Published online Nov 12, 2015. doi: 10.5501/wjv.v4.i4.365
Peer-review started: June 2, 2015
First decision: June 18, 2015
Revised: August 1, 2015
Accepted: September 29, 2015
Article in press: September 30, 2015
Published online: November 12, 2015
Processing time: 167 Days and 13.7 Hours
AIM: To develop a real-time reverse transcription-polymerase chain reaction (RT-PCR) assay to genotype rotavirus (G and P) in Alberta from January 2012 to June 2013.
METHODS: We developed and validated a different approach to perform rotavirus G and P genotyping using a two-step SYBR green RT-PCR (rt-gPCR) by selecting genotype-specific primers of published conventional RT nested PCR (cnRT-PCR) assay and optimizing the amplification conditions. cDNA was first synthesized from total RNA with SuperScript™ II reverse transcriptase kit followed by amplication step using monoplex SYBR green real-time PCR. After the PCR reaction, melting curve analysis was used to determine specific genotype. Sixteen samples previously genotyped using cnRT-PCR were tested using the new assay and the genotyping results were compared as sensitivity analysis. Assay specificity was evaluated by testing other gastroenteritis viruses with the new assay. The amplicon size of each available genotype was determined by gel-electrophoresis and DNA sequences were obtained using Sanger-sequencing method. After validation and optimization, the new assay was used to genotype 122 pediatric clinical stool samples previously tested positive for rotavirus using electron microscopy between January 2012 and June 2013.
RESULTS: The new rt-gPCR assay was validated and optimized. The assay detected G1 to G4, G9, G12 and P[4] and P[8] that were available as positive controls in our laboratory. A single and clear peak of melting curve was generated for each of specific G and P genotypes with a Tm ranging from 80 °C to 82 °C. The sensitivity of rt-gPCR was comparable to cnRT-PCR with 100% correlation of the 16 samples with known G and P genotypes. No cross reaction was found with other gastroenteritis viruses. Using the new rt-gPCR assay, genotypes were obtained for 121 of the 122 pediatric clinical samples tested positive for rotavirus: G1P[8] (42.6%), G2P[4] (4.9%), G3P[8] (10.7%), G9P[8] (10.7%), G9P[4] (6.6%), G12P[8] (23.0%), and unknown GP[8] (0.8%). For the first time, G12 rotavirus strains were found in Alberta and G12 was the second most common genotype during the study period. Gel electrophoresis of all the genotypes showed expected amplicon size for each genotype. The sequence data of the two G12 samples along with other genotypes were blasted in NCBI BLAST or analyzed with Rota C Genotyping tool (http://rotac.regatools.be/). All genotyping results were confirmed to be correct.
CONCLUSION: rt-gPCR is a useful tool for the genotyping and characterization of rotavirus. Monitoring of rotavirus genotypes is important for the identification of emerging strains and ongoing evaluation of rotavirus vaccination programs.
Core tip: Genotyping rotavirus is essential for monitoring strain shifts in rotavirus surveillance and vaccine evaluation. Current conventional semi-nested real-time reverse transcription-polymerase chain reaction (RT-PCR), the most commonly used rotavirus genotyping assay is a labor-intensive, complex multi-step procedure and has long turn around-time. The newly developed SYBR Green real time RT-PCR assay is simple, fast and has comparable sensitivity and specificity as conventional semi-nested RT-PCR. This new assay was used to genotype clinical samples which tested positive for rotavirus from January 2012 to June 2013 and new emerging G12 strains were identified in Alberta, Canada.
- Citation: Tong Y, Lee BE, Pang XL. Rapid genotyping of human rotavirus using SYBR green real-time reverse transcription-polymerase chain reaction with melting curve analysis. World J Virology 2015; 4(4): 365-371
- URL: https://www.wjgnet.com/2220-3249/full/v4/i4/365.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i4.365
Rotavirus group A is a major cause of severe acute gastroenteritis in children worldwide, with most children contracting the infection by five years of age[1]. Rotavirus vaccines are most efficacious in protecting children from severe rotavirus gastroenteritis, especially in areas with very low or low child and adult mortality[2]. Two effective rotavirus vaccines, RotaTeq® (Merck and Co., Inc.), a bovine-human reassortant vaccine covering the G1-G4 genotypes along with P[8] and Rotarix® (GlaxoSmithKline, Inc.), a monovalent attenuated human G1P[8] vaccine, were recommended by the WHO in 2009 for the routine immunization of infant globally[3].
The viral genome consists of 11 double-stranded RNA segments, which encode six structure proteins (VP1-4, VP6 and VP7) and six non-structure proteins (NSP1-6). The traditional classification of rotavirus using serotyping has mostly been replaced with G and P genotyping that are based on the diversity of VP7 and VP4 gene sequences, respectively. Twenty-seven G genotypes (G1-G27) and 35 P genotypes (P[1]-P[35]) have been described and six G (G1-4, G9 and G12) and three P (P[4], P[6], and P[8]) genotypes predominate globally with some regional differences[4,5]. Introduction of vaccines or natural evolution of rotavirus may alter antigenic properties of circulating strains in the regions. The antigenic drift between G and P genotypes could result in a decrease of the effectiveness of vaccines against infection in the near future[6,7]. Therefore, understanding the presence and distribution of G and P genotypes and monitoring the emerging and recombination of rotavirus genotypes are very important prior to and after the introduction of rotavirus vaccines.
Various molecular methods have been developed for rotavirus genotyping. Conventional reverse transcriptase (RT) and nested polymerase chain reaction (cnRT-PCR) broadly used since the 90s[8-10] have been improved with more sensitive and specific primers[11,12]. These improved cnRT-PCR methods recognize a broad cluster of various rotavirus G and P genotypes with good specificity. However, there are several drawbacks of these type of assays including: (1) multi-step maneuvers and the requirement of two runs of PCR reactions and gel electrophoresis; (2) labor intensive protocols and a long turn-around time for final results; (3) difficulty with result interpretations due to non-specific amplicons or multiple amplicons in samples with co-infections; and (4) risk of cross contamination of PCR end-products because of the need for open tube maneuvers. With the 2010 recommendation of use of rotavirus vaccine for healthy infants in Canada[13], a rapid and accurate genotyping assay is needed for the evaluation of the vaccine programs in many provinces.
Recently, real time RT-PCR assays in a closed tube system with their rapid turn around-time, sensitivity and specificity and low risk of cross contamination have been implemented in both research and diagnostic laboratories for the detection of rotavirus in stool samples associated with gastroenteritis[14-16]. To date, rotavirus genotyping using TaqMan real time PCR assay has been reported[17]. The protocol could successfully genotype G1P[8], G2P[4], G3P[8], G4P[8] and G9P[8] with only two multiplex reactions, however the other genotypes and combinations were still unable to be typed. Melting curve analysis uses the melting temperature of double-stranded PCR products to determine the identity of the PCR products and also can detect the presence of nonspecific PCR products or primer-dimers. Melting curve analysis is commonly used in SYBR green RT-PCR (rt-gPCR) to determine PCR products and omits the need for gel electrophoresis. SYBR green real-time PCR with melting curve analysis has been used to type Dengue virus as a genotyping tool[18]. This study is to develop and validate a simple and rapid rotavirus genotyping assay using SYBR Green RT-PCR to detect a broad range of rotavirus strains and to use the new assay to characterize the rotavirus strains circulated in Alberta from January 2012 to June 2013.
Sample preparation, RNA extraction and RT reaction: Archived rotavirus positive pediatric stool samples (n = 16) obtained from the Provincial Laboratory for Public Health (ProvLab) and previously genotyed by the cnRT-PCR[10] were used for the development and validation of the rt-gPCR assay. Viral RNA was extracted from 200 μL of 10% stool suspension (10% W/V with PBS buffer) using MagaZorb® total RNA Mini-Prep kit (Promega, Madison, United States) on an automaton extractor, KingFisher™ mL Magnetic Particle Processors, (Thermo Scientific, Mississauga, Canada) according to manufacturer’s instructions. Using SuperScript™ II Reverse Transcriptase kit and random primer (Life technologies, Ontario, Canada), cDNA was synthesized from 5 μL RNA at 42 °C for 1 h, 75 °C for 15 min after heating 5 min at 97 °C and stored at -20 °C before use.
rt-gPCR: A group of published primers used in cnRT-PCR for genotyping rotavirus G types (G1-4, G8, G9-10) and P types (P[4], P[6], P[8], P[9], P[10], and P[11] ) were selected for the new assay development[8-10] (Table 1). Another two sets of primers for genotyping G8, and G12 which were not previously used in our laboratory was also added in the new rt-gPCR assay[12]. 5 μL of 1:10 diluted cDNA was applied to 20 μL of reaction mixture containing 2 μL of LightCycler® FastStart DNA Master SYBR Green I (Roche Diagnostics, Quebec, Canada), 4 mmol/L MgCl2, 0.2 μmol/L each of VP7-R and specific G-typing primers or 0.4 μmol/L each of VP4-F and specific P-typing primers. The rt-gPCR was performed using LightCylcer® 1.0 (Roche Diagnostics, Quebec, Canada) with four-step experimental run protocol: (1) denaturation program (10 min at 95 °C); (2) 45 cycles of amplification program (10 s at 95 °C; 10 s at 53 °C (P-typing), 57 °C (G-typing), and 25 s of extension at 72 °C); (3) melting curve program (0 s at 95 °C, 120 s at 65 °C and 0 s at 95 °C with ramp rate at 0.1 °C per second); and (4) cooling program down to 40 °C. Since different genotype specific primers would yield amplicons of different size with various GC content percentages, the temperature (Tm) and melting curve of the amplicon of specific rotavirus G or P genotype would be different. Thus melting Tm profiles were used to identify specific rotavirus G or P genotypes in our design. For data analysis, the Tm, fluorescence (d[F1]/dT) under the melting curve window was selected as the parameters for evaluation. A sample would be assigned to a specific genotype when the reaction Tm matched with known genotype controls, and the fluorescence d[F1]/dT was above 1.0. The cycle threshold (Ct) of amplification curve was used to provide a relative quantification. Positive controls of specific G and P genotypes were included in each rt-gPCR run as reference genotype and for quality control.
| Primer | Sequence (5’-3’) | Amplicon size (bp) | Direction | Ref. |
| G-type | ||||
| VP7-R | AAC TTG CCA CCA TTT TTT CC | Antisense | Iturriza-Gómara et al[10] | |
| G1 | CAA GTA CTC AAA TCA ATG ATG G | 618 | Sense | Gouvea et al[8] |
| G2 | CAA TGA TAT TAA CAC ATT TTC TGT G | 521 | Sense | Gouvea et al[8] |
| G3 | ACG AAC TCA ACA CGA GAG G | 682 | Sense | Iturriza-Gómara et al[10] |
| G4 | CGT TTC TGG TGA GGA GTT G | 452 | Sense | Gouvea et al[8] |
| G8 | TTR1 TCG CAC CAT TTG TAA TT | 756 | Sense | Aladin et al[12] |
| G9 | CTT GAT GTG ACT AY1A AAT AC | 179 | Sense | Iturriza-Gómara et al[10] |
| G10 | ATG TCA GAC TAC AR2A TAC TGG | 266 | Sense | Gouvea et al[8] |
| G12 | GGT TAT GTA ATC CGA TGG CG | 396 | Sense | Aladin et al[12] |
| P-type | ||||
| VP4-F | TAT GCT CCA GTN3 AAT TGG | Sense | Simmonds et al[11] | |
| P[4] | CTA TTG TTA GAG GTT AGA GTC | 362 | Antisense | Gentsch et al[9] |
| P[6] | TGT TGA TTA GTT GGA TTC AA | 146 | Antisense | Gentsch et al[9] |
| P[8] | TCT ACT GGR2 TTR2 ACN3 TGC | 224 | Antisense | Iturriza-Gómara et al[10] |
| P[9] | TGA GAC ATG CAA TTG GAC | 270 | Antisense | Gentsch et al[9] |
| P[10] | ATC ATA GTT AGT AGT CGG | 462 | Antisense | Gentsch et al[9] |
| P[11] | GTA AAC ATC CAG AAT GTG | 191 | Antisense | Iturriza-Gómara et al[10] |
Assay sensitivity and specificity: The sensitivity of rt-gPCR assay was compared with cnRT-PCR using ten-fold serial dilutions from neat (undiluted) to 10-6 of samples with known G and P genotypes. Other common gastroenteritis viruses including norovirus, sapovirus, adenovirus, and astrovirus were also tested using the G and P primers to determine the specificity of the rt-gPCR assay.
DNA sequencing: Six G-types including G1 to G4, G9, G12 and two P-types P[4], P[8] detected by the rt-gPCR assay were sequenced with modification as described previously[19]. Briefly, the positive PCR products were run in 2.0% agarose gel and purified with QIAquick Gel Extraction kit (Qiagen, Hilden, Germany) then sequenced using 3730 Genetic Analyzer (Applied biosystems, Foster City, United States) at University of Alberta.
ProvLab provides routine diagnostic testing of stool samples submitted for testing of gastroenteritis viruses using electronic microscope (EM). A total of 122 stool samples with rotavirus identified by EM between January 5, 2012 and June 8, 2013 were genotyped using the validated rt-gPCR assay. The purpose was to determine the performance of the assay for rapid genotyping of rotavirus in the clinical setting.
SD for the Tm of each genotype was calculated to show the variations in Tm. The sequence data of amplicons of G1 to G4, G9, G12, P[4] and P[8] were blasted in NCBI or analyzed with Rota C Genotyping tool (http://rotac.regatools.be/).
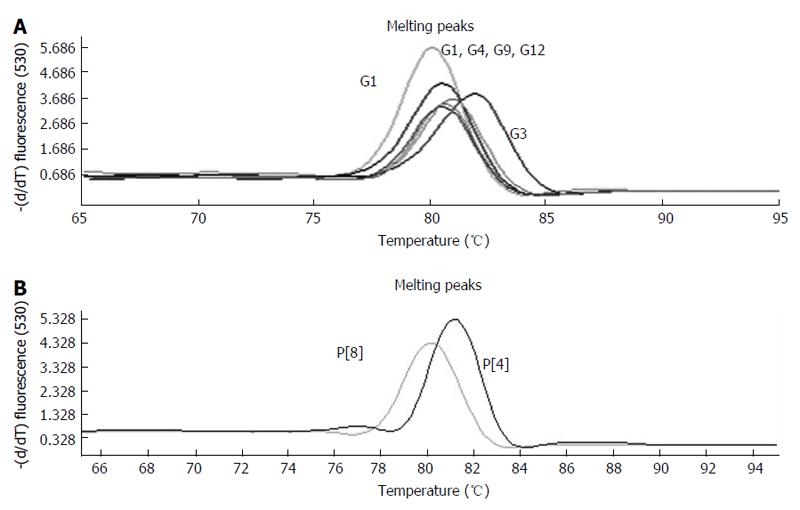
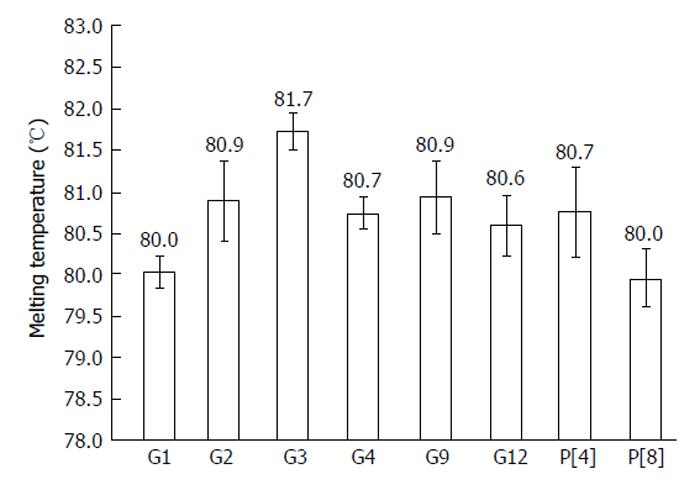
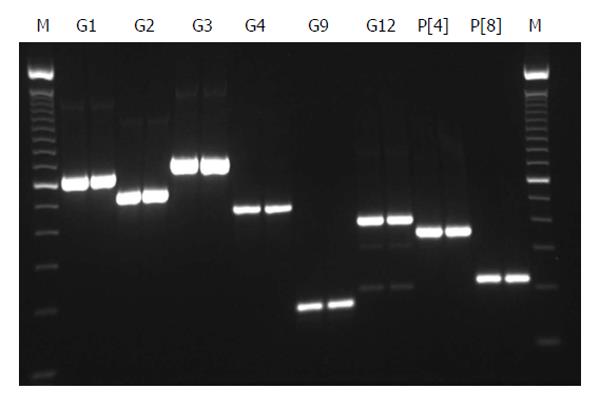
All 16 positive control samples previously genotyped by cnRT-PCR were confirmed using the rt-gPCR assay with 100% concordant results. Using the rt-gPCR assay, a single and clear peak of melting curve was yielded for each of specific G and P genotypes with a Tm ranging from 80 °C to 82 °C (Figure 1). More than 30 of the G1, G3, G9, P[8] and more than 20 of the G2, G4, G12,P[4] replicates and/or different samples at different runs were used to calculate to mean Tm and SD. Each genotype Tm showed very small variation during different PCR runs among different samples. The mean Tm (°C ± SD) for each of the genotypes were: G1 at 80.0 °C± 0.20, G2 at 80.9 °C ± 0.49, G3 at 81.7 °C ± 0.22, G4 at 80.7 °C ± 0.20, G9 at 80.9 °C ± 0.45, G12 at 80.6 °C± 0.37, P[4] at 80.7 °C ± 0.50, and P[8] at 80.0 °C ± 0.34 (Figure 2). Multiplex rt-gPCR could not be performed to simultaneously detect all G or P genotypes because the Tm generated from each genotype was very close. A lower Tm peak (< 77 °C) indicating primer-dimer was observed in the amplification reactions. All rt-gPCR genotype products showed expected amplicon sizes in the gel electrophoresis (Figure 3).
In the sensitivity comparison by serial ten-fold dilution of positive samples, both rt-gPCR and cnRT-PCR detected G2, G3, G4, G9 G12, P[8] at 10-5 dilution. cnRT-PCR detected G1 and P[4] at 10-6 dilution while rt-gPCR detected at 10-5 dilution. For the specificity test, no cross-reaction with other gastroenteritis viruses including norovirus, sapovirus, adenovirus, and astrovirus was observed using the rt-gPCR assay.
The assay was optimized after testing a range of annealing temperatures and primer concentrations. A non-specific amplification was observed for G types at low temperatures (53 °C) but not the P types. The issue with non-specific amplifications was resolved by increasing the annealing temperature to 57 °C and decreasing the concentration of primers to 0.2 μmol/L.
With Rota C Genotyping tool or sequence analysis using NCBI BLAST, all genotyping results generated using rt-gPCR were confirmed to be correct. Two of the G12 positive samples yielded clear reading of 329bp and 335bp respectively had 100% nucleotide identity using NCBI BLAST but could not be directly analyzed using the Rota C Genotyping tool because of the minimal requirement of 500 bp by the tool. The genotyping results of these two samples were further confirmed by analyzing the hits generated using NCBI BLAST with the Rota C tool. By blasting the sequence of these samples in NCBI, the first ten hits showed genotype G12 with 100% nucleotide identity. One of the hits [Rotavirus A strain RVA/Human-wt/ZWE/MRC-DPRU1858/2011/G12P[8] segment 9 capsid glycoprotein VP7 (VP7) gene, complete cds, Genebank: KP753228.1] was further analyzed by the Rota C tool and was confirmed to be G12.
Of the 122 stool samples tested positive for rotavirus by EM, 121 (99%) were characterized by G and/or P genotypes using the rt-gPCR assay except for one sample (1%) that was not typeable. During the study, five G-types were identified, G1, G2, G3, G9 and G12 while only two P types, P[4] and P[8] were found circulating in the province. The predominant G/P combination was G1P[8] at 42.6% (n = 52), followed by G12P[8] (23.0%, n = 28), G3P[8] (10.7%, n = 13), G9P[8] (10.7%, n = 13), G9P[4] (6.6%, n = 8), and G2P[4] (4.9%, n = 6). One sample was typed as P[8] but rt-gPCR for G genotyping did not yield any result. The only untypeable EM positive sample was retested using our in-house rotavirus RT-PCR assay[16] and was found to have a very high Ct of 39 indicating low viral load and possible sample degradation. No mixed infection of different genotypes was found.
We developed a new rt-gPCR assay for genotyping rotavirus and compared the results to the cnRT-PCR assay. The rt-gPCR assay has the same specificity as the cnRT-PCR assay. The sensitivity of rt-gPCR was comparable to cnRT-PCR with 100% correlation of known G and P genotypes samples. The only difference between the two assays was the detection of G1 and P[4] at 10-6 dilution only by cnRT-PCR, which was an expected result as nested PCR could be more sensitive. More importantly, this degree of difference in sensitivity is not significant for clinical applications as high viral load of rotavirus is usually excreted in acute gastroenteritis[15]. Due to the availability of genotypes in our laboratory, the most common G (G1 to G4, G9, G12) and P (P[4] and P[8]) types were validated.
While the rt-qPCR assay has many advantages, including a simpler protocol, shorter turn around-time and lower risk of cross contamination, the multiple monoplex real-time PCR reactions required to genotype respective G and P types have high reagents cost. For cost-saving, a laboratory can design a stepwise testing algorithm to first test for the more common G and P types to reduce reagent and labor costs. Based on our local data and other reported rotavirus genotypes in Canada, we would suggest: For P typing, first test for P[8] and if P[8] is negative, then test for P[4]; for G typing, first test for G1 to be followed by G12 and G9, then G3, G2 +/- G4. This stepwise strategy would cover the range of G and P genotypes for most clinical samples.
The first G12 strain was identified in the Philippines in 1987 followed by reports of sporadic detection in other countries[20-22]. G12 is currently recognized as a globally emerging rotavirus genotype that appears to be spreading more rapidly in recent years[23]. Predominant of G12 was reported in Nepal in 2011, Cameroon in 2010/11 and rotavirus outbreak associated with G12 was found in United States in 2006-07[24-26]. In our study, G12 was detected as the second most prevalent genotype using rt-gPCR in Alberta. G12 could be detected by the new rt-gPCR assay but would be missed by the cnRT-PCR assay. We still believe that G12 was an emerging rotavirus genotype in 2012 as only 1.4% of EM rotavirus positive samples were untypeable from previous nine years in Alberta (data not shown). No G12 has been previously reported in Canada[27]. To our knowledge, this is the first report of G12 genotype detected in Canada. In addition, G9P[4] which was rarely detected in previous years increased dramatically in 2012. These findings emphasize the importance of ongoing surveillance for circulating rotavirus genotypes. In conclusion, the newly developed rt-gPCR assay with optimized primer selection enhanced the detection of broader genotypes which makes this assay a useful tool for the characterization and monitoring of strain shifts in rotavirus surveillance and the evaluation of vaccination program.
We are grateful to ProvLab staff for providing routine diagnostic testing of gastroenteritis viruses by electron microscopy and thank Min Cao for her technical support with the study. We also thank Dr. Jutta K Preiksaitis for scientific discussions and partial financial support.
Genotyping rotavirus is very important for the characterization and monitoring of strain shifts in rotavirus surveillance for the evaluation of vaccination program. The most commonly used rotavirus genotyping assay is a conventional nested reverse transcriptase (RT) polymerase chain reaction (cnRT-PCR) which has been used and revised for more than 30 years. The labor-intensive, complex multi-step procedure and high potential contamination risk due to nested PCR format make this method falling behind current demand.
An accurate, easy-to-perform and rapid rotavirus genotyping tool is needed. Several published studies developed new genotyping PCR assays using multiplex Taqman real time PCR assay to replace conventional nested PCR but has limited sensitivity and can detect only a few G or P types.
Using SYBR Green based RT-PCR with melting curve analysis and melting temperature (Tm) to genotype rotavirus is simple, fast and provides accurate and broad identifications of genotypes. Gel electrophoresis required by traditional conventional nested PCR genotyping assay is not needed and eliminates the risk of contamination from handling post-PCR products. The new assay showed similar sensitivity and specificity as the conventional nested RT-PCR.
The new assay identified common genotypes circulating in Alberta, Canada as well as the emergence of rotavirus G12 in 2012-2013. G12 has never been reported in Canada. During the study, the most predominant rotavirus genotypes were: G1P[8] (42.6%), G12P[8] (23.0%), followed by G3P[8] (10.7%), G9P[8] (10.7%), G9P[4] (6.6%), and G2P[4] (4.9%). These new findings support the importance of ongoing surveillance and characterization of rotavirus genotypes.
Rotavirus G-typing is genotyping viral gene sequences diversity which encoding structure protein VP7; Rotavirus P-typing is genotyping viral gene sequences diversity which encoding structure protein VP4; SYBR green is a commonly used fluorescent DNA binding dye, binds all double-stranded DNA and detection is monitored by measuring the increase in fluorescence throughout the cycle. SYBR Green master mixes are designed for quantitative real-time PCR using a set of two PCR primers that flank the target region; Melting curve analysis is the temperature-dependent dissociation between two DNA-strands can be measured using a DNA-intercalating fluorophore such as SYBR green. Melting Tm can be used for determination of the identity of the target.
The article describes a new assay for genotyping the human rotavirus. The results presented seem pretty convincing and conclusions are valid.
P- Reviewer: Arriagada GL, Giannecchini S, He JY, Pandey KK, Said Z, Song LT, Tetsuya T S- Editor: Tian YL L- Editor: A E- Editor: Liu SQ
| 1. | Parashar UD, Nelson EA, Kang G. Diagnosis, management, and prevention of rotavirus gastroenteritis in children. BMJ. 2013;347:f7204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 2. | Rotavirus vaccines. WHO position paper - January 2013. Wkly Epidemiol Rec. 2013;88:49-64. [PubMed] |
| 3. | Rotavirus vaccines: an update. Wkly Epidemiol Rec. 2009;84:533-540. [PubMed] |
| 4. | Matthijnssens J, Van Ranst M. Genotype constellation and evolution of group A rotaviruses infecting humans. Curr Opin Virol. 2012;2:426-433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 222] [Cited by in RCA: 251] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 5. | Patton JT. Rotavirus diversity and evolution in the post-vaccine world. Discov Med. 2012;13:85-97. [PubMed] |
| 6. | Hull JJ, Teel EN, Kerin TK, Freeman MM, Esona MD, Gentsch JR, Cortese MM, Parashar UD, Glass RI, Bowen MD. United States rotavirus strain surveillance from 2005 to 2008: genotype prevalence before and after vaccine introduction. Pediatr Infect Dis J. 2011;30:S42-S47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 133] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 7. | Gentsch JR, Parashar UD, Glass RI. Impact of rotavirus vaccination: the importance of monitoring strains. Future Microbiol. 2009;4:1231-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 24] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 8. | Gouvea V, Glass RI, Woods P, Taniguchi K, Clark HF, Forrester B, Fang ZY. Polymerase chain reaction amplification and typing of rotavirus nucleic acid from stool specimens. J Clin Microbiol. 1990;28:276-282. [PubMed] |
| 9. | Gentsch JR, Glass RI, Woods P, Gouvea V, Gorziglia M, Flores J, Das BK, Bhan MK. Identification of group A rotavirus gene 4 types by polymerase chain reaction. J Clin Microbiol. 1992;30:1365-1373. [PubMed] |
| 10. | Iturriza-Gómara M, Kang G, Gray J. Rotavirus genotyping: keeping up with an evolving population of human rotaviruses. J Clin Virol. 2004;31:259-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 330] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 11. | Simmonds MK, Armah G, Asmah R, Banerjee I, Damanka S, Esona M, Gentsch JR, Gray JJ, Kirkwood C, Page N. New oligonucleotide primers for P-typing of rotavirus strains: Strategies for typing previously untypeable strains. J Clin Virol. 2008;42:368-373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 12. | Aladin F, Nawaz S, Iturriza-Gómara M, Gray J. Identification of G8 rotavirus strains determined as G12 by rotavirus genotyping PCR: updating the current genotyping methods. J Clin Virol. 2010;47:340-344. [PubMed] |
| 13. | Salvadori M, Le Saux N. Recommendations for the use of rotavirus vaccines in infants. Paediatr Child Health. 2010;15:519-528. [PubMed] |
| 14. | Min BS, Noh YJ, Shin JH, Baek SY, Min KI, Ryu SR, Kim BG, Park MK, Choi SE, Yang EH. Assessment of the quantitative real-time polymerase chain reaction using a cDNA standard for human group A rotavirus. J Virol Methods. 2006;137:280-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 15. | Kang G, Iturriza-Gomara M, Wheeler JG, Crystal P, Monica B, Ramani S, Primrose B, Moses PD, Gallimore CI, Brown DW. Quantitation of group A rotavirus by real-time reverse-transcription-polymerase chain reaction: correlation with clinical severity in children in South India. J Med Virol. 2004;73:118-122. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 112] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 16. | Pang X, Cao M, Zhang M, Lee B. Increased sensitivity for various rotavirus genotypes in stool specimens by amending three mismatched nucleotides in the forward primer of a real-time RT-PCR assay. J Virol Methods. 2011;172:85-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 37] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Kottaridi C, Spathis AT, Ntova CK, Papaevangelou V, Karakitsos P. Evaluation of a multiplex real time reverse transcription PCR assay for the detection and quantitation of the most common human rotavirus genotypes. J Virol Methods. 2012;180:49-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 18. | Chutinimitkul S, Payungporn S, Theamboonlers A, Poovorawan Y. Dengue typing assay based on real-time PCR using SYBR Green I. J Virol Methods. 2005;129:8-15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 19. | Pang XL, Preiksaitis JK, Wong S, Li V, Lee BE. Influence of novel norovirus GII.4 variants on gastroenteritis outbreak dynamics in Alberta and the Northern Territories, Canada between 2000 and 2008. PLoS One. 2010;5:e11599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 20. | Taniguchi K, Urasawa T, Kobayashi N, Gorziglia M, Urasawa S. Nucleotide sequence of VP4 and VP7 genes of human rotaviruses with subgroup I specificity and long RNA pattern: implication for new G serotype specificity. J Virol. 1990;64:5640-5644. [PubMed] |
| 21. | Das S, Varghese V, Chaudhury S, Barman P, Mahapatra S, Kojima K, Bhattacharya SK, Krishnan T, Ratho RK, Chhotray GP. Emergence of novel human group A rotavirus G12 strains in India. J Clin Microbiol. 2003;41:2760-2762. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 77] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 22. | Castello AA, Argüelles MH, Rota RP, Olthoff A, Jiang B, Glass RI, Gentsch JR, Glikmann G. Molecular epidemiology of group A rotavirus diarrhea among children in Buenos Aires, Argentina, from 1999 to 2003 and emergence of the infrequent genotype G12. J Clin Microbiol. 2006;44:2046-2050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 77] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 23. | Soares Lda S, Lobo Pdos S, Mascarenhas JD, Neri DL, Guerra Sde F, de Oliveira Ado S, Maestri RP, Oliveira Dde S, de Menezes EM, Linhares Ada C. Identification of lineage III of G12 rotavirus strains in diarrheic children in the Northern Region of Brazil between 2008 and 2010. Arch Virol. 2012;157:135-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 24. | Ansari S, Sherchand JB, Rijal BP, Parajuli K, Mishra SK, Dahal RK, Shrestha S, Tandukar S, Chaudhary R, Kattel HP. Characterization of rotavirus causing acute diarrhoea in children in Kathmandu, Nepal, showing the dominance of serotype G12. J Med Microbiol. 2013;62:114-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 29] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 25. | Ndze VN, Papp H, Achidi EA, Gonsu KH, László B, Farkas S, Kisfali P, Melegh B, Esona MD, Bowen MD. One year survey of human rotavirus strains suggests the emergence of genotype G12 in Cameroon. J Med Virol. 2013;85:1485-1490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 26. | Mijatovic-Rustempasic S, Teel EN, Kerin TK, Hull JJ, Roy S, Weinberg GA, Payne DC, Parashar UD, Gentsch JR, Bowen MD. Genetic analysis of G12P[8] rotaviruses detected in the largest U.S. G12 genotype outbreak on record. Infect Genet Evol. 2014;21:214-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 27. | McDermid A, Le Saux N, Grudeski E, Bettinger JA, Manguiat K, Halperin SA, Macdonald L, Déry P, Embree J, Vaudry W. Molecular characterization of rotavirus isolates from select Canadian pediatric hospitals. BMC Infect Dis. 2012;12:306. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |