ICP0 is highly phosphorylated. On two-dimensional gel electrophoreses, ICP0 phosphorylation status changes along with the progression of infection[6]. The phosphorylation sites on ICP0 has been mapped to three phosphor-clusters by tandem mass spectrometry. Cluster 1 is at residues 222-250, cluster 2 is at residues 356-386, and cluster 3 is at residues 505-528[145] (Figure 1). Davido and colleagues showed that serine/threonine mutations in these clusters demolished the transactivation activity of ICP0 and reduced the viral replication in mice[145,146]. Viral protein UL13 was found important for ICP0 phosphorylation[147]. However, how ICP0 phosphorylation coordinates with ICP0 localizations or ICP0 protein-protein interactions to affect the infection is not yet known.
Other modifications of ICP0 are understudied. ICP0 is believed to be nucleotidylated because it can be radiolabeled in infected cells cultured with [α-32P]GTP or [2-3H]ATP containing medium[5]. ICP0 may also be ubiquitinated because it is found to autoubiquitinate itself in in vitro polyubiquitination assays[8].
At least in the infection of HSV-1 (strain F), ICP0 undergoes a rapid degradation at early infection in both proteasome dependent and proteasome independent manners. The protein is then stabilized at late infection[9]. The proteasome independent cleavage occurs in the central region of ICP0 and the rapid turnover depends on the cis presence of an active RING finger as well as the phosphorylation status of ICP0[9,10].
Like all herpesvirus family members, HSV-1 establishes latent infection. The peculiar life cycle of HSV-1 necessitates a close interaction and a delicate balance between the virus and its host. ICP0 of HSV-1, a unique multifunctional protein, plays a key regulatory role to enhance gene expression in lytic infection and to reactivate virion production from latent infection. This protein is tightly regulated on transcriptional, post-transcriptional and post-translational levels. Through its intrinsic functional domains and its ability to interact with a wide range of binding partners, ICP0 can target many cellular protein for proteasomal degradation and regulate various cell pathways via protein-protein interactions.
To achieve its multiple functions, ICP0 undergoes modification and subcellular translocation. Early in infection, ICP0 is immediately imported into the nucleus upon synthesis. Once inside the nucleus, it is recruited to adhere at and then fuse with ND10 to co-mingle with ND10 components. The ND10-fusion process ensures ICP0 to quickly access large amounts of PML and Sp100 for degradation and to extensively interact with many of the regulatory factors located within ND10. This early step in HSV-1 infection is vital for the outcome of a productive infection, not only by destroying and dispersing the repressive factors but also by capturing favorable factors that help establishing replication compartment. Upon viral DNA entering the nucleus, host cell attempt to silence the foreign intrusion by: (1) forming ND10 bodies near viral DNA[148]; (2) recruiting chromatin repressors[149]; and (3) stimulating IFN responses[45]. In a way, HSV-1 deploys ICP0 to approach ND10 is a “smart” move because ND10 serves as a molecular hub for many cellular pathways and it is able to recruit component factors upon specific stimulations[78]. Therefore, adopting factors recruited to ND10 during infection while destroying and repelling restrictive components is an effective strategy to boost viral replication. In fact, various cellular check point proteins such as USP7, CoREST, Cyclin D3, BMAL1 and CLOCK are all recruited to ND10 upon infection and they are found in HSV-1 replication compartments[47,135,141,149]. In fact, HSV-1 replication compartments are established at the sites where ND10 loci have been located before their dispersal[149]. ICP0 interacting with the molecular hub ND10 is a major adaptation to coordinate the multi-tasking of ICP0 functions. Likely the sequential steps of ICP0-ND10 interaction, ND10-adhesion, ND10-fusion, ND10-retention[53], play important roles in achieving the “destroy and then take over” strategy.
Once the replication compartments are set up in the infected cells, ICP0 may have additional functions in a diffused pattern in nucleus and then in the cytoplasm. Whether the trafficking of ICP0 is regulated by post-translational modification or proteolytic processing is currently unknown. Solving the road map of ICP0 being in the right place at the right time will be a continuous interest in the near future for herpes virology field.
ICP0 is required for latency reactivation[24]. The subtle balance of ICP0 level in latent infection may be achieved by microRNA regulation. The rapid turnover of ICP0 on the protein level may also be essential for the maintenance and reactivation of latent infection. After all, one good way to achieve massive spreading is to keep the sporadic but not severe recurrent infections.
P- Reviewer: Diefenbach R, Li QH, Rajcani J, Zheng CF S- Editor: Kong JX L- Editor: A E- Editor: Li D
| 1. | Roizman B, Knipe DM. Whitley RJ. Herpes Simplex Viruses, in Fields Virology, 6 Edition, Lippincott Williams & Wilkins. 2013;1823-1897. |
| 2. | Honess RW, Roizman B. Proteins specified by herpes simplex virus. XI. Identification and relative molar rates of synthesis of structural and nonstructural herpes virus polypeptides in the infected cell. J Virol. 1973;12:1347-1365. [PubMed] |
| 3. | Honess RW, Roizman B. Regulation of herpesvirus macromolecular synthesis. I. Cascade regulation of the synthesis of three groups of viral proteins. J Virol. 1974;14:8-19. [PubMed] |
| 4. | Ackermann M, Braun DK, Pereira L, Roizman B. Characterization of herpes simplex virus 1 alpha proteins 0, 4, and 27 with monoclonal antibodies. J Virol. 1984;52:108-118. [PubMed] |
| 5. | Blaho JA, Mitchell C, Roizman B. Guanylylation and adenylylation of the alpha regulatory proteins of herpes simplex virus require a viral beta or gamma function. J Virol. 1993;67:3891-3900. [PubMed] |
| 6. | Advani SJ, Hagglund R, Weichselbaum RR, Roizman B. Posttranslational processing of infected cell proteins 0 and 4 of herpes simplex virus 1 is sequential and reflects the subcellular compartment in which the proteins localize. J Virol. 2001;75:7904-7912. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 7. | Weber PC, Spatz SJ, Nordby EC. Stable ubiquitination of the ICP0R protein of herpes simplex virus type 1 during productive infection. Virology. 1999;253:288-298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Canning M, Boutell C, Parkinson J, Everett RD. A RING finger ubiquitin ligase is protected from autocatalyzed ubiquitination and degradation by binding to ubiquitin-specific protease USP7. J Biol Chem. 2004;279:38160-38168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 113] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Gu H, Poon AP, Roizman B. During its nuclear phase the multifunctional regulatory protein ICP0 undergoes proteolytic cleavage characteristic of polyproteins. Proc Natl Acad Sci USA. 2009;106:19132-19137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 10. | Zhu Z, Du T, Zhou G, Roizman B. The stability of herpes simplex virus 1 ICP0 early after infection is defined by the RING finger and the UL13 protein kinase. J Virol. 2014;88:5437-5443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 11. | Everett RD. ICP0, a regulator of herpes simplex virus during lytic and latent infection. Bioessays. 2000;22:761-770. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Hagglund R, Roizman B. Role of ICP0 in the strategy of conquest of the host cell by herpes simplex virus 1. J Virol. 2004;78:2169-2178. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 184] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 13. | Roizman B, Gu H, Mandel G. The first 30 minutes in the life of a virus: unREST in the nucleus. Cell Cycle. 2005;4:1019-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 14. | Boutell C, Everett RD. Regulation of alphaherpesvirus infections by the ICP0 family of proteins. J Gen Virol. 2013;94:465-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 174] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 15. | Everett RD, Boutell C, Hale BG. Interplay between viruses and host sumoylation pathways. Nat Rev Microbiol. 2013;11:400-411. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 132] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 16. | Zhou G, Du T, Roizman B. The role of the CoREST/REST repressor complex in herpes simplex virus 1 productive infection and in latency. Viruses. 2013;5:1208-1218. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 17. | Gelman IH, Silverstein S. Identification of immediate early genes from herpes simplex virus that transactivate the virus thymidine kinase gene. Proc Natl Acad Sci USA. 1985;82:5265-5269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 207] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 18. | Quinlan MP, Knipe DM. Stimulation of expression of a herpes simplex virus DNA-binding protein by two viral functions. Mol Cell Biol. 1985;5:957-963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 77] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 19. | Myers RM, Rio DC, Robbins AK, Tjian R. SV40 gene expression is modulated by the cooperative binding of T antigen to DNA. Cell. 1981;25:373-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 259] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 20. | Nevins JR. Mechanism of activation of early viral transcription by the adenovirus E1A gene product. Cell. 1981;26:213-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 428] [Cited by in RCA: 513] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 21. | Dixon RA, Schaffer PA. Fine-structure mapping and functional analysis of temperature-sensitive mutants in the gene encoding the herpes simplex virus type 1 immediate early protein VP175. J Virol. 1980;36:189-203. [PubMed] |
| 22. | DeLuca NA, McCarthy AM, Schaffer PA. Isolation and characterization of deletion mutants of herpes simplex virus type 1 in the gene encoding immediate-early regulatory protein ICP4. J Virol. 1985;56:558-570. [PubMed] |
| 23. | Stow ND, Stow EC. Isolation and characterization of a herpes simplex virus type 1 mutant containing a deletion within the gene encoding the immediate early polypeptide Vmw110. J Gen Virol. 1986;67:2571-2585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 325] [Cited by in RCA: 350] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 24. | Leib DA, Coen DM, Bogard CL, Hicks KA, Yager DR, Knipe DM, Tyler KL, Schaffer PA. Immediate-early regulatory gene mutants define different stages in the establishment and reactivation of herpes simplex virus latency. J Virol. 1989;63:759-768. [PubMed] |
| 25. | Purifoy DJ, Powell KL. DNA-binding proteins induced by herpes simplex virus type 2 in HEp-2 cells. J Virol. 1976;19:717-731. [PubMed] |
| 26. | Wilcox KW, Kohn A, Sklyanskaya E, Roizman B. Herpes simplex virus phosphoproteins. I. Phosphate cycles on and off some viral polypeptides and can alter their affinity for DNA. J Virol. 1980;33:167-182. [PubMed] |
| 27. | Nabel GJ, Rice SA, Knipe DM, Baltimore D. Alternative mechanisms for activation of human immunodeficiency virus enhancer in T cells. Science. 1988;239:1299-1302. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 203] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 28. | Gius D, Laimins LA. Activation of human papillomavirus type 18 gene expression by herpes simplex virus type 1 viral transactivators and a phorbol ester. J Virol. 1989;63:555-563. [PubMed] |
| 29. | Kwun HJ, Han HJ, Lee WJ, Kim HS, Jang KL. Transactivation of the human endogenous retrovirus K long terminal repeat by herpes simplex virus type 1 immediate early protein 0. Virus Res. 2002;86:93-100. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 49] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 30. | Chen JX, Zhu XX, Silverstein S. Mutational analysis of the sequence encoding ICP0 from herpes simplex virus type 1. Virology. 1991;180:207-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 31. | Everett RD. A detailed mutational analysis of Vmw110, a trans-acting transcriptional activator encoded by herpes simplex virus type 1. EMBO J. 1987;6:2069-2076. [PubMed] |
| 32. | Everett R, O’Hare P, O’Rourke D, Barlow P, Orr A. Point mutations in the herpes simplex virus type 1 Vmw110 RING finger helix affect activation of gene expression, viral growth, and interaction with PML-containing nuclear structures. J Virol. 1995;69:7339-7344. [PubMed] |
| 33. | Freemont PS, Hanson IM, Trowsdale J. A novel cysteine-rich sequence motif. Cell. 1991;64:483-484. [PubMed] |
| 34. | Everett RD, Barlow P, Milner A, Luisi B, Orr A, Hope G, Lyon D. A novel arrangement of zinc-binding residues and secondary structure in the C3HC4 motif of an alpha herpes virus protein family. J Mol Biol. 1993;234:1038-1047. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 91] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 35. | Barlow PN, Luisi B, Milner A, Elliott M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J Mol Biol. 1994;237:201-211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 206] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 37. | Metzger MB, Pruneda JN, Klevit RE, Weissman AM. RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta. 2014;1843:47-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 373] [Cited by in RCA: 460] [Article Influence: 38.3] [Reference Citation Analysis (0)] |
| 38. | Boutell C, Sadis S, Everett RD. Herpes simplex virus type 1 immediate-early protein ICP0 and is isolated RING finger domain act as ubiquitin E3 ligases in vitro. J Virol. 2002;76:841-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 305] [Cited by in RCA: 319] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 39. | Hagglund R, Van Sant C, Lopez P, Roizman B. Herpes simplex virus 1-infected cell protein 0 contains two E3 ubiquitin ligase sites specific for different E2 ubiquitin-conjugating enzymes. Proc Natl Acad Sci USA. 2002;99:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 88] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 40. | Chelbi-Alix MK, de Thé H. Herpes virus induced proteasome-dependent degradation of the nuclear bodies-associated PML and Sp100 proteins. Oncogene. 1999;18:935-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 276] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 41. | Everett RD, Parada C, Gripon P, Sirma H, Orr A. Replication of ICP0-null mutant herpes simplex virus type 1 is restricted by both PML and Sp100. J Virol. 2008;82:2661-2672. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 177] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 42. | Lilley CE, Chaurushiya MS, Boutell C, Landry S, Suh J, Panier S, Everett RD, Stewart GS, Durocher D, Weitzman MD. A viral E3 ligase targets RNF8 and RNF168 to control histone ubiquitination and DNA damage responses. EMBO J. 2010;29:943-955. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 140] [Cited by in RCA: 147] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 43. | Chaurushiya MS, Lilley CE, Aslanian A, Meisenhelder J, Scott DC, Landry S, Ticau S, Boutell C, Yates JR, Schulman BA. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol Cell. 2012;46:79-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 63] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 44. | Orzalli MH, DeLuca NA, Knipe DM. Nuclear IFI16 induction of IRF-3 signaling during herpesviral infection and degradation of IFI16 by the viral ICP0 protein. Proc Natl Acad Sci USA. 2012;109:E3008-E3017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 322] [Cited by in RCA: 354] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 45. | Orzalli MH, Conwell SE, Berrios C, DeCaprio JA, Knipe DM. Nuclear interferon-inducible protein 16 promotes silencing of herpesviral and transfected DNA. Proc Natl Acad Sci USA. 2013;110:E4492-E4501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 151] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 46. | Kawaguchi Y, Van Sant C, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 interacts with and stabilizes the cell cycle regulator cyclin D3. J Virol. 1997;71:7328-7336. [PubMed] |
| 47. | Everett RD, Meredith M, Orr A, Cross A, Kathoria M, Parkinson J. A novel ubiquitin-specific protease is dynamically associated with the PML nuclear domain and binds to a herpesvirus regulatory protein. EMBO J. 1997;16:1519-1530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 319] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 48. | Kawaguchi Y, Tanaka M, Yokoymama A, Matsuda G, Kato K, Kagawa H, Hirai K, Roizman B. Herpes simplex virus 1 alpha regulatory protein ICP0 functionally interacts with cellular transcription factor BMAL1. Proc Natl Acad Sci USA. 2001;98:1877-1882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 49. | Gu H, Roizman B. Herpes simplex virus-infected cell protein 0 blocks the silencing of viral DNA by dissociating histone deacetylases from the CoREST-REST complex. Proc Natl Acad Sci USA. 2007;104:17134-17139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 158] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 50. | Wadsworth S, Jacob RJ, Roizman B. Anatomy of herpes simplex virus DNA. II. Size, composition, and arrangement of inverted terminal repetitions. J Virol. 1975;15:1487-1497. [PubMed] |
| 51. | Perry LJ, Rixon FJ, Everett RD, Frame MC, McGeoch DJ. Characterization of the IE110 gene of herpes simplex virus type 1. J Gen Virol. 1986;67:2365-2380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 156] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 52. | Poon AP, Silverstein SJ, Roizman B. An early regulatory function required in a cell type-dependent manner is expressed by the genomic but not the cDNA copy of the herpes simplex virus 1 gene encoding infected cell protein 0. J Virol. 2002;76:9744-9755. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 53. | Gu H, Zheng Y, Roizman B. Interaction of herpes simplex virus ICP0 with ND10 bodies: a sequential process of adhesion, fusion, and retention. J Virol. 2013;87:10244-10254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 54. | Natarajan R, Deshmane S, Valyi-Nagy T, Everett R, Fraser NW. A herpes simplex virus type 1 mutant lacking the ICP0 introns reactivates with normal efficiency. J Virol. 1991;65:5569-5573. [PubMed] |
| 55. | Weber PC, Wigdahl B. Identification of dominant-negative mutants of the herpes simplex virus type 1 immediate-early protein ICP0. J Virol. 1992;66:2261-2267. [PubMed] |
| 56. | Weber PC, Kenny JJ, Wigdahl B. Antiviral properties of a dominant negative mutant of the herpes simplex virus type 1 regulatory protein ICP0. J Gen Virol. 1992;73:2955-2961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 57. | Everett RD, Cross A, Orr A. A truncated form of herpes simplex virus type 1 immediate-early protein Vmw110 is expressed in a cell type dependent manner. Virology. 1993;197:751-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 94] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 58. | Stevens JG, Wagner EK, Devi-Rao GB, Cook ML, Feldman LT. RNA complementary to a herpesvirus alpha gene mRNA is prominent in latently infected neurons. Science. 1987;235:1056-1059. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 679] [Cited by in RCA: 673] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 59. | Rock DL, Nesburn AB, Ghiasi H, Ong J, Lewis TL, Lokensgard JR, Wechsler SL. Detection of latency-related viral RNAs in trigeminal ganglia of rabbits latently infected with herpes simplex virus type 1. J Virol. 1987;61:3820-3826. [PubMed] |
| 60. | Cui C, Griffiths A, Li G, Silva LM, Kramer MF, Gaasterland T, Wang XJ, Coen DM. Prediction and identification of herpes simplex virus 1-encoded microRNAs. J Virol. 2006;80:5499-5508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 61. | Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR. MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature. 2008;454:780-783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 550] [Cited by in RCA: 539] [Article Influence: 31.7] [Reference Citation Analysis (0)] |
| 62. | Jiang X, Brown D, Osorio N, Hsiang C, Li L, Chan L, Ben–Mohamed L, Wechsler SL. A herpes simplex virus type 1 mutant disrupted for microRNA H2 with increased neurovirulence and rate of reactivation. J Neurovirol. 2015;21:199-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 63. | Lium EK, Silverstein S. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential alpha27 gene. J Virol. 1997;71:8602-8614. [PubMed] |
| 64. | Everett RD. Analysis of the functional domains of herpes simplex virus type 1 immediate-early polypeptide Vmw110. J Mol Biol. 1988;202:87-96. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 111] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 65. | Everett RD. Construction and characterization of herpes simplex virus type 1 mutants with defined lesions in immediate early gene 1. J Gen Virol. 1989;70:1185-1202. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 150] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 66. | Tyers M, Willems AR. One ring to rule a superfamily of E3 ubiquitin ligases. Science. 1999;284:601, 603-604. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 120] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 67. | Joazeiro CA, Weissman AM. RING finger proteins: mediators of ubiquitin ligase activity. Cell. 2000;102:549-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 940] [Cited by in RCA: 965] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 68. | Gu H, Roizman B. The degradation of promyelocytic leukemia and Sp100 proteins by herpes simplex virus 1 is mediated by the ubiquitin-conjugating enzyme UbcH5a. Proc Natl Acad Sci USA. 2003;100:8963-8968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 110] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 69. | Vanni E, Gatherer D, Tong L, Everett RD, Boutell C. Functional characterization of residues required for the herpes simplex virus 1 E3 ubiquitin ligase ICP0 to interact with the cellular E2 ubiquitin-conjugating enzyme UBE2D1 (UbcH5a). J Virol. 2012;86:6323-6333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 70. | Maul GG, Negorev D, Bell P, Ishov AM. Review: properties and assembly mechanisms of ND10, PML bodies, or PODs. J Struct Biol. 2000;129:278-287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 219] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 71. | Everett RD, Chelbi-Alix MK. PML and PML nuclear bodies: implications in antiviral defence. Biochimie. 2007;89:819-830. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 358] [Cited by in RCA: 351] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 72. | Van Damme E, Laukens K, Dang TH, Van Ostade X. A manually curated network of the PML nuclear body interactome reveals an important role for PML-NBs in SUMOylation dynamics. Int J Biol Sci. 2010;6:51-67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 140] [Cited by in RCA: 158] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 73. | Zhong S, Salomoni P, Pandolfi PP. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85-E90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 436] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 74. | Cohen N, Sharma M, Kentsis A, Perez JM, Strudwick S, Borden KL. PML RING suppresses oncogenic transformation by reducing the affinity of eIF4E for mRNA. EMBO J. 2001;20:4547-4559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 141] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 75. | Wang ZG, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi PP. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 402] [Cited by in RCA: 407] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 76. | Bernardi R, Pandolfi PP. Role of PML and the PML-nuclear body in the control of programmed cell death. Oncogene. 2003;22:9048-9057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 77. | Carbone R, Pearson M, Minucci S, Pelicci PG. PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene. 2002;21:1633-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 135] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 78. | Geoffroy MC, Chelbi-Alix MK. Role of promyelocytic leukemia protein in host antiviral defense. J Interferon Cytokine Res. 2011;31:145-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 156] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 79. | Everett RD, Freemont P, Saitoh H, Dasso M, Orr A, Kathoria M, Parkinson J. The disruption of ND10 during herpes simplex virus infection correlates with the Vmw110- and proteasome-dependent loss of several PML isoforms. J Virol. 1998;72:6581-6591. [PubMed] |
| 80. | Everett RD, Rechter S, Papior P, Tavalai N, Stamminger T, Orr A. PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J Virol. 2006;80:7995-8005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 277] [Cited by in RCA: 273] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 81. | Chee AV, Lopez P, Pandolfi PP, Roizman B. Promyelocytic leukemia protein mediates interferon-based anti-herpes simplex virus 1 effects. J Virol. 2003;77:7101-7105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 155] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 82. | Parkinson J, Lees-Miller SP, Everett RD. Herpes simplex virus type 1 immediate-early protein vmw110 induces the proteasome-dependent degradation of the catalytic subunit of DNA-dependent protein kinase. J Virol. 1999;73:650-657. [PubMed] |
| 83. | Everett RD, Earnshaw WC, Findlay J, Lomonte P. Specific destruction of kinetochore protein CENP-C and disruption of cell division by herpes simplex virus immediate-early protein Vmw110. EMBO J. 1999;18:1526-1538. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 200] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 84. | Lomonte P, Sullivan KF, Everett RD. Degradation of nucleosome-associated centromeric histone H3-like protein CENP-A induced by herpes simplex virus type 1 protein ICP0. J Biol Chem. 2001;276:5829-5835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 133] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 85. | Boutell C, Canning M, Orr A, Everett RD. Reciprocal activities between herpes simplex virus type 1 regulatory protein ICP0, a ubiquitin E3 ligase, and ubiquitin-specific protease USP7. J Virol. 2005;79:12342-12354. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 104] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 86. | Grady SL, Hwang J, Vastag L, Rabinowitz JD, Shenk T. Herpes simplex virus 1 infection activates poly(ADP-ribose) polymerase and triggers the degradation of poly(ADP-ribose) glycohydrolase. J Virol. 2012;86:8259-8268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 59] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 87. | Conwell SE, White AE, Harper JW, Knipe DM. Identification of TRIM27 as a novel degradation target of herpes simplex virus 1 ICP0. J Virol. 2015;89:220-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 88. | Kalamvoki M, Gu H, Roizman B. Overexpression of the ubiquitin-specific protease 7 resulting from transfection or mutations in the ICP0 binding site accelerates rather than depresses herpes simplex virus 1 gene expression. J Virol. 2012;86:12871-12878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 89. | Gu H, Roizman B. The two functions of herpes simplex virus 1 ICP0, inhibition of silencing by the CoREST/REST/HDAC complex and degradation of PML, are executed in tandem. J Virol. 2009;83:181-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 90. | Mullen MA, Ciufo DM, Hayward GS. Mapping of intracellular localization domains and evidence for colocalization interactions between the IE110 and IE175 nuclear transactivator proteins of herpes simplex virus. J Virol. 1994;68:3250-3266. [PubMed] |
| 91. | Everett RD, Maul GG. HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 1994;13:5062-5069. [PubMed] |
| 92. | Lopez P, Van Sant C, Roizman B. Requirements for the nuclear-cytoplasmic translocation of infected-cell protein 0 of herpes simplex virus 1. J Virol. 2001;75:3832-3840. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 84] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 93. | Kalamvoki M, Roizman B. Role of herpes simplex virus ICP0 in the transactivation of genes introduced by infection or transfection: a reappraisal. J Virol. 2010;84:4222-4228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 94. | Zhu Z, Cai W, Schaffer PA. Cooperativity among herpes simplex virus type 1 immediate-early regulatory proteins: ICP4 and ICP27 affect the intracellular localization of ICP0. J Virol. 1994;68:3027-3040. [PubMed] |
| 95. | Tanaka M, Kato A, Satoh Y, Ide T, Sagou K, Kimura K, Hasegawa H, Kawaguchi Y. Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J Virol. 2012;86:5264-5277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 96. | Kawaguchi Y, Bruni R, Roizman B. Interaction of herpes simplex virus 1 alpha regulatory protein ICP0 with elongation factor 1delta: ICP0 affects translational machinery. J Virol. 1997;71:1019-1024. [PubMed] |
| 97. | Paladino P, Collins SE, Mossman KL. Cellular localization of the herpes simplex virus ICP0 protein dictates its ability to block IRF3-mediated innate immune responses. PLoS One. 2010;5:e10428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 86] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 98. | Yao F, Courtney RJ. Association of ICP0 but not ICP27 with purified virions of herpes simplex virus type 1. J Virol. 1992;66:2709-2716. [PubMed] |
| 99. | Loret S, Guay G, Lippé R. Comprehensive characterization of extracellular herpes simplex virus type 1 virions. J Virol. 2008;82:8605-8618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 304] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 100. | Sedlackova L, Rice SA. Herpes simplex virus type 1 immediate-early protein ICP27 is required for efficient incorporation of ICP0 and ICP4 into virions. J Virol. 2008;82:268-277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 101. | Delboy MG, Siekavizza-Robles CR, Nicola AV. Herpes simplex virus tegument ICP0 is capsid associated, and its E3 ubiquitin ligase domain is important for incorporation into virions. J Virol. 2010;84:1637-1640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 102. | Delboy MG, Nicola AV. A pre-immediate-early role for tegument ICP0 in the proteasome-dependent entry of herpes simplex virus. J Virol. 2011;85:5910-5918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 103. | Liang Y, Kurakin A, Roizman B. Herpes simplex virus 1 infected cell protein 0 forms a complex with CIN85 and Cbl and mediates the degradation of EGF receptor from cell surfaces. Proc Natl Acad Sci USA. 2005;102:5838-5843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 36] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 104. | Liang Y, Roizman B. State and role of SRC family kinases in replication of herpes simplex virus 1. J Virol. 2006;80:3349-3359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 105. | Zheng Y, Gu H. Identification of three redundant segments responsible for herpes simplex virus 1 ICP0 to fuse with ND10 nuclear bodies. J Virol. 2015;89:4214-4226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 106. | Hay RT. SUMO: a history of modification. Mol Cell. 2005;18:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1249] [Cited by in RCA: 1328] [Article Influence: 66.4] [Reference Citation Analysis (0)] |
| 107. | Geiss-Friedlander R, Melchior F. Concepts in sumoylation: a decade on. Nat Rev Mol Cell Biol. 2007;8:947-956. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1291] [Cited by in RCA: 1426] [Article Influence: 79.2] [Reference Citation Analysis (0)] |
| 108. | Wimmer P, Schreiner S, Dobner T. Human pathogens and the host cell SUMOylation system. J Virol. 2012;86:642-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 109. | Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci USA. 2004;101:14373-14378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 453] [Cited by in RCA: 482] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 110. | Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I. Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem. 2006;281:16117-16127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 433] [Cited by in RCA: 414] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 111. | Sun H, Leverson JD, Hunter T. Conserved function of RNF4 family proteins in eukaryotes: targeting a ubiquitin ligase to SUMOylated proteins. EMBO J. 2007;26:4102-4112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 243] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 112. | Prudden J, Pebernard S, Raffa G, Slavin DA, Perry JJ, Tainer JA, McGowan CH, Boddy MN. SUMO-targeted ubiquitin ligases in genome stability. EMBO J. 2007;26:4089-4101. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 283] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 113. | Boutell C, Cuchet-Lourenço D, Vanni E, Orr A, Glass M, McFarlane S, Everett RD. A viral ubiquitin ligase has substrate preferential SUMO targeted ubiquitin ligase activity that counteracts intrinsic antiviral defence. PLoS Pathog. 2011;7:e1002245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 144] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 114. | Everett RD, Boutell C, Pheasant K, Cuchet-Lourenço D, Orr A. Sequences related to SUMO interaction motifs in herpes simplex virus 1 protein ICP0 act cooperatively to stimulate virus infection. J Virol. 2014;88:2763-2774. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 115. | Ciufo DM, Mullen MA, Hayward GS. Identification of a dimerization domain in the C-terminal segment of the IE110 transactivator protein from herpes simplex virus. J Virol. 1994;68:3267-3282. [PubMed] |
| 116. | Lium EK, Panagiotidis CA, Wen X, Silverstein SJ. The NH2 terminus of the herpes simplex virus type 1 regulatory protein ICP0 contains a promoter-specific transcription activation domain. J Virol. 1998;72:7785-7795. [PubMed] |
| 117. | Meredith M, Orr A, Elliott M, Everett R. Separation of sequence requirements for HSV-1 Vmw110 multimerisation and interaction with a 135-kDa cellular protein. Virology. 1995;209:174-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 71] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 118. | Perusina Lanfranca M, Mostafa HH, Davido DJ. Two overlapping regions within the N-terminal half of the herpes simplex virus 1 E3 ubiquitin ligase ICP0 facilitate the degradation and dissociation of PML and dissociation of Sp100 from ND10. J Virol. 2013;87:13287-13296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 119. | Meredith M, Orr A, Everett R. Herpes simplex virus type 1 immediate-early protein Vmw110 binds strongly and specifically to a 135-kDa cellular protein. Virology. 1994;200:457-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 120. | Li M, Chen D, Shiloh A, Luo J, Nikolaev AY, Qin J, Gu W. Deubiquitination of p53 by HAUSP is an important pathway for p53 stabilization. Nature. 2002;416:648-653. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 758] [Cited by in RCA: 799] [Article Influence: 34.7] [Reference Citation Analysis (0)] |
| 121. | Huang Z, Wu Q, Guryanova OA, Cheng L, Shou W, Rich JN, Bao S. Deubiquitylase HAUSP stabilizes REST and promotes maintenance of neural progenitor cells. Nat Cell Biol. 2011;13:142-152. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 139] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 122. | Song MS, Salmena L, Carracedo A, Egia A, Lo-Coco F, Teruya-Feldstein J, Pandolfi PP. The deubiquitinylation and localization of PTEN are regulated by a HAUSP-PML network. Nature. 2008;455:813-817. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 445] [Cited by in RCA: 437] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 123. | Pfoh R, Lacdao IK, Georges AA, Capar A, Zheng H, Frappier L, Saridakis V. Crystal Structure of USP7 Ubiquitin-like Domains with an ICP0 Peptide Reveals a Novel Mechanism Used by Viral and Cellular Proteins to Target USP7. PLoS Pathog. 2015;11:e1004950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 57] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 124. | Pozhidaeva AK, Mohni KN, Dhe-Paganon S, Arrowsmith CH, Weller SK, Korzhnev DM, Bezsonova I. Structural Characterization of Interaction between Human Ubiquitin-specific Protease 7 and Immediate-Early Protein ICP0 of Herpes Simplex Virus-1. J Biol Chem. 2015;290:22907-22918. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 125. | Gu H, Liang Y, Mandel G, Roizman B. Components of the REST/CoREST/histone deacetylase repressor complex are disrupted, modified, and translocated in HSV-1-infected cells. Proc Natl Acad Sci USA. 2005;102:7571-7576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 164] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 126. | Andrés ME, Burger C, Peral-Rubio MJ, Battaglioli E, Anderson ME, Grimes J, Dallman J, Ballas N, Mandel G. CoREST: a functional corepressor required for regulation of neural-specific gene expression. Proc Natl Acad Sci USA. 1999;96:9873-9878. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 347] [Cited by in RCA: 384] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 127. | Qureshi IA, Gokhan S, Mehler MF. REST and CoREST are transcriptional and epigenetic regulators of seminal neural fate decisions. Cell Cycle. 2010;9:4477-4486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 114] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 128. | Ferenczy MW, Ranayhossaini DJ, Deluca NA. Activities of ICP0 involved in the reversal of silencing of quiescent herpes simplex virus 1. J Virol. 2011;85:4993-5002. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 129. | Everett RD. Depletion of CoREST does not improve the replication of ICP0 null mutant herpes simplex virus type 1. J Virol. 2010;84:3695-3698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 130. | Liang Y, Vogel JL, Narayanan A, Peng H, Kristie TM. Inhibition of the histone demethylase LSD1 blocks alpha-herpesvirus lytic replication and reactivation from latency. Nat Med. 2009;15:1312-1317. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 215] [Cited by in RCA: 238] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 131. | Sherr CJ. D-type cyclins. Trends Biochem Sci. 1995;20:187-190. [PubMed] |
| 132. | Malumbres M, Barbacid M. Cell cycle, CDKs and cancer: a changing paradigm. Nat Rev Cancer. 2009;9:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2982] [Cited by in RCA: 2821] [Article Influence: 176.3] [Reference Citation Analysis (0)] |
| 133. | DeCaprio JA. How the Rb tumor suppressor structure and function was revealed by the study of Adenovirus and SV40. Virology. 2009;384:274-284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 134. | Van Sant C, Kawaguchi Y, Roizman B. A single amino acid substitution in the cyclin D binding domain of the infected cell protein no. 0 abrogates the neuroinvasiveness of herpes simplex virus without affecting its ability to replicate. Proc Natl Acad Sci USA. 1999;96:8184-8189. [PubMed] |
| 135. | Kalamvoki M, Roizman B. ICP0 enables and monitors the function of D cyclins in herpes simplex virus 1 infected cells. Proc Natl Acad Sci USA. 2009;106:14576-14580. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 136. | Van Sant C, Lopez P, Advani SJ, Roizman B. Role of cyclin D3 in the biology of herpes simplex virus 1 ICPO. J Virol. 2001;75:1888-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 137. | Advani SJ, Brandimarti R, Weichselbaum RR, Roizman B. The disappearance of cyclins A and B and the increase in activity of the G(2)/M-phase cellular kinase cdc2 in herpes simplex virus 1-infected cells require expression of the alpha22/U(S)1.5 and U(L)13 viral genes. J Virol. 2000;74:8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 72] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 138. | Schang LM, Rosenberg A, Schaffer PA. Transcription of herpes simplex virus immediate-early and early genes is inhibited by roscovitine, an inhibitor specific for cellular cyclin-dependent kinases. J Virol. 1999;73:2161-2172. [PubMed] |
| 139. | Davido DJ, Von Zagorski WF, Maul GG, Schaffer PA. The differential requirement for cyclin-dependent kinase activities distinguishes two functions of herpes simplex virus type 1 ICP0. J Virol. 2003;77:12603-12616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 140. | Glossop NR, Hardin PE. Central and peripheral circadian oscillator mechanisms in flies and mammals. J Cell Sci. 2002;115:3369-3377. [PubMed] |
| 141. | Kalamvoki M, Roizman B. Circadian CLOCK histone acetyl transferase localizes at ND10 nuclear bodies and enables herpes simplex virus gene expression. Proc Natl Acad Sci USA. 2010;107:17721-17726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 142. | Panier S, Durocher D. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amst). 2009;8:436-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 143] [Cited by in RCA: 143] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 143. | Luijsterburg MS, van Attikum H. Close encounters of the RNF8th kind: when chromatin meets DNA repair. Curr Opin Cell Biol. 2012;24:439-447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 144. | Taylor KE, Mossman KL. Cellular Protein WDR11 Interacts with Specific Herpes Simplex Virus Proteins at the trans-Golgi Network To Promote Virus Replication. J Virol. 2015;89:9841-9852. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 145. | Davido DJ, von Zagorski WF, Lane WS, Schaffer PA. Phosphorylation site mutations affect herpes simplex virus type 1 ICP0 function. J Virol. 2005;79:1232-1243. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 146. | Mostafa HH, Thompson TW, Kushnir AS, Haenchen SD, Bayless AM, Hilliard JG, Link MA, Pitcher LA, Loveday E, Schaffer PA. Herpes simplex virus 1 ICP0 phosphorylation site mutants are attenuated for viral replication and impaired for explant-induced reactivation. J Virol. 2011;85:12631-12637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 147. | Ogle WO, Ng TI, Carter KL, Roizman B. The UL13 protein kinase and the infected cell type are determinants of posttranslational modification of ICP0. Virology. 1997;235:406-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 148. | Maul GG, Ishov AM, Everett RD. Nuclear domain 10 as preexisting potential replication start sites of herpes simplex virus type-1. Virology. 1996;217:67-75. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 222] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 149. | Gu H, Roizman B. Engagement of the lysine-specific demethylase/HDAC1/CoREST/REST complex by herpes simplex virus 1. J Virol. 2009;83:4376-4385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 3.2] [Reference Citation Analysis (0)] |