Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.219
Peer-review started: December 6, 2014
First decision: December 26, 2014
Revised: January 24, 2015
Accepted: April 27, 2015
Article in press: April 29, 2015
Published online: August 12, 2015
Processing time: 249 Days and 22 Hours
While human immunodeficiency virus 1 (HIV-1) infection is controlled through continuous, life-long use of a combination of drugs targeting different steps of the virus cycle, HIV-1 is never completely eradicated from the body. Despite decades of research there is still no effective vaccine to prevent HIV-1 infection. Therefore, the possibility of an RNA interference (RNAi)-based cure has become an increasingly explored approach. Endogenous gene expression is controlled at both, transcriptional and post-transcriptional levels by non-coding RNAs, which act through diverse molecular mechanisms including RNAi. RNAi has the potential to control the turning on/off of specific genes through transcriptional gene silencing (TGS), as well as fine-tuning their expression through post-transcriptional gene silencing (PTGS). In this review we will describe in detail the canonical RNAi pathways for PTGS and TGS, the relationship of TGS with other silencing mechanisms and will discuss a variety of approaches developed to suppress HIV-1 via manipulation of RNAi. We will briefly compare RNAi strategies against other approaches developed to target the virus, highlighting their potential to overcome the major obstacle to finding a cure, which is the specific targeting of the HIV-1 reservoir within latently infected cells.
Core tip: The lack of progress in developing an effective human immunodeficiency virus 1 (HIV-1) vaccine has motivated the pressing need for alternate therapies to cure HIV. RNAi therapeutics represent an alternate approach to a functional cure by offering specific targeting of the HIV-1 latent reservoir with the significant advantage of allowing cessation of combination antiretroviral therapy.
- Citation: Méndez C, Ahlenstiel CL, Kelleher AD. Post-transcriptional gene silencing, transcriptional gene silencing and human immunodeficiency virus. World J Virology 2015; 4(3): 219-244
- URL: https://www.wjgnet.com/2220-3249/full/v4/i3/219.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i3.219
Human immunodeficiency virus 1 (HIV-1) infection can be successfully controlled by combination antiretroviral therapy (cART). However, the development of an effective vaccine or an alternative therapy remains the ideal solution since cART has several disadvantages. Adverse effects[1], high costs of therapy, emergence of resistant viruses[2,3] and in particular, the fact that life-long continuous treatment is required[4-6] are just a few examples. Years of research pursuing an HIV-1 vaccine have shown how challenging this task continues to be, with even the most promising trials showing only marginal efficacy[7,8].
Two main obstacles must be overcome to obtain either a vaccine or a cure. First, the high mutation rate of the virus allows extensive accumulation of genetic changes. These genetic changes generate variation with minimal compromise of the virus identity[9-11]; Second, the virus is never eradicated from the body, even after prolonged therapy[6]. While cART has been largely able to deal with the variability of the virus by simultaneously targeting multiple key steps of its replication cycle, it has no direct effect upon latently infected cells[11,12]. The latter, commonly known as latent reservoirs, includes very long-lived resting memory CD4+ T cells[13], macrophages and other cell types[14,15], all of which carry latent proviruses. Provirus refers to the viral form that has been integrated into the cell’s genome and is inherited through each cell division. Latent means it is transcriptionally inactive, but is able to re-activate after stimulation[16-19] and is capable of causing substantial viremia when therapy ceases[20,21].
The viral reservoir, a term used to refer to the latently infected cells as a whole, is maintained throughout the life span of an infected individual. During episodes of low-level viremia and/or homeostatic proliferation of T cells the reservoir seems to be replenished, but contribution of each of these processes is still disputed[21-24].
Latently infected cells are considered the major obstacle to a cure for HIV. They remain immunologically and biochemically silent, becoming invisible to the immune system with no expression of viral antigens on their surface. The only known difference between latently infected cells and un-infected cells is a newly integrated “gene”: the genome of the HIV provirus.
Considerable effort has been put into understanding the molecular mechanisms of latency in order to develop strategies that specifically target either the latently infected cells or directly target the provirus within them. The establishment of latency results from a variety of molecular mechanisms, mainly transcriptional interference and epigenetic mechanisms. It is believed that there is a repressive epigenetic component in most of the inducible proviruses. This component is facultative heterochromatin, a compact yet dynamic state of chromatin that impedes proviral transcription[25-27]. Opposing approaches, which aim to modify the repressive epigenetic profile established at the HIV promoter, have been developed. These either activate proviral transcription by inducing chromatin relaxation or obstruct transcription through stabilization of heterochromatin.
The first strategy has already been tested in cells from HIV infected (+) patients and is currently being tested in a number of clinical trials (http://aidsinfo.nih.gov/clinical-trials/search/b/0/reservoirs and http://aidsinfo.nih.gov/clinical-trials/search/b/0/vorinostat), using pharmacological drugs or cytokines that directly and/or indirectly induce activation of HIV provirus through a variety of cellular pathways[28-30]. However, while viral transcripts from apparently latently infected cells have been detected, no significant change or reduction in the size of the latent reservoir-proviral integrated DNA-has been observed[30,31]. There is currently a debate as to whether these cell associated viral RNA transcripts represent transcripts driven by the endogenous HIV promoter, the 5’LTR or whether these are so called “read through transcripts” which arise from altered expression from the promoter of the parent gene into which HIV has integrated[32]. The results of further trials of these agents are awaited.
The second strategy is based on RNAi and has the advantage of being specifically directed to viral mRNAs or the provirus regardless of the cell type infected. Aiming to target persistent infection in the first place, most RNAi approaches are designed to directly cleave HIV mRNAs and were first designed in the early 2000s. Significant advances have transpired in the field, beginning from those manipulating PTGS to target viral mRNAs and cellular cofactors that support HIV replication, to those using TGS to induce heterochromatin at the HIV promoter. In this review we will discuss both PTGS and TGS RNAi based approaches for HIV, and provide a brief commentary on other gene therapy alternatives currently under development.
RNAi is an evolutionarily conserved mechanism that is present from lower eukaryotes through to mammals. Because it is beyond the scope of this review to discuss each of these, we will mainly focus on the mammalian RNAi pathways. However, we will also include some other species-specific examples to illustrate pertinent points.
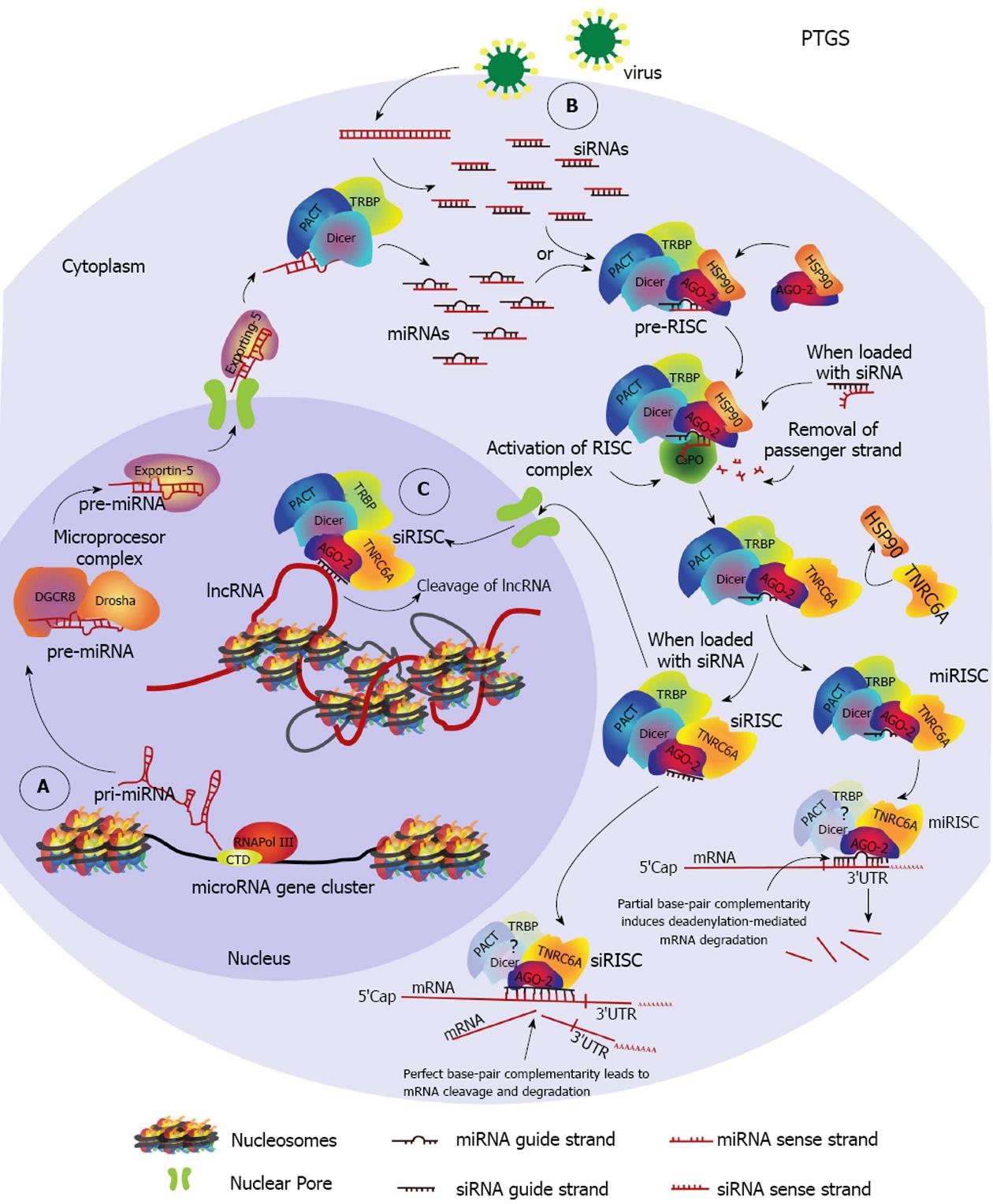
The first evidence of RNAi was reported in transgenic tobacco plants expressing antisense or sense RNAs from the coat-protein gene of the tobacco etch virus (TEV)[33,34]. The plants did not show evidence of infection after challenged with TEV, suggesting the presence of a protective nucleic acid-dependent mechanism that was later proved to spread throughout the plant in a systemic way (reviewed in[35]). The precise mechanism was described in the worm Caenorhabditis elegans (C. elegans), in which interference of endogenous gene expression through inoculation of homologous dsRNA molecules was demonstrated and the involvement of a catalytic and an amplification event was suggested[36-38]. It was further demonstrated that RNA interference, as it began to be known, resulted in genetic silencing and co-suppression of the targeted gene[37,38]. Following this discovery, vast exploitation of RNAi for discovery of gene function in reverse genetics of mammalian cells began and soon after was developed as a therapeutic tool, with several clinical trials currently underway for a variety of human diseases (http://www.clinicaltrials.gov/ct2/results?term=RNAi&Search=Search)[39]. This RNAi pathway is known as PTGS (Figure 1). It functions in the cytoplasm and impedes translation of an mRNA into protein, by direct cleavage or by initiating degradation of the targeted mRNA sequence.
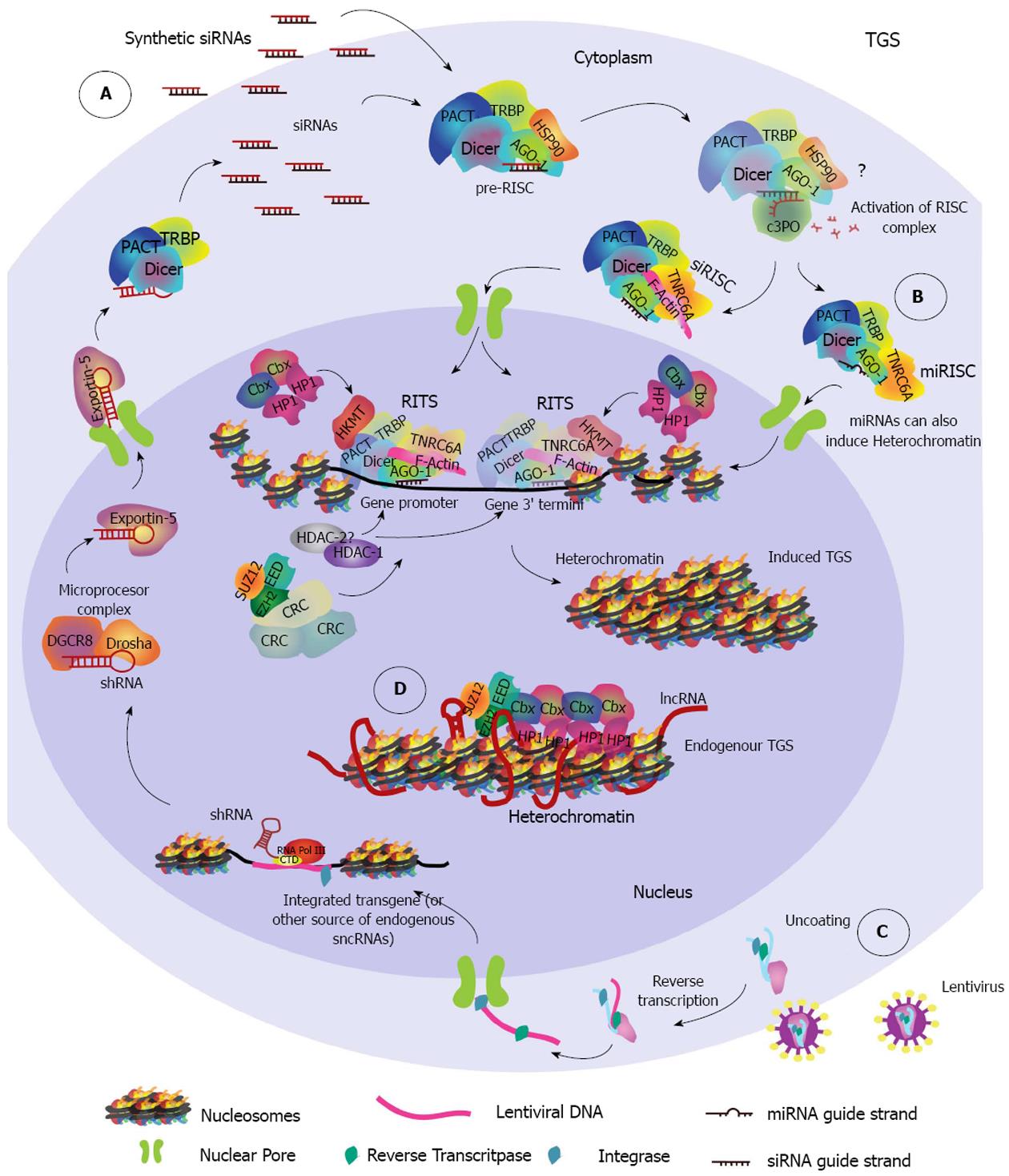
It was not until 2004 that the nuclear RNAi pathway TGS, involving chromatin compaction, was identified[40] (Figure 2). Presently, both PTGS and TGS have been found to be functional in the nucleus of mammalian cells, but only TGS seems to repress gene transcription directly through chromatin remodeling.
During RNAi small non-coding RNAs (sncRNAs) are used as guides through sequence homology to target either mRNA transcripts or gene promoters[41-43]. These sncRNAs are loaded into Argonaute proteins forming the main effector complex; however, other cellular cofactors are required for the process to occur. There are three major kinds of sncRNAs involved in RNAi: small interfering RNAs (endogenous- and exogenous-siRNAs), microRNAs (miRNAs) and piwi-associated RNAs (piRNAs)[44], which we will describe briefly in the next section. The Argonaute proteins are further subdivided into the Argonaute subfamily (AGO-1, AGO-2, AGO-3 and AGO-4, in humans), and the Piwi subfamily (HILI or PIWIL2, HIWI1 or PIWIL1, HIWI2 or PIWIL4 and HIWI3, in humans)[44,45]. There are also species-specific AGO proteins that we will not discuss (recently reviewed in[46]), with the exception of specific examples.
Recently, many novel non-canonical sncRNAs involved in RNAi have been discovered[47-49]. However we will only focus on the three major classes previously mentioned. SncRNAs are generally classified depending on their biogenesis (Dicer/Drosha dependent or independent), their size (about 21-30 nt) and the Argonaute protein they bind (AGO 1-4). They can be endogenous or exogenous depending on their origin. The endogenous sncRNAs are produced from transcription units (Figure 1A), protein coding genes (exons and introns), convergent promoters, long non-coding RNAs (lncRNAs), gene clusters, repetitive elements or retro-elements, such as transposons, while the exogenous sncRNAs are either synthetic or of viral origin (Figure 1B).
SiRNAs: Exo-siRNAs and endo-siRNAs: SiRNAs are about 21-nt long duplexes generated in the cytoplasm by cleavage of endogenous or exogenous long dsRNA precursors (e.g., lncRNA) by the endonuclease Dicer (Figure 1B). These siRNAs are then loaded onto a specific AGO protein. When exogenous, synthetic siRNAs may be delivered to the cells by transfection/nucleofection protocols or may originate from expression of artificial integrated constructs (lentivirus transduction), such as short-hairpin RNAs (shRNAs). Naturally occurring exo-siRNAs in mammalian cells were not discovered until very recently, and were found to originate from dsRNA intermediates of viral replication in mouse embryonic stem cells[50]. On the other hand, mammalian endo-siRNAs were identified in somatic tissue and found to be processed through a non-canonical Drosha-independent mechanism, from Dicer cleavage of a long nuclear hairpin RNA expressed from short interspersed nuclear elements (SINEs)[51].
Generally, siRNAs direct the cleavage of their cognate mRNA through PTGS when they mutually base pair with perfect complementarity[52-54]. Mutations in the siRNA or in the target region within the mRNA sequence usually reduce or abolish silencing, which is why RNAi is considered a very specific mechanism[55,56]. In addition, siRNAs can also induce TGS, a nuclear RNAi pathway, whenever they target a complementary sequence within the promoter or the 3’ end of a gene[57,58]. Additionally, siRNA-directed transcriptional gene activation (TGA) has also been reported for several genes[57,59].
miRNAs: In their canonical pathway miRNAs are first transcribed as primary-miRNAs (pri-RNA) by RNA Pol II, and are then processed by the nuclear RNAse III protein Drosha - an RNAse type III enzyme (Figure 1A). Drosha and co-factor DiGeorge syndrome critical region gene 8 (DGCR8) form the Microprocessor complex. This complex generates precursor-miRNAs (pre-miRNA) that are further processed, exported to the cytoplasm by Exportin 5, and cleaved by Dicer. Dicer generates 22-nt miRNA duplexes that are analogous to siRNA duplexes. Non-canonical pathways exist which are Drosha, Dicer or DGCR8 independent. Importantly, unlike siRNA-duplexes, miRNA-duplexes frequently contain mismatches and about the first 2-7 nts at the 5’ end of the guide strand, known as the seed region, may target the 3’ untranslated (3’ UTR) region of multiple mRNAs. Based on this multiple targeting ability, other biochemical characteristics and evolutionary conservation, microRNAs are clustered into families (http://www.mirbase.org/index.shtml). In miRNAs, complete base pair complementarity with target mRNA is found within this seed region, which allows for mismatches towards the 3’ end. MiRNAs predominantly direct deadenylation-dependent mRNA-decay that results in translational repression, but they are also able to induce sequestration. While common in plants, in mammals on rare occasions when miRNAs show complete complementarity to the target region they can induce cleavage of the mRNA[60-62]. Deadenylation and other ways of translational repression and sequestration result from partial complementarity between miRNA and the target mRNA[44,47,63,64]. In a similar way to siRNAs, when mature miRNAs show complete homology to a promoter region, they are able to induce TGS[65-67] (Figure 1C). However, this miRNA pathway is not well described due to the few cases that have been reported.
piRNAs: PiRNAs are longer (about 25-31 nt) than siRNAs or mature miRNAs and have fundamental roles in maintenance of stemness, transgenerational inheritance and genome instability through targeting of repetitive sequences (e.g., endogenous retroviruses and transposon elements), among other functions (reviewed in[68]). In mammals, piRNAs are expressed in germ cells and somatic germ cells (SGC), but their role in somatic stem cells, such as hematopoietic stem cells, remains controversial[69,70]. Even though there is expression of piwi-pathway-specific AGO proteins in human CD34+ stem cells[71], further evidence is still required to confirm a functional piRNA pathway in somatic stem cells different to SGCs.
Interestingly, piRNAs direct specific genome rearrangements in ciliates and this precise genome editing results in either somatic elimination[72] or retention[73], indicating the versatility of this particular RNAi pathway. PiRNAs can target mRNAs through a PTGS-like mechanism, however they may also induce TGS by directing heterochromatin formation at the target regions[74,75]. Intriguingly, members of the piRNA pathway are highly expressed in certain human cancer cells (reviewed in[76]), though it is still unknown whether they are the cause or the effect. To our knowledge there are no reports regarding the use of synthetic piRNAs and since they have not been manipulated for human therapy we will not explore these further. However, the ability of piRNAs to establish a permanent, stable and inheritable silencing through directed epigenetic chromatin modifications and other mechanisms makes them of great interest for future study, especially since silencing mediated through their activities is inherited to every single cell of a multicellular organism.
PTGS: In humans, RNAi induced mRNA cleavage is directed only by AGO-2[54,77]. Loading of the sncRNA duplex onto the AGO proteins is well described for AGO-2 and involves the heat-shock protein 90 (HSP90) (Figure 1). HSP90 aids in the recruitment[78] and stabilization[79] of unloaded AGO within processing bodies (P-bodies). Inhibition of HSP90 results in unpaired siRNA- and/or miRNA-dependent silencing, respectively. These P-bodies are cytoplasmic structures that contain mRNA decay factors, untranslated mRNA, translational repressors and RNAi related factors[80]. The active silencing complex is named RISC or miRISC, depending on the type of sncRNA (siRNAs or miRNAs, respectively) that is loaded onto the AGO protein. We will refer to both as RISC, unless specified.
The sncRNA-duplex/AGO-2 complex is called pre-RISC (pre-RNA-induced silencing complex) and requires the removal of one of the strands of the RNA, the passenger strand, in order to become an active RISC complex[81,82] (Figure 1). The strand that remains in RISC is known as the guide strand. Passenger/guide strand selection depends on the individual thermodynamic properties of each sncRNA molecule within the duplex; these properties generally create energetic asymmetry between the duplex ends, allowing differentiation and selection of the guide strand[83]. Asymmetry means that the duplex is energetically less stable at one 5’ end and causes unwinding to begin at this site. As a result, the strand whose 5’ end lies in the less stable end of the duplex will be loaded onto the AGO protein, becoming the guide strand. Whenever the energetic difference between the duplex ends is small or negligible, both strands may be randomly loaded[83,84].
Dicer seems to play a role in sensing and positioning the guide strand, facilitating removal of the passenger strand. This ability appears to be activated through its interaction with Transactivation Response (TAR) RNA-Binding Protein (TRBP) and Protein Kinase RNA (PKR) Activator (PACT), both double-stranded RNA binding proteins (dsRBP)[85] (Figure 1). However, there is contradictory evidence regarding this role for Dicer[85,86], and further research may be needed to clarify these observations. Nonetheless, it is important to mention that proper selection of the guide ensures specificity towards silencing the intended target.
Pre-RISC activation requires the slicer activity of AGO-2, specifically the nicking of the passenger strand[87-89]. After nicking, the endonuclease component 3 promoter of RISC complex (C3PO), composed of Trax and Translin proteins in humans, is able to cleave and remove the passenger strand[87,90,91]. This results in activation of pre-RISC into the RISC complex. Within RISC, the guide strand is used to scan mRNAs for a region with full or partial base pair complementarity. Once the region is found the mRNA is either cleaved, deadenylated or stored during translational repression[63,92,93] (Figure 1). Storage and repression of translation may have a role in gene regulation of processes that require a quick response, as translation can be initiated from stored transcripts rather than relying on de novo transcription[94].
While AGO-2 can direct either cleavage or translational repression of mRNAs, non-catalytic AGO proteins like AGO-1 seem mostly involved in translational repression, since they are unable to cleave mRNA transcripts[46]. Furthermore, it is currently unknown how RISC activation occurs for non-catalytic AGO proteins (AGO-1, 3 and 4). However, owing to their inability to cleave mRNAs, activation of the silencing complex must either be different or rely on help from additional cofactors.
For silencing to occur, human AGO-2 requires direct binding with TNRC6A, also known as GW182, a mRNA binding protein rich in glycine/tryptophan repeats[95,96]. Interaction between GW182 and AGO-2 proteins is crucial for miRNA-mediated silencing and appears to take place directly after the passenger strand is removed by C3PO[96,97]. Both AGO-2 and GW182/TNRC6A have been shown to co-localize with siRNA and miRNAs within GW bodies (GWB)[98], another term for P-bodies. Based on these observations, it has been proposed that silencing by miRNAs requires an effector complex formed of at least one AGO and one GW182/TNRC6A protein[99] (Figure 1).
Interestingly, human GW182/TNRC6A was found to transport AGO-2 proteins to the nucleus during miRNA-induced silencing of a nuclear non-coding RNA[100]. The latter constitutes evidence of a nuclear PTGS pathway (Figure 1C). Indeed, increasing evidence supports a functional nuclear PTGS pathway, with a recent study demonstrating not only the presence of PTGS related proteins in the nucleus of mammalian cells, but an active AGO-2-RISC complex able to efficiently cleave two nuclear lncRNAs, Malat1 and Neat1[101]. These studies add to previous evidence indicating the existence of a nuclear RNAi pathway and suggest that PTGS and TGS may be closely related.
We have aimed to provide a detailed overview of the molecular mechanism of PTGS in order to understand the unknowns of TGS and further compare the manipulation of PTGS or TGS for HIV gene therapy. PTGS pathways have been exploited for HIV therapeutics, and several clinical trials are currently testing PTGS-based gene therapy approaches directed to cellular and viral transcripts. A major disadvantage of using PTGS to treat HIV is that PTGS requires viral transcription because it acts on mRNAs. First, this gives the virus the chance to evolve resistance mutations and escape silencing; Second, latent proviruses will not be targeted since they are not undergoing active transcription. Improvements in siRNA/miRNA design and expression have been developed aimed at overcoming these caveats and will be discussed in the HIV-1 section.
TGS: TGS is a conserved mechanism of gene regulation across species and has been extensively studied in the plant model Arabidopsis thaliana (A. thaliana), the worm model Caenorhabditis elegans (C. elegans), and the fission yeast Schizosaccharomyceae pombe (S.pombe). The first evidence for TGS was observed in plants, and it was found to require siRNA-induced DNA methylation for heterochromatin formation (recently reviewed in[102]). However, the mechanism in S.pombe shed insight on the identification of TGS in mammals. In this microorganism, siRNAs generated from centromeric repeats are first processed by Dicer, then loaded onto AGO-1, and together with the proteins Chp1 and Tas3 form the silencing complex, namely the RNA-induced initiator of transcriptional gene silencing (RITS) complex. This RITS complex is analogous to the RISC complex from PTGS. RITS is then directed through siRNA base pair complementarity to a specific locus at which it induces recruitment of Clr4 (histone methyltransferase) and Swi6 (chromo domain binding protein) in order to establish and spread heterochromatin domains[103-105].
Human sncRNA-directed TGS is mainly, but not exclusively directed by AGO-1 rather than AGO-2, and is generally triggered by promoter-targeted sncRNAs (Figure 2). Recent evidence suggests that it may be also triggered by sncRNAs that target the 3’ termini of genes[58,106]. While increasing evidence suggests a role for AGO-2 in nuclear gene silencing[100], it seems to be predominantly through a nuclear PTGS that involves RNA cleavage[101], with only few described exceptions[107]. There is also evidence of RNA-induced nuclear silencing without heterochromatin formation, involving both AGO1 and AGO2[108], and there seems to be various RNA-directed nuclear-pathways that control transcription at different stages[109]. However, it is generally accepted that heterochromatin and its associated markers (i.e., histone methylation and deacetylation) is a characteristic feature of TGS. Therefore, for this review we will focus on the different endogenous TGS mechanisms that involve heterochromatin formation induced by sncRNAs loaded into an AGO protein.
Heterochromatin is considered a hallmark of repressive silent chromatin, ubiquitous in eukaryotic organisms. In mammals, its establishment at a particular locus is a result of protein interactions and cross talk with multiple silencing mechanisms such as DNA methylation, genomic imprinting and Polycomb group of proteins (PcG)[65,103,110]. The epigenetic profiles across mammalian genomes are very heterogeneous and show a wide range of silencing dynamics. Silencing extends from permanent and inheritable to inducible, dynamic silencing. The former is mainly but not restricted to, constitutive heterochromatin and is found in centromeres and telomeres[111]; while the latter, predominantly within facultative heterochromatin, controls specific gene expression during differentiation and development[112].
SncRNA-directed TGS in mammalian cells has been a controversial topic since its discovery, nearly a decade ago, with some still doubting its existence. These doubts have relied on the inability to explain in detail the molecular mechanisms driving TGS. In particular, the much awaited identification and characterization of a functional nuclear mammalian RITS complex, because there are apparently no RNAi proteins with homology to Tas3 and Chp1 present in the nucleus and AGO-1 is non-catalytic. At present, most of the evidence of mammalian sncRNA-AGO-1 directed TGS relies on synthetic siRNAs or shRNAs driving TGS to control infectious agents, such as HIV-1[113], or cellular genes that support viral replication[114]. Nonetheless, the relatively slow accumulation of evidence has supported the existence of this functional pathway, with evidence for miRNA-induced TGS in senescence[107] and in differentiation[65]. We will explain the basis for the doubts and show the recent evidence supporting mammalian sncRNA-directed TGS.
The breakthrough proving the existence of a TGS mechanism in mammalian cells came with the identification of the human ortholog for Clr4, known as Suppressor of variegation (Su(var)3-9) in D. melanogaster and Su(var)39H in humans; and then with the ortholog for Swi6, known as Histone Protein 1 - alpha (HP1-α) (in both D. melanogaster and humans)[115-117]. However, no human orthologs for Chp1 and Tas3 proteins from fission yeast RITS complex have been yet identified. At present, there are more questions than answers about the series of events in humans that result in siRNA-AGO-1 mediated heterochromatin formation and activation of the RITS complex. It is possible that both PTGS and TGS share a core multi-protein complex, which may differ in accessory subcellular or pathway-specific co-factors, because the initial steps of TGS may potentially resemble those of PTGS.
There is also controversy regarding the activation of the RITS complex during TGS. It is assumed that removal of the passenger strand occurs during TGS to allow the RITS complex to scan for the target sequence that is complementary to the guide strand. However, since AGO-1 lacks the catalytic amino acid tetrad DEDH responsible for the slicing function, it is not clear how this process occurs[118]. AGO-1 needs to nick the passenger strand from the siRNA duplex, so C3PO or a similar complex would be able to remove the passenger strand.
On one side, it was shown in vitro from bacterially expressed human AGO proteins, that AGO-1 is able to cleave the passenger strand, but requires assistance for removal of the cleaved fragments[119]. This has been interpreted as non-catalytic AGO proteins being very inefficient catalysts and having an extremely low nickase activity.
This is in agreement with findings in mouse embryonic stem cells, in which the absence of the four mammalian AGO proteins resulted in apoptosis, but the expression of any one of the other AGO proteins in isolation, was enough to rescue the cells and restore a functional RNAi pathway, showing evidence for functional redundancy[120]. In addition, another study showed that non-catalytic AGO proteins are loaded within the duplex but removal of passenger strand takes place approximately 2 to 3 d[121]. The process of passenger strand removal is currently unknown.
In contrast, the crystallographic structures of human AGO-1 in association with endogenous RNA (1.75 Å) and in association with Let-7 miRNA (2.5 Å) were used to show that while highly similar to hAGO-2-RNA structures, there was an absolute requirement for the introduction of the catalytic tetrad by introduction of a single point mutation as well as the reconstitution of a loop called PL3, in order to restore the slicer functionality of AGO-1[122]. These observations argue against a catalytic role for AGO-1.
It seems more likely that other proteins aid non-catalytic AGOs during this step. These cofactors would be present in the AGO knockout mice study and in the cells used to show removal of passenger strand after a few days, but not in the bacterial system, in which cleaved fragments remained loaded to the AGO proteins. Comprehensive studies are required to address this question definitively.
An increasing number of studies have found PTGS-related proteins in the nucleus of mammalian cells, such as GW182/TRNC6A and the endonucleases hC3PO and Dicer[100,123-125]. These proteins appear to have functions related to both to the mechanisms underpinning PTGS in the nucleus and to the regulation of chromatin and transcription.
For example, human Dicer has been shown to interact with NU153, a non-canonical nuclear transport nucleoporin, as demonstrated by co-localization within the nucleus[125]. In addition, human Dicer has been shown to associate with the chromatin structures of ribosomal DNA[124]. It also has a role in termination of transcription[126], in regulation of intergenic transcription in the human β-globin gene cluster[127] and in regulation of nuclear receptor (NR) signaling, as evidenced by direct binding of Dicer to NR promoter regions[128]. Further, Dicer has been reported to be required in heterochromatin formation in fission yeast[129] and in vertebrates[130], suggesting its presence in the nucleus of human cells could be due to an as yet unidentified role in mammalian TGS (Figure 2B).
We previously mentioned that GW182/TNRC6A shuffles AGO-2 proteins between the nucleus and cytoplasm through a non-canonical nuclear localization signal[100]. Additionally, GW182/TNRC6 associates with all four RNA loaded-AGO proteins during PTGS. Therefore, it is a possibility that Dicer is contained within a loaded AGO-1-TRNC6A complex during the nuclear shuffling that occurs during TGS[131] (Figure 2). Furthermore, the interaction between GW182/TNRC6 and AGO-1 occurs through binding of the GW repeats of GW182/TNRC6 to the Piwi domain of AGO-1[77]. This is intriguing because the fission yeast RITS member protein, Tas3, has a GW-repeat-containing motif and interacts with AGO-1 to promote TGS[132]. It is therefore possible that, the Tas3/AGO-1 interaction in fission yeast could be analogous, not homologous, to the AGO-1 and GW182/TNRC6 interaction in humans. Consistent with this hypothesis, the plant specific PTGS-related GW protein NERD was found to be involved in TGS in A. thaliana[133]. Thus, there is evolutionary evidence supporting the likelihood of a link between the two mammalian pathways in the nucleus.
Protein complexes containing AGO-2, TNRC6A, Dicer and TRBP have been immunoprecipitated from human isolated cell nuclei. These protein complexes were able to induce PTGS, with the specific cleavage of four different nuclear lncRNAs mediated by corresponding siRNAs[101]. Similar complexes were immunoprecipitated with nuclear AGO-1 and found to harbor the same PTGS proteins, supporting the notion of a core complex for both pathways. However, this study did not identify proteins that have been implicated in the loading of sncRNA onto AGO proteins, such as C3PO and HSP90 within mammalian cell nuclei. In previous studies the identification of these proteins could have been the result of contamination from cytoplasmic remnants. This study highlighted the importance of ensuring that isolated nuclei are free from endoplasmic reticulum (ER) to avoid contamination with cytoplasmic AGO-containing complexes. Recently, a comprehensive protocol was developed to ensure that purified nuclei are free from ER contamination[134].
It is important to note that the majority of studies aimed at understanding the mechanisms of loading and activation of silencing complexes incorporating non-catalytic AGO proteins have done it in the context of PTGS, either in the cytoplasm or in the nucleus. These studies have not specifically targeted genes embedded in chromatin. Therefore, a possibility remains that siRNAs or miRNAs that are only homologous to specific regions such as promoter regions, can be identified and differentially processed. In this way, complexes could share a common core, but would vary in accessory proteins that modify their function to induce either TGS or PTGS.
Consistent with this model, a recent study unveiled a sorting mechanism in humans, which directs differential loading of AGO-1 proteins for unique sncRNAs in the setting of a viral infection[135]. However the determinants of this selection remain unknown. Nonetheless, most sncRNAs were loaded in equivalent ratios to AGO-1 and AGO-2 proteins and thus these unsorted sncRNAs may be used to scan targets in both cellular compartments. Therefore we hypothesize that when there are targets in both compartments, both pathways are likely to occur, depending how efficient each of these sncRNAs is for the pathway.
While the understanding of the molecular mechanisms of PTGS is reasonably complete, and there is some evidence of commonalities with TGS, there are far many more uncertainties in the TGS mechanism. Several important early steps in the TGS mechanism remain to be fully deciphered, including the precise mechanism that determines RITS recognition of target, the characteristics or type of target and the determinants of induction of different epigenetic heterochromatin profiles. In addition, while human TGS can be thought of at a single cell level, its implication needs to be considered within the context of a multicellular organism. Many changes or epigenetic check points occur early during embryogenesis and development or during cell differentiation. While some changes are dynamic allowing differentiation of cells down different pathways, once certain check points are reached epigenetic profiles are more stable and are inherited to daughter cells through multiple cell divisions.
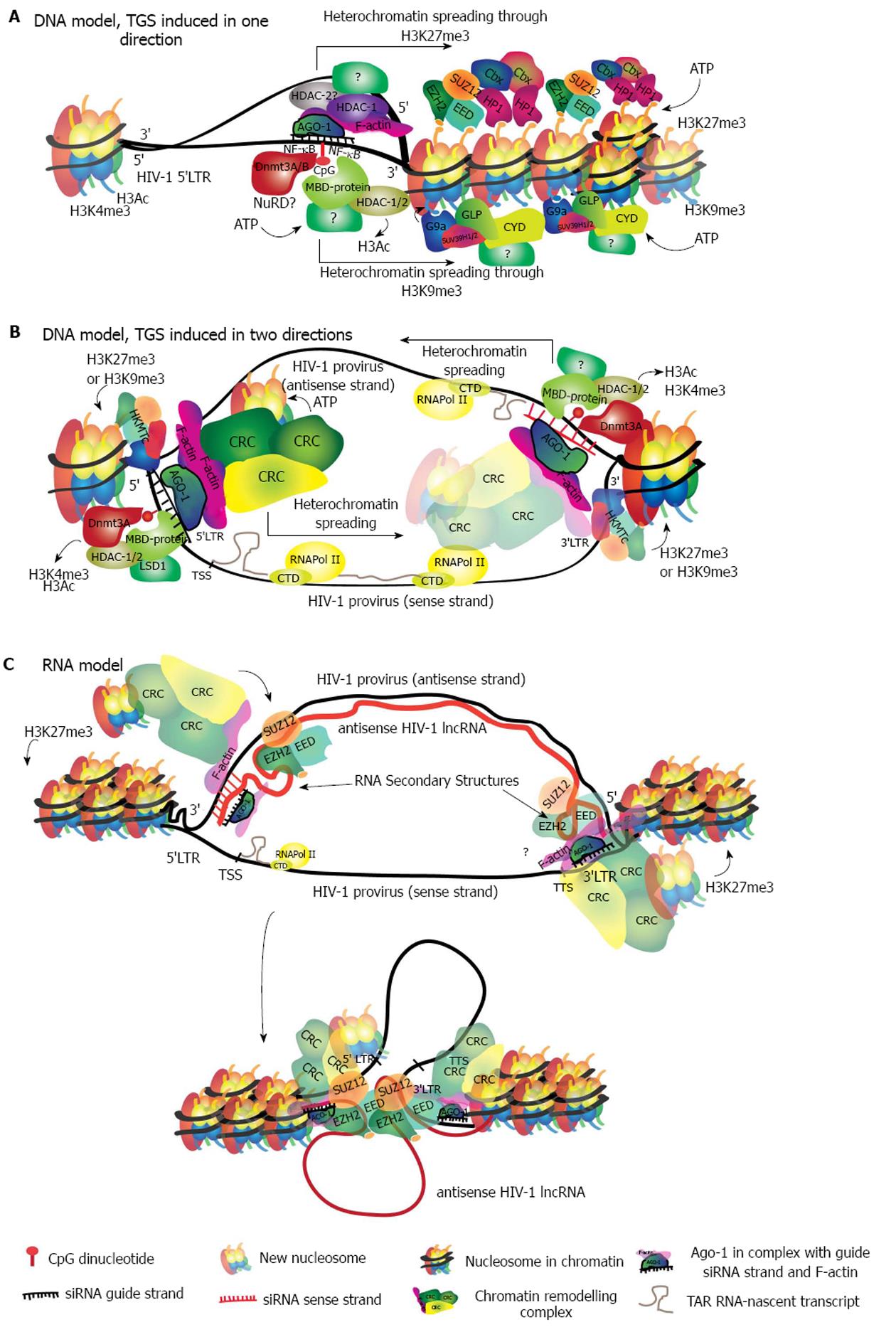
At present, there is evidence supporting two main models describing target recognition. The first is a siRNA/DNA-binding model[65,136], during which the RITS complex binds directly to chromatin. This binding seems to be dependent on the interaction between the siRNA and its DNA-target sequence. Once the interaction has taken place it triggers the in situ recruitment of chromatin remodeling factors that induce heterochromatin and establish silencing (Figure 3A). We previously introduced the unresolved question of how the passenger strand is removed. In the TGS model however, each strand of the duplex will find a target on DNA, in the same location but on different DNA strands. Therefore, for the sake of identifying the target region, both strands are potentially useful. In HIV-1, a siRNA guide-strand targeting a promoter region will find two target sites. One on the 5’LTR of the sense strand, and the other in the antisense strand in the region that is complementary to the 3’LTR of the sense strand (Figure 3B).
In the second model the RITS complex binds to an RNA intermediate, finding its target in either an antisense transcript or in a sense nascent transcript (recently reviewed in[59] and in[137]). In this model, only one strand of the duplex acts as the guide strand (Figure 3C). Presently, there is more experimental evidence supporting the RNA model given that, owing to its similarity with lncRNAs silencing mechanisms, more studies have tested this hypothesis. Though, there are still critical gaps in the data and more evidence is required to further evaluate the DNA model. It is possible that each of the models occur under particular conditions and potentially a variety of mechanisms control the diverse and precise regulation of gene expression in humans.
Establishment of heterochromatin is a progressive process. Once the RITS complex has found its target region a series of events follow, which generally initiate with removal and or replacement of specific histone-tail post-translational modifications to alter the biochemistry and structure of the associated chromatin (Table 1). Numerous histone modifications important for histone structure and gene regulation have been described[138], however we will only be discussing canonical acetylation and methylation marks that have been related to TGS and HIV-1. The different histone tail modifications are generated and recognized by histone deacetylases (HDACs), histone and DNA methyltransferases (HMTs and DNMTs, respectively), and chromatin modifying complexes. Ultimately, the combination of histone tail modifications and the recruitment of protein complexes make up a pattern that relates to the specific transcription state of a gene (a recent review can be found in[139]).
| Histone residue | Modification | Function | Writers | Erasers | Readers | Reviewed in |
| H3K4 | Ac | Transcription activation | [228] | |||
| me1 (enhancer sequences)me2/me3 (regulatory elements at the 5' end of active genes, and in poised genes) | Transcription activationTranscription activation, resolution of bivalency from poised genes | SET1 (tri)[229], SET7 (mono)[230], MLL[231], SMYD2[232] | LSD1 (mono and di)[233], JARID1A/KDM5A JARID1B/KDM5B (di and tri)[234] | CHD1[235], RAG2[236], TAF3[237], BPTF[238], BHC80[239], ING FAMILY[240], PYGO2[241] | [166,242] | |
| H3T6 | Phosphorylation | Transcription activation | PKC B | LSD1 | [243] | |
| H3K9 | Acme1/me2me3 (non-genic regions, centromeric heterochromatin, satellite sequences, long terminal repeats) | Transcription activation, histone deposition | GCN5/PCAF[244] | SIRT6[245] | BRD4[246] | [247] |
| Transcritional silencing, heterochromatin | SUV39H1/2[143], G9a[248], SETDB1[249] | JMJD1A/KDM3A[250], JMJD1B/KDM3B[251], JMJD1C/TRIP8, JMJD2A/KDM4A (B/C/D)[252] | HP1[253], EED 17406994), TDRD7[254], MPP8[255], UHRF1/2[256], GLP[248], CDY FAMILY[257] | |||
| H3K27 | me1/me2/me3, heterochromatin and facultative heterochromatin | Transcritional silencing, heterochromatin, poised genes | EZH2, EZH1[258] | JMJD1A/KDM3A, JMJD1B/KDM3B, KDM6A/UTX, JMJD3/KDM68, JMJD3/KDM6B[259] | Cbx proteins[165], EED[260] | [166,261] |
| H3K36 | Ac | Transcription activation | GCN5, PCAF[244] | [262] | ||
| me1/me2 (in the body and 3' end of genes)me2/me3 (gene bodies) | Transcription elongation | NSD1, NSD2[263], SET2[264], SMYD2[232], MMSET[265] | ASH1[266], JHDM1[267], JHDM1A/KDM2A, JHDM1B/KDM2B[268] | ISW1B[269] | ||
| H4K20 | me1 me2me3 (non-genic regions, centromeric heterochromatin, satellite sequences, long terminal repeats | Transcritional silencing, heterochromatin, repression of proinflammatory genes | PR-SET7/SET8[270] SUV420H1, SUV420H2[274]SUV420H2[274], SMYD5[275] | PHF8[271]PHF2[275]PHF2[275] | L3MBTL1[272]PHF20[276], L3MBTL1[277]NcoR[275] | [273] |
HDACs are required early in heterochromatin formation and remove acetylation (Ac) marks that are frequently found in actively transcribing chromatin. HDACs appear to be continuously recruited to epigenetically repressed loci[140], however, in very robust silencing, HDACs may not be continuously recruited. HDACs are recruited to chromatin by different mechanisms that are in some cases dependent on DNA methylation in CpG islands (discussed below). This differential recruitment is attributed to HDACs being able to form higher order complexes that may or may not include methyl-CpG-binding domain (MBD)-containing proteins[141].
The removal of Ac marks is necessary for the establishment of methylation repressive marks and chromatin compaction[142]. Several lysine residues from histone tails can be methylated by specific histone lysine methyltransferases (HKMTs) in order to repress chromatin (Table 1). Methylated residues are recognized by HP1 and HKMTs, both of which bind to chromatin and dimerize to induce chromatin compaction[143]. Nucleosome compaction exposes hidden lysine residues that become accessible to further methylation by HKMTs. Progressive methylation recruits more HP1-α and chromatin remodeling complexes. Chromatin remodeling complexes promote the establishment and spread of heterochromatin through a positive feedback loop with HP1[144] (Figure 3A).
Heterochromatin is also the final outcome of DNA methylation, genomic imprinting[145] and Polycomb (PcG) mediated silencing[65,146]. Therefore, RNAi-induced TGS has the potential to induce a variety of epigenetic profiles.
CpG islands (CGIs) are genomic regions that are unusually high in their CG or CpG content when compared to the genomic average of these nucleotides. CGIs are predominantly found in promoter regions and are demethylated during active gene transcription[40]. Conversely, methylation of promoter CGIs is associated with epigenetic gene repression. Thus, DNA methylation accounts for another layer of control of gene expression. It is well known that DNA-nucleotide-methyl-transferases (DNMT) methylate CpG residues[147] and seem to catalyse the reverse reaction[148]. However, the Ten-Eleven-Translocation enzymes (TET) are considered the main CpG DNA demethylases[149] while proteins containing DNA-methyl-CpG-binding domain (MBD) recognize the methylated status[150] in order to induce heterochromatin. However, it is not known how methylation is selectively established at precise promoters.
Genomic DNA methylation of CpG islands is fundamental for the programmed repression of genes during embryogenesis in mammalian cells. The methylation pattern is erased in the early embryo in order to establish the totipotent state, but is re-established during implantation with pluripotency genes being methylated and thus repressed[151,152]. Methylation of CGIs is then recognized by HKMTs that contain a methyl-binding domain (MBD) domain, in this case G9a. G9a establishes H3K9me3 and recruits HDACs, inducing HP1-α binding and local heterochromatinization. Heterochromatinization of HP1 promotes de novo DNA methylation by DNMT3 and further spreads silencing by repeating the loop[151] (Figure 3A). In humans, DNMT3 establishes de novo methylation and is responsible for tissue-specific DNA methylation patterns[153,154].
In the case of TGS, recent studies have shown that DNA methylation of CGIs is not required for siRNA-guided heterochromatin formation in fission yeast, as was initially described[155]. Similarly, the signatures of TGS in mammals appear to be somewhat diverse and may require DNA methylation in some cases. Interestingly, there is an RNAi-directed DNA methylation process that triggers TGS in plants[156], which is reminiscence of a mechanism in mammalian cells: piRNAs are known to direct DNA methylation in the male germ line in order to repress expression of transposable elements, but a similar mechanism has not been described on somatic cells[157]. However, there is some indirect evidence of a similar mechanism in mammalian somatic cells when transduced with lentiviral vectors. In fact, reduced expression of the introduced transgene was observed during differentiation in a murine model and silencing was found to be the result of DNA methylation of the promoter of the lentivirus driven gene[158]. Furthermore, it is well known that a considerable amount of integrated vectors become silent[159], and this effect seems to be dependent on the promoter chosen to drive the ectopic expression of the gene[160,161]. These observations could be related to ubiquitous RNA guided-DNA methylation pathway mechanism in mammalian cells aimed at controlling endogenous retroviruses. It is clear though, that de novo DNA methylation can provide stability for the inheritance of gene repression patterns through generations[151]. In this instance, TGS involving DNA methylation is likely to characterize robust silencing of a gene.
The PcG defines a group of genes that play a fundamental role in development and whose deletion results in early embryonic lethality in mice[162]. The PcG perform an antagonistic role to the trithorax group (TrxG) of proteins by inducing epigenetic gene repression. Both, PcG and TrxG, ensure the maintenance of proper expression patterns throughout the life span of a multicellular organism. There are two main repressive multi-subunit complexes formed by PcG: Polycomb-repressive complexes 1 and 2 (PCR1 and PCR2)[163,164].
PCR1 efficiently compacts chromatin through a variety of subunits that either identify and bind to H3K27me3, or mono-ubiquitilate Lys119 of histone 2 variant 2 (H2A), both of which promote nucleosome compaction. PCR1 is actually a group of functionally related but diverse protein complexes made up of different subunits that vary its function[165]. In addition to its role in development, roles in senescence, self-renewal, cancer and even gene activation have been recently identified for PCR1[165]. Interestingly, both complexes appear related, with PCR1 eventually acting downstream of PCR2 on certain loci.
PCR2 establishes the repressive epigenetic signature, H3K27me2/3 through its enhancer of zeste 1 and 2 subunits (EZH1, EZH2)[164] and induces chromatin compaction. In addition to H3K27me3, the activation mark H3K4me3 is also established by PCR2. Characteristically, genes co-expressing both, K3K37me3 and H3K4me3 epigenetic marks, are poised for transcription in undifferentiated cells. This state of epigenetic bivalency is resolved by the exclusive expression of H3K4me3 in transcriptionally active loci or H3K27me3 in transcriptionally repressed loci[166].
A direct link between PCR2 and TGS during regulation of granulopoyesis was elegantly demonstrated. Further this process was shown to be fundamental in driving progenitor lineage decisions at checkpoints of differentiation, in particular at the NF1-A gene. In this study, miRNA-223 directly bound to the NFI-A promoter region through its seed region and induced TGS of this gene through recruitment of the PcG proteins, YY1 and SUZ12, along with AGO-1 and Dicer[65]. This evidence supports previous findings of a siRNA-directed TGS, involving AGO-1, recruitment of EZH2, induction of H3K9me2 and the PTGS protein TRBP2[114]. Furthermore, the primary miRNA-208b has recently been shown to interact with EZH2, a Polycomb-group protein associated with gene silencing through chromatin remodeling[146]. Together, these studies clearly show that not only siRNAs, but also endogenous promoter-targeted miRNAs are able to trigger TGS in mammalian cells through recruitment of PcG proteins.
Interestingly, genes that are repressed by PcG express short-RNAs (about 50-200 nts) that interact with PCR2 to promote silencing[167]. However, no AGO proteins are involved in this case and the mechanism resembles that of X-chromosome inactivation (Xi) (explained in the next section), with SUZ12 subunit of PCR2 binding to a short RNA-stem loop from the BSN gene that mimics Xist A-Repeat (RepA) stem-loop. The important concept to highlight is that short RNAs can be transcribed from repressed loci and are used to guide repressor complexes to maintain these loci in a silent state.
Genomic imprinting is the mechanism by which parental-origin specific expression of imprinted genes is controlled in somatic cells (reviewed in[168]). It requires the DNA methylation of a region within the imprinting control region (ICR) that lies in the cluster of imprinted genes. This ICR is only demethylated in the germ cells but is then specifically re-methylated during fertilization depending on whether the maternal or the paternal allele is to be expressed in the somatic cells[169]. It is considered to be a very strong and stable silencing.
A well-studied case, that would be an example for the second TGS model, is Xi. During Xi, expression of the lncRNA Xist represses transcription from the paternal chromosome[110]. However, Xist is further regulated by the antisense lncRNA Tsix. After transcription, lncRNA Tsix induces silencing of Xist by recruiting PCR2, establishing H3K27me3 marks and enhancing de novo hyper methylation by DNMT3A[170]. The crucial link between RNAi and genomic imprinting in Xist regulation seems to be in the cleavage of the Xist-Tsix duplex by Dicer, which generates siRNAs targeting Xist leading to heterochromatin formation. These siRNAs in turn silence Xist and in this system deletion of Dicer appears to abolish silencing[145]. Currently, there is a dispute regarding the role of Dicer in this process and thus of RNAi in Xi, because Dicer knockout embryonic stem cells have shown contrasting results with either a defect in Xi (arguing in favor) or no defect at all (arguing against). A very detailed discussion about these contrasting results can be read in[171]. It is worth noting that other nuclear endonucleases could potentially induce cleavage in the absence of Dicer. However, recent findings showed that depletion of Dicer in human female cells has no effect in the epigenetic silencing of Xi, but results in up-regulation of X-linked genes, indicating that Dicer may be important for dosage compensation of those genes in differentiated cells[172].
Xi is just one of several examples of genomic imprinting during which specific DNA methylation and a lncRNA drive long-range epigenetic heterochromatic silencing through recruitment of PcG (Figure 2D). Because genomic imprinting involves recruitment of PcG proteins to an RNA intermediate, establishment of epigenetic repressive marks and short RNAs derived from the targeted genes, it supports the model of an RNA intermediate in sncRNA-directed TGS.
All these endogenous silencing mechanisms are an example of the different possibilities that may result when inducing TGS through sncRNAs (Figures 2 and 3). TGS is part of an enormous gene regulation network that involves a wide variety of mechanisms and protein interactions, whose combination yield diverse specific gene silencing outcomes. While we do not know yet how to induce each of these different epigenetic profiles, this mechanism has the power to silence the HIV-1 promoter in an inheritable, stable and permanent fashion, which we have reported through siRNA-induced TGS.
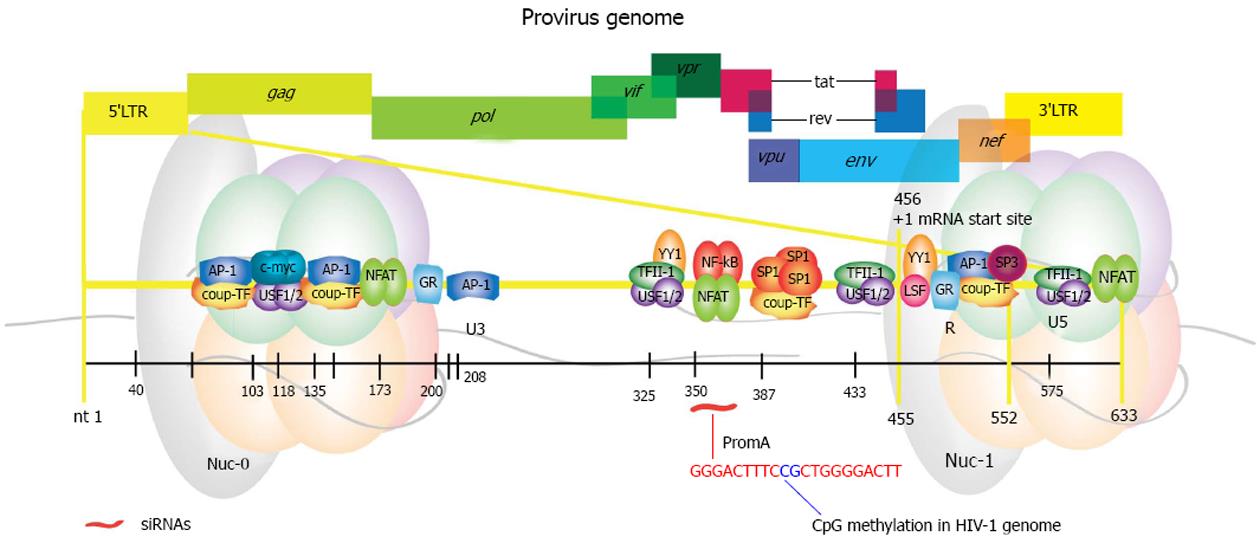
HIV establishes a long-term infection in dividing and non-dividing cells. The integrated proviral form is flanked by two long terminal repeats (LTRs) that originate from reverse transcription and are fundamental for viral replication[173]. HIV provirus behaves like a cellular gene; it has its own promoter located in the 5’ LTR and is rich in responsive elements for binding of several cellular transcription factors (Figure 4). It also has a 3’ LTR, which ensures the viral mRNAs are polyadenylated and capped mimicking cellular transcripts[174]. Of note, both LTRs have the same sequence and the 3’LTR is transcribed into the 3’UTR of the viral transcripts.
Upon integration, the provirus goes through an initial phase of abortive transcription. This phase is characterized by the presence of a non-processive RNA Pol II at the promoter region that is overcome upon expression of the viral trans-activating protein (Tat). Tat is imported back to the nucleus and binds the trans-activator response element (TAR), an RNA hairpin structure coded by the HIV promoter, greatly enhancing transcription[175]. Although most integrated proviruses are able to overcome abortive transcription, some become latent[27].
HIV latency is an interesting model to study because it is likely to be the result of various endogenous TGS mechanisms. Studies have described a variety of epigenetic profiles at the HIV promoter some of which are associated with extremely robust silencing such that reactivation of HIV is resistant in the face of substantial cell activation.
Generally, H3K9me3 is considered to be mutually exclusive with H3K27me3, and are found in different loci. More specifically, H3K9me3 is associated with silencing of endogenous retroviruses and retro-transposons, and is also enriched in constitutive heterochromatin regions and pericentromeric heterochromatin[176]. On the other hand, H3K27me3 is associated with a more dynamic silencing of varying strengths, which may depend on the presence of the H3K4me3 activation mark, as well as other undefined factors.
In HIV-1 infection, H3K27me3 has been found enriched in the 5’LTR promoter in cell line models of latent infection in which the virus reactivates upon stimulation[177]. This is consistent with H3K27me3 being generally a more flexible epigenetic repressive mark and with the likelihood that most of the inducible latent provirus is silenced through pathways involving H3K27me3, rather than H3K9me3. H3K9me3 has only been found in a few HIV-1 latency studies and re-activation of latent provirus carrying this mark has either not been observed after strong stimulation (with Phorbol-Myristate-Acetate treatment) or has required silencing of HP1-γ or other factors through RNAi[178,179]. This supports H3K9me3 as a more robust repressive epigenetic mark.
Similarly, Suv39H1, another HKMT responsible for H3K9me3, has been found to be recruited to latent HIV promoter in microglial cells[180], while in a different T-cell latency model, G9a, another HKMT responsible for H3K9 methylation), was found to be a determinant of proviral latency[181]. Moreover, the HKMT LSD1 is also recruited to the HIV promoter by the cofactor CTIP2 and establishes H3K9me3 to promote latency, rather than activation[178]. Additionally, EZH2, one of the PCR2 subunits that establish H3K27me3, has been found to be present at the LTR of latent provirus. Knockdown of EZH2 resulted in higher transcriptional activation of the HIV promoter than when knocking down Suv39H1[177], indicating that the former is associated with a more responsive epigenetic silencing.
Recently, a nuclear lncRNA expressed as an antisense transcript initiated from the viral 3’LTR, was found to modulate HIV-1 replication[182]. This lncRNA was further shown to exert epigenetic modulation of the 5’LTR HIV promoter by recruiting both DNMT3 and EZH2, resembling a genomic imprinting mechanism[183]. These observations are consistent with HIV CpG islands being methylated in a latency model[184]. It has been described that transcriptional silencing by Xist requires RepA, which is a short RNA transcript containing the A-repeat that forms an RNA secondary structure to which EZH2 and other PcG members bind, and whose deletion prevents silencing[185]. Given the similarity of the HIV antisense lncRNA mechanism to that of Xist, the TAR RNA-loop secondary structure fundamental for HIV transcription could potentially be involved in an interaction with EZH2. While the latter statement is hypothetical, the evidence thus far points towards a robust silencing of HIV by this lncRNA. The scope of this discovery may be extrapolated to the barriers to achieving reactivation of latent provirus as a therapeutic approach. Reactivation strategies to purge the latent reservoir, such as the use of histone deacetylase inhibitors (HDACis) have not been successful, despite using a variety of agents like Vorinostat and Panabinostat, with different potencies and specificities in inducing HIV specific chromatin relaxation[32]. The mechanism by which this HIV antisense lncRNA maintains latency might explain in part this difficulty, because a very robust and deep silencing may be established in a great deal of latent proviruses that make up the reservoir. Moreover, it could be potentially harmful to aim at disrupting this HIV lncRNA silencing because strategies directed to it could have an impact on other genomic regions strongly repressed by similar mechanisms.
Pan-HDACis have been developed that target more than one class of HDACs and the development of HDACis with isozyme specificity are on the scope[186]. However, HDACis will not specifically target only HIV, instead these drugs induce general chromatin relaxation on cellular genes and so have effects that are no HIV-specific. In addition, given the evident epigenetic complexity of HIV latency, more than one type of enzyme involved in epigenetic silencing will be needed to fully disrupt the latent provirus.
Collectively, the characteristic heterogeneity observed in the studies describing either HIV latency or on those aimed at re-activation of the latent provirus may be explained by the considerable density of binding sites for cellular transcription factors within the 5’LTR (Figure 4 and Table 2), in conjunction with the modulation executed by the HIV antisense lncRNA. Thus, it is possible that inducing TGS through siRNAs/shRNAs that target different regions within these DNA binding elements could result in the establishment of varied epigenetic profiles.
| Name | Position1 | Function | Cell type | Notes | Ref. |
| Nuc-0 | About 40-200 | Structural | Consistent across different cell types | Stable. Stability seems independent of transcription | [278] |
| AP-1/COUP-TF | About 103 | Activation/Repression | [279] | ||
| c-myc/RBF-2 (USF1/2) | 118-124 | Repression/Activation | HeLa-CAT-CD4 and J-Lat J89 (Jurkat) | Binds the sequence CACTGAC in HIV promoter, but the canonical sequence is CACGTGAC | [280,281] |
| Recruited by Sp1, can bind directly to the promoter to recruit HDAC1 | |||||
| RBF-2 can potentially bind to the CTGAC of this motif. | |||||
| AP-1/COUP-TF | About 135 | Repression/Activation | Cell type variation | COUP-TF binds to the nuclear responsive element | [180,279] |
| NFAT | 173 | Activation | Consistent across different cell lines | NFAT consensus sequence TGGAAA maps on antisense strand | [282] |
| GRE-I | 192-197 | Repression/Activation | Cell type variation | GRE-like element AGAACA | [283-285] |
| AP-1 | About 208 | AP-1 recently found to be crucial for latency | [286] | ||
| YY1/RBF-2 | About 336 | Repression/Activation | Jurkat, HeLa | Putative E-box element RBEIII. Sequence overlaps YY1, RBF-2/TFII-I and AP-1 binding sites | [281,287,288] |
| NFAT/NF-kB | 350 | Activation/Repression | Consistent across different cell types | Two shared in-tandem binding sites for each transcription factor. NF-kB in the sense strand, NFAT in the antisense | [289-291] |
| COUP-TF/Sp1/CTIP-2 | About 388 | Activation/Repression | Microglial, Oligodendrocytes, T lymphocytes | COUP-TF synergises and interacts with SP1 to activate, while CTIP2 directly binds to SP1 and represses transcription | [279,292,293] |
| Nuc-1 | 450-610 | Structural | Consistent across different cell types | This nucleosome is remodelled to induce HIV latency or transcriptional gene silencing | [278] |
| RBF-2/AP-4 | 435-440 | Activation/Repression | HEK293T, Jurkat | Both bind the E-box element CAGCTG, which has been named RBEI | [288,294-296] |
| GRE-II | 450-455 | Activation/Repression | Cell type variation | GRE-like element TGTACT | [283-285] |
| LSF/YY1 | about 440-483 | Repression | HeLa | LSF recruits YY1. This interaction recruits HDCA1 to initiate repression | [281,297,298] |
| GRE-III | 471-476 | Repression/Activation | Cell type variation | GRE-like element AGACCA | [283-285] |
| COUP-TF/AP-1/SP3 | About 485 | Repression/Activation | Microglial | Synergises and interacts with SP3 | [180,279] |
| RBF-2 | About 576 | Activation/Repression | Jurkat | Binds an atypical RBEIII element: ACTGCTGA | [288,294] |
| NFAT | 618 | Activation | Consistent across different cell lines | NFAT consensus sequence TGGAAA maps on sense strand | [291] |
Initial applications of RNAi to HIV were designed to target viral mRNA transcripts through the PTGS pathway[187]. These first attempts used transfection of one siRNA directed against important viral transcripts such as gag[187], env[188] and rev[189], and also cellular genes important for HIV-1 replication cycle, such as CD4[190] and CCR5 or CXCR4 chemokine receptors[191]. Suppression was not sustained whenever only viral mRNAs were targeted due to the emergence of resistant variants[192-194]. It became clear that a combinatorial RNAi against HIV would provide better protection and this correlated with delayed viral escape[195]. Further analysis of resistant viruses was useful to guide the design of more effective shRNAs[194]. Indeed, escape-proof shRNAs were identified that exerted potent and prolonged HIV suppression[196]. However, this approach was not completely robust as escape was observed from combinatorial shRNAs despite these being specifically designed to target previously characterized resistant viral variants[197]. Since then, multiple design approaches have been developed using a variety of strategies in search of the best combination of siRNA/shRNAs molecules that might prevent viral escape[198,199].
Following these findings, shRNAs targeting both conserved viral genes and host cellular genes required for viral replication became the preferred way to overcome this problem. Indeed, targeting only cellular genes such as CD4[190] and CXCR4 and particularly the CCR5 chemokine receptor dramatically reduced the emergence of resistant viruses[200]. Currently, PTGS is not envisaged as a stand-alone strategy for treating HIV. Rather its putative use is in combination with other types of gene therapy technologies, which we will discuss in the section for alternative gene therapy approaches.
The field of sncRNA-induced TGS for HIV therapeutics is less developed and has been hampered by the doubts regarding the existence of the pathway in mammalian nuclei. Nonetheless, siRNA and shRNA approaches have been efficiently developed that achieve long-term in vitro suppression of HIV replication, accompanied by epigenetic profiles which resemble those described in studies of the latent form of HIV-1.
We designed a siRNA, designated PromA, directed to the tandem repeat of NF-κB binding sites found in the HIV promoter (Figure 4). It can induce prolonged suppression of active HIV-1 infection in vitro and induces methylation of the CpG dinucleotide that maps to the sequence linking NF-κB tandem sites[201]. This HIV suppression was associated with recruitment of AGO-1 and HDAC1, and increased presence of H3K9me2 at the HIV promoter and involved nucleosome remodeling[202]. Later, long-term suppression (about 90 d) in conjunction with enrichment of H3K27me3 was observed when using stable expression of a shRNA targeting the same region[203]. H3K9me2 and H3K9me3 were also enriched but at much lower levels (H3K27me3 >>> H3K9me2 > H3K9me3). Suppression was then proved to be specific, as mutations in the shRNA sequence impaired virus suppression[204]. Interestingly, we identified F-actin as a key player in nuclear transportation of promoter-targeted siRNAs in mammalian cells, using the same siRNA constructs[205]. Results from this study are consistent with selective transport of promoter-targeted sncRNAs, which has also been shown for AGO-1 by other groups[135], as mentioned earlier.
Using a TGS-based gene therapy for treating HIV infection has several advantages over other therapies. First, TGS acts directly at the HIV promoter giving the virus virtually no opportunity to develop resistance; Second, it is likely able to act on latent provirus, whereby it potentially locks the virus in the latent state impeding future re-activation; Third, small doses of the effector molecules are sufficient to induce silencing since integrated provirus in a clinical setting is limited to less than 2 to 3 copies per cell[206]; And fourth, the silencing could potentially be inherited, though this remains to be definitely demonstrated.
Furthermore, an interesting point to note is that since the 5’LTR promoter contains the same sequence as the 3’LTR, a siRNA/shRNA designed to target the promoter region will also have a second target in the proviral 3’LTR. This could be potentially beneficial, as heterochromatin could be induced from both ends of the provirus (Figure 3B). Other potential targets are the 3’UTRs of viral mRNAs, whose targeting mainly depends on the efficiency of a siRNA to induce PTGS, or both PTGS and TGS simultaneously. In the latter case, PTGS would function until TGS is established, impeding transcription of viral mRNAs. However, an efficient siRNA/shRNA targeting both PTGS and TGS pathways has not yet been identified. Indeed, our siRNA PromA targeting the NF-κB did not show a significant PTGS effect on viral mRNAs[202] when we measured the effect in a setting mimicking an active HIV transcription owing to its clinical relevance, rather than using a weak promoter. In addition, the 1-LTR and 2-LTR circle intermediates of abortive HIV integration, which reside within the nucleus, may be targeted as well. While transcription and translation of viral genes from these unintegrated DNA forms has been observed, the contribution of these to actual infection is not clear[207]. And lastly, the linear DNA intermediate, that is synthesized in the cytoplasm by the RT enzyme and will become integrated as provirus, also contains the two viral LTRs, and several host proteins are known to interact with it[207]. While PTGS acts only post-HIV integration on viral mRNAs, rather than on incoming viral RNA genomes[208], the effect of promoter-targeted siRNAs in the incoming reverse-transcribed HIV genome and other unintegrated DNA forms has not been investigated.
Essentially, if sequence complementarity and/or sequence features of the promoter-targeted siRNA are the main determinant for target binding, then an activated RITS complex could potentially bind to any type of molecule containing the target sequence.
Hope for an HIV cure re-emerged after the successful bone marrow transplantation of Timothy Ray Brown-the leukemia patient known as the Berlin patient - with stem cells homozygous for the Δ32 deletion in the CCR5 gene (CCR5Δ32)[209]. This gene encodes an important co-receptor used by the virus to enter the host cells and individuals carrying the homozygous mutation have proven resistant to HIV infection by CCR5-tropic viruses[210]. Timothy was cured from both leukemia and HIV. Years after the transplant, he remains virus-free even when no longer under cART[211]. Since then, researchers have been developing various strategies to transform hematopoietic stem cells (CD34+) into HIV resistant cells, with the aim of reproducing this outcome.
Consequently, CCR5 has become the favorite cellular factor to target, especially since HIV CCR5-tropic strains are predominantly present during early stages of the disease and often persist into later stages[212,213]. Moreover, individuals with this mutation appear to be otherwise healthy apart from an as yet unconfirmed increase in susceptibility to West Nile infection[214] and hepatitis B virus infection[215]. These statements have raised the concern of whether CCR5 is implicated in immune system-related diseases[216]. An interesting discussion in this topic can be read in[217]. Thus, the effect of knocking down CCR5 could results in unpredicted effects.
Presently, different genetic therapy technologies are being tested for their in vivo ability to generate HIV resistant cells. From combined PTGS approaches, to genome editing with Zinc finger nucleases (ZFN), transcription activator-like effector nucleases (TALENs) or clustered regularly interspaced short palindromic repeats elements (CRISPR) associated caspase 9 (Cas9).
The most recent strategies involving PTGS use triple combination vectors. For example, a viral vector expressing shRNA against CCR5, an shRNA against TRIM5α isoform and a TAR-decoy against HIV[218] was successfully tested in a humanized NOD-RAG-/-IL2rγ-/- knockout mouse model. Similarly, a strategy using a viral vector expressing an shRNA against HIV tat/rev, a TAR-decoy element and ribozyme against CCR5[219] was initially tested using modified autologous CD4+ T cells in HIV positive patients who had failed therapy (NTC01153646), and is now been tested as an adjunct therapy using modified CD34+ T cells in patients with acquired-immune deficiency syndrome (AIDS)-related non-Hodgkin Lymphoma (NHL) (NCT01961063) and in patients with AIDS-related NHL requiring stem cell transplantation (NCT00569985). Importantly, long-term expression of the effector molecules from this construct has been detected in multiple cell lineages from treated patients, in which a combination of transduced and untransduced CD34+ cells were used[220].
ZFN strategies predominantly target CCR5. Recently, a phase I clinical trial (NTC00842634) testing the transfusion of CCR5 ZFN-modified autologous CD4+ T cells into HIV positive patients[221] showed that the procedure was feasible and safe. During an anti viral therapy treatment interruption the modified cells had a higher survival over non-modified cells. Also, patients showed decreased HIV DNA levels in blood. Currently, the effect of repeated doses of the ZFN-modified CD4+ T cells is being tested (NCT02225665). Although, these clinical trials use modified CD4+ T cells rather than CD34+ cells, recent studies in a humanized mice model showed low engraftment, but proper multi-lineage differentiation of the CCR5-ZFN CD34+ cells[222].
TALENs and CRISPR have not yet been trialed in humans. However, the results from in vitro studies are very promising[223], with CRISPR editing able to excise the provirus from infected cells, and thus able to target latent proviruses[224]. ZFNs have also been used to target the provirus, using lentivirus to achieve stable expression of the nucleases[225]. However, the above-mentioned ZFN-related clinical trials used adenovirus vectors. Generally, genome-editing approaches use non-integrative adenoviral vectors. Adenoviral vectors are diluted after each cell division and direct transient expression of the editing nuclease. Transient expression has been the choice for genome-editing approaches on the grounds that a continuous expression of a selected editing nuclease could be potentially risky as it may result in off-target genome editing. To date, it remains to be addressed if ZFN/TALEN/CRISPR genetically modified CD34+ are safe to use in humans and whether they are feasible approaches towards a functional cure.
Presently, a variety of strategies are being tested in order to breakthrough this highly challenging treatment barrier. There are still several large hurdles to be surmounted. Currently there is a lack of adequate delivery systems for targeting cells with HIV infection and the latent reservoir. Further TGS/PTGS approaches require stable expression from vectors, such as lentiviral vectors but this must be combined with high transduction and engraftment rates, for therapy to be effective. In the same way, genome-editing approaches rely on vectors that drive transient expression of the editing enzyme, but get diluted after each cell division. Thus, achieving high genome editing efficiency is one of the limitations.
Importantly, TGS and CRISPR genome editing have the potential to target proviruses directly, and therefore could be effective in targeting latent provirus. Yet this strength may also be an inherent weakness and thus a careful selection of the targeted sequences of HIV-1 is fundamental. Unfortunately, 5’LTR sequences from proven replication competent proviruses are the least represented in curated databases in comparison to other HIV genomic regions. Nonetheless, combinatorial strategies are also an option within these therapies, and may be designed to target an additional host factor as well.
Gene therapy technologies that target only CCR5 may be unable to target latent provirus that is already present. In addition, they may select HIV-1 viruses with tropism for the CXCR4 co-receptor, allowing escape and potentially more rapid disease progression. This evolution is more likely if latent provirus remains in untargeted compartments.
The combinatorial strategies from PTGS, which target the virus and a host factor such as CCR5, provide an additional mechanism that directly restricts the virus and could possibly delay or imped viral evolution. In this regard, it could potentially provide some protection from CXCR4-tropic emerging viruses or re-activating from latent proviruses.
Basically, with present technologies none of the effector molecules for these therapies can be directly administered to an infected patient. Rather, autologous cells are obtained, genetically modified, and then transferred back to the patient. Generally, these therapies aim at modifying CD34+ cells in order to develop multi-lineage HIV resistance and thus long-term protection to the infection. Indeed, the limitation of most of these therapies relies on the efficiency of several steps throughout the complete intervention process. For instance, the efficiency or success to which the autologous cells are first, modified ex vivo; Second, re-mobilized or transplanted; third, engrafted within the bone marrow; and fourth, either achieve a sustained and prolonged multi-lineage expression of the modified trait/gene or achieve a certain percentage of modified cells from all the lineages enough to provide protection. Furthermore, the engrafted modified cells will share a niche with the wild-type cells, unless ablation of the immune system is performed before. Therefore, understanding the interactions and signaling between these two populations sharing a niche could give us a better prediction of the long-term success of these therapies. Factors such as symmetric and asymmetric cell division[226], unidentified endogenous mechanisms of genomic mosaicism detection in stem cells[227] and other cellular and molecular pathways may play an important role. For instance, if it is confirmed that Piwi proteins are expressed in hematopoietic stem cells, this could potentially have an impact in those therapies that rely on integrative gene therapy vectors.
Finally, other concerns remain such as the worldwide implementation of these gene-therapy strategies and their cost, particularly in developing countries. Consequently, the development of delivery methods that facilitate the clinical application of these therapies is an important quest.
The various RNAi strategies to target HIV reviewed here provide a potential alternate approach to combating HIV infection and the latent reservoir, with the results of current and future RNAi therapeutic trials poised to reveal whether this approach represents a possible pathway towards a functional HIV cure.
P- Reviewer: Arriagada GL, Zou C S- Editor: Tian YL L- Editor: A E- Editor: Yan JL
| 1. | Reust CE. Common adverse effects of antiretroviral therapy for HIV disease. Am Fam Physician. 2011;83:1443-1451. [PubMed] |
| 2. | Cortez KJ, Maldarelli F. Clinical management of HIV drug resistance. Viruses. 2011;3:347-378. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 3. | Sarmento-Castro R, Vasconcelos C, Aguas MJ, Marques R, Oliveira J. Virologic suppression in treatment-experienced patients after virologic rebound or failure of therapy. Curr Opin HIV AIDS. 2011;6 Suppl 1:S12-S20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 4. | Hamlyn E, Ewings FM, Porter K, Cooper DA, Tambussi G, Schechter M, Pedersen C, Okulicz JF, McClure M, Babiker A. Plasma HIV viral rebound following protocol-indicated cessation of ART commenced in primary and chronic HIV infection. PLoS One. 2012;7:e43754. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 5. | Steingrover R, Pogány K, Fernandez Garcia E, Jurriaans S, Brinkman K, Schuitemaker H, Miedema F, Lange JM, Prins JM. HIV-1 viral rebound dynamics after a single treatment interruption depends on time of initiation of highly active antiretroviral therapy. AIDS. 2008;22:1583-1588. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 6. | Chun TW, Justement JS, Murray D, Hallahan CW, Maenza J, Collier AC, Sheth PM, Kaul R, Ostrowski M, Moir S. Rebound of plasma viremia following cessation of antiretroviral therapy despite profoundly low levels of HIV reservoir: implications for eradication. AIDS. 2010;24:2803-2808. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 238] [Cited by in RCA: 234] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 7. | O’Connell RJ, Kim JH, Corey L, Michael NL. Human immunodeficiency virus vaccine trials. Cold Spring Harb Perspect Med. 2012;2:a007351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Robb ML, Rerks-Ngarm S, Nitayaphan S, Pitisuttithum P, Kaewkungwal J, Kunasol P, Khamboonruang C, Thongcharoen P, Morgan P, Benenson M. Risk behaviour and time as covariates for efficacy of the HIV vaccine regimen ALVAC-HIV (vCP1521) and AIDSVAX B/E: a post-hoc analysis of the Thai phase 3 efficacy trial RV 144. Lancet Infect Dis. 2012;12:531-537. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 178] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 9. | Ndung’u T, Weiss RA. On HIV diversity. AIDS. 2012;26:1255-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 10. | Araújo LA, Almeida SE. HIV-1 diversity in the envelope glycoproteins: implications for viral entry inhibition. Viruses. 2013;5:595-604. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 11. | Folks TM. Mechanisms and strategies of viral antigenic variation. Ann N Y Acad Sci. 1994;730:37-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 12. | Sharkey M. Tracking episomal HIV DNA: implications for viral persistence and eradication of HIV. Curr Opin HIV AIDS. 2013;8:93-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Chun TW, Fauci AS. HIV reservoirs: pathogenesis and obstacles to viral eradication and cure. AIDS. 2012;26:1261-1268. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 139] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 14. | Coleman CM, Wu L. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology. 2009;6:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 178] [Cited by in RCA: 175] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 15. | Wu L. The role of monocyte-lineage cells in human immunodeficiency virus persistence: mechanisms and progress. Wei Sheng Wu Yu Gan Ran. 2011;6:129-132. [PubMed] |
| 16. | Mehla R, Bivalkar-Mehla S, Zhang R, Handy I, Albrecht H, Giri S, Nagarkatti P, Nagarkatti M, Chauhan A. Bryostatin modulates latent HIV-1 infection via PKC and AMPK signaling but inhibits acute infection in a receptor independent manner. PLoS One. 2010;5:e11160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 175] [Cited by in RCA: 193] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 17. | Shirakawa K, Chavez L, Hakre S, Calvanese V, Verdin E. Reactivation of latent HIV by histone deacetylase inhibitors. Trends Microbiol. 2013;21:277-285. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 18. | Fernandez G, Zeichner SL. Cell line-dependent variability in HIV activation employing DNMT inhibitors. Virol J. 2010;7:266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 19. | Victoriano AF, Okamoto T. Transcriptional control of HIV replication by multiple modulators and their implication for a novel antiviral therapy. AIDS Res Hum Retroviruses. 2012;28:125-138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Sharkey M, Babic DZ, Greenough T, Gulick R, Kuritzkes DR, Stevenson M. Episomal viral cDNAs identify a reservoir that fuels viral rebound after treatment interruption and that contributes to treatment failure. PLoS Pathog. 2011;7:e1001303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 21. | Castro P, Plana M, González R, López A, Vilella A, Nicolas JM, Gallart T, Pumarola T, Bayas JM, Gatell JM. Influence of episodes of intermittent viremia (“blips”) on immune responses and viral load rebound in successfully treated HIV-infected patients. AIDS Res Hum Retroviruses. 2013;29:68-76. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 22. | Pasternak AO, de Bruin M, Jurriaans S, Bakker M, Berkhout B, Prins JM, Lukashov VV. Modest nonadherence to antiretroviral therapy promotes residual HIV-1 replication in the absence of virological rebound in plasma. J Infect Dis. 2012;206:1443-1452. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 23. | Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15:893-900. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1487] [Cited by in RCA: 1431] [Article Influence: 89.4] [Reference Citation Analysis (0)] |
| 24. | Bosque A, Famiglietti M, Weyrich AS, Goulston C, Planelles V. Homeostatic proliferation fails to efficiently reactivate HIV-1 latently infected central memory CD4+ T cells. PLoS Pathog. 2011;7:e1002288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 138] [Cited by in RCA: 168] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 25. | Shan L, Yang HC, Rabi SA, Bravo HC, Shroff NS, Irizarry RA, Zhang H, Margolick JB, Siliciano JD, Siliciano RF. Influence of host gene transcription level and orientation on HIV-1 latency in a primary-cell model. J Virol. 2011;85:5384-5393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 26. | Coiras M, López-Huertas MR, Pérez-Olmeda M, Alcamí J. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol. 2009;7:798-812. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 217] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 27. | Mbonye U, Karn J. Control of HIV latency by epigenetic and non-epigenetic mechanisms. Curr HIV Res. 2011;9:554-567. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 28. | van Praag RM, Prins JM, Roos MT, Schellekens PT, Ten Berge IJ, Yong SL, Schuitemaker H, Eerenberg AJ, Jurriaans S, de Wolf F. OKT3 and IL-2 treatment for purging of the latent HIV-1 reservoir in vivo results in selective long-lasting CD4+ T cell depletion. J Clin Immunol. 2001;21:218-226. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 106] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 29. | Wang FX, Xu Y, Sullivan J, Souder E, Argyris EG, Acheampong EA, Fisher J, Sierra M, Thomson MM, Najera R. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest. 2005;115:128-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 125] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 30. | Sagot-Lerolle N, Lamine A, Chaix ML, Boufassa F, Aboulker JP, Costagliola D, Goujard C, Pallier C, Delfraissy JF, Lambotte O. Prolonged valproic acid treatment does not reduce the size of latent HIV reservoir. AIDS. 2008;22:1125-1129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 123] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 31. | Archin NM, Liberty AL, Kashuba AD, Choudhary SK, Kuruc JD, Crooks AM, Parker DC, Anderson EM, Kearney MF, Strain MC. Administration of vorinostat disrupts HIV-1 latency in patients on antiretroviral therapy. Nature. 2012;487:482-485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 872] [Cited by in RCA: 989] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 32. | Bullen CK, Laird GM, Durand CM, Siliciano JD, Siliciano RF. New ex vivo approaches distinguish effective and ineffective single agents for reversing HIV-1 latency in vivo. Nat Med. 2014;20:425-429. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 367] [Cited by in RCA: 420] [Article Influence: 38.2] [Reference Citation Analysis (0)] |
| 33. | Lindbo JA, Dougherty WG. Untranslatable transcripts of the tobacco etch virus coat protein gene sequence can interfere with tobacco etch virus replication in transgenic plants and protoplasts. Virology. 1992;189:725-733. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 200] [Cited by in RCA: 205] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 34. | Lindbo JA, Dougherty WG. Pathogen-derived resistance to a potyvirus: immune and resistant phenotypes in transgenic tobacco expressing altered forms of a potyvirus coat protein nucleotide sequence. Mol Plant Microbe Interact. 1992;5:144-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 99] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Melnyk CW, Molnar A, Baulcombe DC. Intercellular and systemic movement of RNA silencing signals. EMBO J. 2011;30:3553-3563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 219] [Article Influence: 15.6] [Reference Citation Analysis (0)] |
| 36. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10118] [Article Influence: 374.7] [Reference Citation Analysis (1)] |
| 37. | Montgomery MK, Fire A. Double-stranded RNA as a mediator in sequence-specific genetic silencing and co-suppression. Trends Genet. 1998;14:255-258. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 149] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 38. | Montgomery MK, Xu S, Fire A. RNA as a target of double-stranded RNA-mediated genetic interference in Caenorhabditis elegans. Proc Natl Acad Sci USA. 1998;95:15502-15507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 422] [Cited by in RCA: 409] [Article Influence: 15.1] [Reference Citation Analysis (0)] |
| 39. | Mantha N, Das SK, Das NG. RNAi-based therapies for Huntington’s disease: delivery challenges and opportunities. Ther Deliv. 2012;3:1061-1076. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 40. | Morris KV, Chan SW, Jacobsen SE, Looney DJ. Small interfering RNA-induced transcriptional gene silencing in human cells. Science. 2004;305:1289-1292. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 679] [Cited by in RCA: 678] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 41. | Hutvágner G, Zamore PD. RNAi: nature abhors a double-strand. Curr Opin Genet Dev. 2002;12:225-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 325] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 42. | Sigova A, Rhind N, Zamore PD. A single Argonaute protein mediates both transcriptional and posttranscriptional silencing in Schizosaccharomyces pombe. Genes Dev. 2004;18:2359-2367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 108] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 43. | Han J, Kim D, Morris KV. Promoter-associated RNA is required for RNA-directed transcriptional gene silencing in human cells. Proc Natl Acad Sci USA. 2007;104:12422-12427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 217] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 44. | Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2362] [Cited by in RCA: 2454] [Article Influence: 153.4] [Reference Citation Analysis (0)] |
| 45. | Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 367] [Cited by in RCA: 391] [Article Influence: 23.0] [Reference Citation Analysis (0)] |
| 46. | Meister G. Argonaute proteins: functional insights and emerging roles. Nat Rev Genet. 2013;14:447-459. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 756] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 47. | Li L, Liu Y. Diverse small non-coding RNAs in RNA interference pathways. Methods Mol Biol. 2011;764:169-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 48. | Castel SE, Martienssen RA. RNA interference in the nucleus: roles for small RNAs in transcription, epigenetics and beyond. Nat Rev Genet. 2013;14:100-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 713] [Cited by in RCA: 695] [Article Influence: 57.9] [Reference Citation Analysis (0)] |
| 49. | Polikepahad S, Corry DB. Profiling of T helper cell-derived small RNAs reveals unique antisense transcripts and differential association of miRNAs with argonaute proteins 1 and 2. Nucleic Acids Res. 2013;41:1164-1177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 50. | Maillard PV, Ciaudo C, Marchais A, Li Y, Jay F, Ding SW, Voinnet O. Antiviral RNA interference in mammalian cells. Science. 2013;342:235-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 317] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 51. | Castellano L, Stebbing J. Deep sequencing of small RNAs identifies canonical and non-canonical miRNA and endogenous siRNAs in mammalian somatic tissues. Nucleic Acids Res. 2013;41:3339-3351. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 90] [Cited by in RCA: 86] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 52. | Ameres SL, Horwich MD, Hung JH, Xu J, Ghildiyal M, Weng Z, Zamore PD. Target RNA-directed trimming and tailing of small silencing RNAs. Science. 2010;328:1534-1539. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 514] [Cited by in RCA: 460] [Article Influence: 30.7] [Reference Citation Analysis (0)] |
| 53. | Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 295] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 54. | Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature. 2001;411:494-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6971] [Cited by in RCA: 7007] [Article Influence: 292.0] [Reference Citation Analysis (0)] |
| 55. | Holen T, Amarzguioui M, Wiiger MT, Babaie E, Prydz H. Positional effects of short interfering RNAs targeting the human coagulation trigger Tissue Factor. Nucleic Acids Res. 2002;30:1757-1766. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 504] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 56. | Nowotny M, Yang W. Structural and functional modules in RNA interference. Curr Opin Struct Biol. 2009;19:286-293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 38] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 57. | Chu Y, Yue X, Younger ST, Janowski BA, Corey DR. Involvement of argonaute proteins in gene silencing and activation by RNAs complementary to a non-coding transcript at the progesterone receptor promoter. Nucleic Acids Res. 2010;38:7736-7748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 58. | Younger ST, Corey DR. Transcriptional regulation by miRNA mimics that target sequences downstream of gene termini. Mol Biosyst. 2011;7:2383-2388. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 59. | Gagnon KT, Corey DR. Argonaute and the nuclear RNAs: new pathways for RNA-mediated control of gene expression. Nucleic Acid Ther. 2012;22:3-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 54] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 60. | Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1287] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 61. | Ameres SL, Zamore PD. Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol. 2013;14:475-488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 865] [Cited by in RCA: 913] [Article Influence: 76.1] [Reference Citation Analysis (0)] |
| 62. | Bracken CP, Szubert JM, Mercer TR, Dinger ME, Thomson DW, Mattick JS, Michael MZ, Goodall GJ. Global analysis of the mammalian RNA degradome reveals widespread miRNA-dependent and miRNA-independent endonucleolytic cleavage. Nucleic Acids Res. 2011;39:5658-5668. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 63. | Shukla GC, Singh J, Barik S. MicroRNAs: Processing, Maturation, Target Recognition and Regulatory Functions. Mol Cell Pharmacol. 2011;3:83-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 16] [Reference Citation Analysis (0)] |
| 64. | Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011;12:19-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 511] [Article Influence: 34.1] [Reference Citation Analysis (0)] |
| 65. | Zardo G, Ciolfi A, Vian L, Starnes LM, Billi M, Racanicchi S, Maresca C, Fazi F, Travaglini L, Noguera N. Polycombs and microRNA-223 regulate human granulopoiesis by transcriptional control of target gene expression. Blood. 2012;119:4034-4046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 124] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 66. | Kim DH, Saetrom P, Snøve O, Rossi JJ. MicroRNA-directed transcriptional gene silencing in mammalian cells. Proc Natl Acad Sci USA. 2008;105:16230-16235. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 564] [Cited by in RCA: 528] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 67. | Huang V, Li LC. miRNA goes nuclear. RNA Biol. 2012;9:269-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 68. | Ross RJ, Weiner MM, Lin H. PIWI proteins and PIWI-interacting RNAs in the soma. Nature. 2014;505:353-359. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 332] [Cited by in RCA: 319] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 69. | Nolde MJ, Cheng EC, Guo S, Lin H. Piwi genes are dispensable for normal hematopoiesis in mice. PLoS One. 2013;8:e71950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 70. | Jacobs JE, Wagner M, Dhahbi J, Boffelli D, Martin DI. Deficiency of MIWI2 (Piwil4) induces mouse erythroleukemia cell differentiation, but has no effect on hematopoiesis in vivo. PLoS One. 2013;8:e82573. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 71. | Sharma AK, Nelson MC, Brandt JE, Wessman M, Mahmud N, Weller KP, Hoffman R. Human CD34(+) stem cells express the hiwi gene, a human homologue of the Drosophila gene piwi. Blood. 2001;97:426-434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 162] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 72. | Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228-2241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 108] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 73. | Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243-1255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 110] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 74. | Pal-Bhadra M, Bhadra U, Birchler JA. RNAi related mechanisms affect both transcriptional and posttranscriptional transgene silencing in Drosophila. Mol Cell. 2002;9:315-327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 293] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 75. | Huang XA, Yin H, Sweeney S, Raha D, Snyder M, Lin H. A major epigenetic programming mechanism guided by piRNAs. Dev Cell. 2013;24:502-516. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 185] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 76. | Mei Y, Clark D, Mao L. Novel dimensions of piRNAs in cancer. Cancer Lett. 2013;336:46-52. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 95] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 77. | Behm-Ansmant I, Rehwinkel J, Doerks T, Stark A, Bork P, Izaurralde E. mRNA degradation by miRNAs and GW182 requires both CCR4: NOT deadenylase and DCP1: DCP2 decapping complexes. Genes Dev. 2006;20:1885-1898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 716] [Cited by in RCA: 724] [Article Influence: 38.1] [Reference Citation Analysis (0)] |
| 78. | Pare JM, Tahbaz N, López-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol Biol Cell. 2009;20:3273-3284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 119] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 79. | Johnston M, Geoffroy MC, Sobala A, Hay R, Hutvagner G. HSP90 protein stabilizes unloaded argonaute complexes and microscopic P-bodies in human cells. Mol Biol Cell. 2010;21:1462-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 140] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 80. | Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol Cell Biol. 2007;27:3970-3981. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 529] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 81. | Kim K, Lee YS, Carthew RW. Conversion of pre-RISC to holo-RISC by Ago2 during assembly of RNAi complexes. RNA. 2007;13:22-29. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 82. | Sakurai K, Amarzguioui M, Kim DH, Alluin J, Heale B, Song MS, Gatignol A, Behlke MA, Rossi JJ. A role for human Dicer in pre-RISC loading of siRNAs. Nucleic Acids Res. 2011;39:1510-1525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 83. | Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1969] [Cited by in RCA: 1954] [Article Influence: 88.8] [Reference Citation Analysis (0)] |
| 84. | Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1801] [Cited by in RCA: 1792] [Article Influence: 81.5] [Reference Citation Analysis (0)] |
| 85. | Noland CL, Ma E, Doudna JA. siRNA repositioning for guide strand selection by human Dicer complexes. Mol Cell. 2011;43:110-121. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 111] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 86. | Betancur JG, Tomari Y. Dicer is dispensable for asymmetric RISC loading in mammals. RNA. 2012;18:24-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Ye X, Huang N, Liu Y, Paroo Z, Huerta C, Li P, Chen S, Liu Q, Zhang H. Structure of C3PO and mechanism of human RISC activation. Nat Struct Mol Biol. 2011;18:650-657. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 112] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 88. | Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD. Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell. 2005;123:607-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 785] [Cited by in RCA: 819] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 89. | Rand TA, Petersen S, Du F, Wang X. Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell. 2005;123:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 540] [Cited by in RCA: 535] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 90. | Liu Y, Ye X, Jiang F, Liang C, Chen D, Peng J, Kinch LN, Grishin NV, Liu Q. C3PO, an endoribonuclease that promotes RNAi by facilitating RISC activation. Science. 2009;325:750-753. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 205] [Cited by in RCA: 187] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 91. | Parizotto EA, Lowe ED, Parker JS. Structural basis for duplex RNA recognition and cleavage by Archaeoglobus fulgidus C3PO. Nat Struct Mol Biol. 2013;20:380-386. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 92. | Eulalio A, Huntzinger E, Nishihara T, Rehwinkel J, Fauser M, Izaurralde E. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21-32. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 303] [Cited by in RCA: 318] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 93. | Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877-6888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1033] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 94. | Crist CG, Montarras D, Buckingham M. Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell. 2012;11:118-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 221] [Cited by in RCA: 256] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 95. | Lian SL, Li S, Abadal GX, Pauley BA, Fritzler MJ, Chan EK. The C-terminal half of human Ago2 binds to multiple GW-rich regions of GW182 and requires GW182 to mediate silencing. RNA. 2009;15:804-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 115] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 96. | Takimoto K, Wakiyama M, Yokoyama S. Mammalian GW182 contains multiple Argonaute-binding sites and functions in microRNA-mediated translational repression. RNA. 2009;15:1078-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 97. | Ding L, Han M. GW182 family proteins are crucial for microRNA-mediated gene silencing. Trends Cell Biol. 2007;17:411-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 91] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 98. | Ikeda K, Satoh M, Pauley KM, Fritzler MJ, Reeves WH, Chan EK. Detection of the argonaute protein Ago2 and microRNAs in the RNA induced silencing complex (RISC) using a monoclonal antibody. J Immunol Methods. 2006;317:38-44. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 72] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 99. | Eulalio A, Huntzinger E, Izaurralde E. GW182 interaction with Argonaute is essential for miRNA-mediated translational repression and mRNA decay. Nat Struct Mol Biol. 2008;15:346-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 314] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 100. | Nishi K, Nishi A, Nagasawa T, Ui-Tei K. Human TNRC6A is an Argonaute-navigator protein for microRNA-mediated gene silencing in the nucleus. RNA. 2013;19:17-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 146] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 101. | Gagnon KT, Li L, Chu Y, Janowski BA, Corey DR. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211-221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 295] [Article Influence: 26.8] [Reference Citation Analysis (0)] |
| 102. | Zhang H, Zhu JK. RNA-directed DNA methylation. Curr Opin Plant Biol. 2011;14:142-147. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 180] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 103. | Djupedal I, Ekwall K. Epigenetics: heterochromatin meets RNAi. Cell Res. 2009;19:282-295. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 104. | Creamer KM, Partridge JF. RITS-connecting transcription, RNA interference, and heterochromatin assembly in fission yeast. Wiley Interdiscip Rev RNA. 2011;2:632-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 105. | Grewal SI, Elgin SC. Transcription and RNA interference in the formation of heterochromatin. Nature. 2007;447:399-406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 311] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 106. | Yue X, Schwartz JC, Chu Y, Younger ST, Gagnon KT, Elbashir S, Janowski BA, Corey DR. Transcriptional regulation by small RNAs at sequences downstream from 3’ gene termini. Nat Chem Biol. 2010;6:621-629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 88] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 107. | Benhamed M, Herbig U, Ye T, Dejean A, Bischof O. Senescence is an endogenous trigger for microRNA-directed transcriptional gene silencing in human cells. Nat Cell Biol. 2012;14:266-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 200] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 108. | Jiang G, Zheng L, Pu J, Mei H, Zhao J, Huang K, Zeng F, Tong Q. Small RNAs targeting transcription start site induce heparanase silencing through interference with transcription initiation in human cancer cells. PLoS One. 2012;7:e31379. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 109. | Ameyar-Zazoua M, Rachez C, Souidi M, Robin P, Fritsch L, Young R, Morozova N, Fenouil R, Descostes N, Andrau JC. Argonaute proteins couple chromatin silencing to alternative splicing. Nat Struct Mol Biol. 2012;19:998-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 218] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 110. | Kanduri C, Whitehead J, Mohammad F. The long and the short of it: RNA-directed chromatin asymmetry in mammalian X-chromosome inactivation. FEBS Lett. 2009;583:857-864. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 111. | Almouzni G, Probst AV. Heterochromatin maintenance and establishment: lessons from the mouse pericentromere. Nucleus. 2011;2:332-338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 75] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 112. | Trojer P, Reinberg D. Facultative heterochromatin: is there a distinctive molecular signature? Mol Cell. 2007;28:1-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 342] [Cited by in RCA: 358] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 113. | Suzuki K, Hattori S, Marks K, Ahlenstiel C, Maeda Y, Ishida T, Millington M, Boyd M, Symonds G, Cooper DA. Promoter Targeting shRNA Suppresses HIV-1 Infection In vivo Through Transcriptional Gene Silencing. Mol Ther Nucleic Acids. 2013;2:e137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 114. | Kim DH, Villeneuve LM, Morris KV, Rossi JJ. Argonaute-1 directs siRNA-mediated transcriptional gene silencing in human cells. Nat Struct Mol Biol. 2006;13:793-797. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 317] [Cited by in RCA: 325] [Article Influence: 17.1] [Reference Citation Analysis (0)] |
| 115. | Lorentz A, Ostermann K, Fleck O, Schmidt H. Switching gene swi6, involved in repression of silent mating-type loci in fission yeast, encodes a homologue of chromatin-associated proteins from Drosophila and mammals. Gene. 1994;143:139-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 123] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 116. | Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation, and mitotic progression. Mol Cell Biol. 2000;20:3728-3741. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 148] [Cited by in RCA: 155] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 117. | Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh PB. Functional mammalian homologues of the Drosophila PEV-modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923-1938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 328] [Cited by in RCA: 348] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 118. | Nakanishi K, Ascano M, Gogakos T, Ishibe-Murakami S, Serganov AA, Briskin D, Morozov P, Tuschl T, Patel DJ. Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Rep. 2013;3:1893-1900. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 119. | Wang B, Li S, Qi HH, Chowdhury D, Shi Y, Novina CD. Distinct passenger strand and mRNA cleavage activities of human Argonaute proteins. Nat Struct Mol Biol. 2009;16:1259-1266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 120. | Su H, Trombly MI, Chen J, Wang X. Essential and overlapping functions for mammalian Argonautes in microRNA silencing. Genes Dev. 2009;23:304-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 189] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 121. | Petri S, Dueck A, Lehmann G, Putz N, Rüdel S, Kremmer E, Meister G. Increased siRNA duplex stability correlates with reduced off-target and elevated on-target effects. RNA. 2011;17:737-749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 122. | Faehnle CR, Elkayam E, Haase AD, Hannon GJ, Joshua-Tor L. The making of a slicer: activation of human Argonaute-1. Cell Rep. 2013;3:1901-1909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 107] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 123. | Cho YS, Chennathukuzhi VM, Handel MA, Eppig J, Hecht NB. The relative levels of translin-associated factor X (TRAX) and testis brain RNA-binding protein determine their nucleocytoplasmic distribution in male germ cells. J Biol Chem. 2004;279:31514-31523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 38] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 124. | Sinkkonen L, Hugenschmidt T, Filipowicz W, Svoboda P. Dicer is associated with ribosomal DNA chromatin in mammalian cells. PLoS One. 2010;5:e12175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 67] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 125. | Ando Y, Tomaru Y, Morinaga A, Burroughs AM, Kawaji H, Kubosaki A, Kimura R, Tagata M, Ino Y, Hirano H. Nuclear pore complex protein mediated nuclear localization of dicer protein in human cells. PLoS One. 2011;6:e23385. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 126. | Castel SE, Ren J, Bhattacharjee S, Chang AY, Sánchez M, Valbuena A, Antequera F, Martienssen RA. Dicer promotes transcription termination at sites of replication stress to maintain genome stability. Cell. 2014;159:572-583. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 93] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 127. | Haussecker D, Proudfoot NJ. Dicer-dependent turnover of intergenic transcripts from the human beta-globin gene cluster. Mol Cell Biol. 2005;25:9724-9733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Redfern AD, Colley SM, Beveridge DJ, Ikeda N, Epis MR, Li X, Foulds CE, Stuart LM, Barker A, Russell VJ. RNA-induced silencing complex (RISC) Proteins PACT, TRBP, and Dicer are SRA binding nuclear receptor coregulators. Proc Natl Acad Sci USA. 2013;110:6536-6541. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 78] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 129. | Emmerth S, Schober H, Gaidatzis D, Roloff T, Jacobeit K, Bühler M. Nuclear retention of fission yeast dicer is a prerequisite for RNAi-mediated heterochromatin assembly. Dev Cell. 2010;18:102-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 130. | Giles KE, Ghirlando R, Felsenfeld G. Maintenance of a constitutive heterochromatin domain in vertebrates by a Dicer-dependent mechanism. Nat Cell Biol. 2010;12:94-99; sup pp 1-6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 42] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 131. | Baillat D, Shiekhattar R. Functional dissection of the human TNRC6 (GW182-related) family of proteins. Mol Cell Biol. 2009;29:4144-4155. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 132. | Partridge JF, DeBeauchamp JL, Kosinski AM, Ulrich DL, Hadler MJ, Noffsinger VJ. Functional separation of the requirements for establishment and maintenance of centromeric heterochromatin. Mol Cell. 2007;26:593-602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 133. | Pontier D, Picart C, Roudier F, Garcia D, Lahmy S, Azevedo J, Alart E, Laudié M, Karlowski WM, Cooke R. NERD, a plant-specific GW protein, defines an additional RNAi-dependent chromatin-based pathway in Arabidopsis. Mol Cell. 2012;48:121-132. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 102] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 134. | Gagnon KT, Li L, Janowski BA, Corey DR. Analysis of nuclear RNA interference in human cells by subcellular fractionation and Argonaute loading. Nat Protoc. 2014;9:2045-2060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 157] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 135. | Yamakawa N, Okuyama K, Ogata J, Kanai A, Helwak A, Takamatsu M, Imadome K, Takakura K, Chanda B, Kurosaki N. Novel functional small RNAs are selectively loaded onto mammalian Ago1. Nucleic Acids Res. 2014;42:5289-5301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 136. | Nakama M, Kawakami K, Kajitani T, Urano T, Murakami Y. DNA-RNA hybrid formation mediates RNAi-directed heterochromatin formation. Genes Cells. 2012;17:218-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 137. | Morris KV. siRNA-mediated transcriptional gene silencing: the potential mechanism and a possible role in the histone code. Cell Mol Life Sci. 2005;62:3057-3066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 62] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 138. | Kimura H. Histone modifications for human epigenome analysis. J Hum Genet. 2013;58:439-445. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 331] [Article Influence: 27.6] [Reference Citation Analysis (0)] |
| 139. | Rothbart SB, Strahl BD. Interpreting the language of histone and DNA modifications. Biochim Biophys Acta. 2014;1839:627-643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 501] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 140. | Gurard-Levin ZA, Almouzni G. Histone modifications and a choice of variant: a language that helps the genome express itself. F1000Prime Rep. 2014;6:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 36] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 141. | Verdin E. Histone Deacetylases. New Jersey: Humana Press 2006; . [DOI] [Full Text] |
| 142. | Zentner GE, Henikoff S. Regulation of nucleosome dynamics by histone modifications. Nat Struct Mol Biol. 2013;20:259-266. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 643] [Cited by in RCA: 636] [Article Influence: 53.0] [Reference Citation Analysis (0)] |
| 143. | Krouwels IM, Wiesmeijer K, Abraham TE, Molenaar C, Verwoerd NP, Tanke HJ, Dirks RW. A glue for heterochromatin maintenance: stable SUV39H1 binding to heterochromatin is reinforced by the SET domain. J Cell Biol. 2005;170:537-549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 144. | Maison C, Almouzni G. HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol. 2004;5:296-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 452] [Cited by in RCA: 462] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 145. | Ogawa Y, Sun BK, Lee JT. Intersection of the RNA interference and X-inactivation pathways. Science. 2008;320:1336-1341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 232] [Cited by in RCA: 218] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 146. | Mathiyalagan P, Okabe J, Chang L, Su Y, Du XJ, El-Osta A. The primary microRNA-208b interacts with Polycomb-group protein, Ezh2, to regulate gene expression in the heart. Nucleic Acids Res. 2014;42:790-803. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 147. | Jurkowska RZ, Jurkowski TP, Jeltsch A. Structure and function of mammalian DNA methyltransferases. Chembiochem. 2011;12:206-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 499] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 148. | Chen CC, Wang KY, Shen CK. DNA 5-methylcytosine demethylation activities of the mammalian DNA methyltransferases. J Biol Chem. 2013;288:9084-9091. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 149. | Delatte B, Fuks F. TET proteins: on the frenetic hunt for new cytosine modifications. Brief Funct Genomics. 2013;12:191-204. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 150. | Zou X, Ma W, Solov’yov IA, Chipot C, Schulten K. Recognition of methylated DNA through methyl-CpG binding domain proteins. Nucleic Acids Res. 2012;40:2747-2758. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 99] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 151. | Feldman N, Gerson A, Fang J, Li E, Zhang Y, Shinkai Y, Cedar H, Bergman Y. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006;8:188-194. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 503] [Cited by in RCA: 477] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 152. | Gidekel S, Bergman Y. A unique developmental pattern of Oct-3/4 DNA methylation is controlled by a cis-demodification element. J Biol Chem. 2002;277:34521-34530. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 98] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 153. | Yu DH, Ware C, Waterland RA, Zhang J, Chen MH, Gadkari M, Kunde-Ramamoorthy G, Nosavanh LM, Shen L. Developmentally programmed 3’ CpG island methylation confers tissue- and cell-type-specific transcriptional activation. Mol Cell Biol. 2013;33:1845-1858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 154. | Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99:247-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4452] [Cited by in RCA: 4279] [Article Influence: 164.6] [Reference Citation Analysis (0)] |
| 155. | Capuano F, Mülleder M, Kok R, Blom HJ, Ralser M. Cytosine DNA methylation is found in Drosophila melanogaster but absent in Saccharomyces cerevisiae, Schizosaccharomyces pombe, and other yeast species. Anal Chem. 2014;86:3697-3702. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 167] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 156. | Matzke MA, Mosher RA. RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet. 2014;15:394-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 966] [Cited by in RCA: 1015] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 157. | Mann JR, Mattiske DM. RNA interference in mammalian DNA methylation. Biochem Cell Biol. 2012;90:70-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 158. | Herbst F, Ball CR, Tuorto F, Nowrouzi A, Wang W, Zavidij O, Dieter SM, Fessler S, van der Hoeven F, Kloz U. Extensive methylation of promoter sequences silences lentiviral transgene expression during stem cell differentiation in vivo. Mol Ther. 2012;20:1014-1021. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 88] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 159. | Mok HP, Javed S, Lever A. Stable gene expression occurs from a minority of integrated HIV-1-based vectors: transcriptional silencing is present in the majority. Gene Ther. 2007;14:741-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 160. | Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167-176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 125] [Cited by in RCA: 138] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 161. | Qin JY, Zhang L, Clift KL, Hulur I, Xiang AP, Ren BZ, Lahn BT. Systematic comparison of constitutive promoters and the doxycycline-inducible promoter. PLoS One. 2010;5:e10611. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 318] [Cited by in RCA: 377] [Article Influence: 25.1] [Reference Citation Analysis (0)] |
| 162. | Pasini D, Bracken AP, Hansen JB, Capillo M, Helin K. The polycomb group protein Suz12 is required for embryonic stem cell differentiation. Mol Cell Biol. 2007;27:3769-3779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 544] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 163. | Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 902] [Cited by in RCA: 1026] [Article Influence: 64.1] [Reference Citation Analysis (0)] |
| 164. | Margueron R, Reinberg D. The Polycomb complex PRC2 and its mark in life. Nature. 2011;469:343-349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2760] [Cited by in RCA: 2510] [Article Influence: 179.3] [Reference Citation Analysis (0)] |
| 165. | Gil J, O’Loghlen A. PRC1 complex diversity: where is it taking us? Trends Cell Biol. 2014;24:632-641. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 111] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 166. | Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev. 2013;27:1318-1338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 583] [Cited by in RCA: 617] [Article Influence: 51.4] [Reference Citation Analysis (0)] |
| 167. | Kanhere A, Viiri K, Araújo CC, Rasaiyaah J, Bouwman RD, Whyte WA, Pereira CF, Brookes E, Walker K, Bell GW. Short RNAs are transcribed from repressed polycomb target genes and interact with polycomb repressive complex-2. Mol Cell. 2010;38:675-688. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 304] [Article Influence: 20.3] [Reference Citation Analysis (0)] |
| 168. | Li X. Genomic imprinting is a parental effect established in mammalian germ cells. Curr Top Dev Biol. 2013;102:35-59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 169. | Barlow DP. Genomic imprinting: a mammalian epigenetic discovery model. Annu Rev Genet. 2011;45:379-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 189] [Cited by in RCA: 189] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 170. | Nesterova TB, Popova BC, Cobb BS, Norton S, Senner CE, Tang YA, Spruce T, Rodriguez TA, Sado T, Merkenschlager M. Dicer regulates Xist promoter methylation in ES cells indirectly through transcriptional control of Dnmt3a. Epigenetics Chromatin. 2008;1:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 72] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 171. | Kota SK. RNAi in X inactivation: contrasting findings on the role of interference. Bioessays. 2009;31:1280-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 172. | Kota SK, Roy Chowdhury D, Rao LK, Padmalatha V, Singh L, Bhadra U. Uncoupling of X-linked gene silencing from XIST binding by DICER1 and chromatin modulation on human inactive X chromosome. Chromosoma. 2015;124:249-262. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 173. | Hu WS, Hughes SH. HIV-1 reverse transcription. Cold Spring Harb Perspect Med. 2012;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 283] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 174. | Wilusz J. Putting an ‘End’ to HIV mRNAs: capping and polyadenylation as potential therapeutic targets. AIDS Res Ther. 2013;10:31. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 17] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 175. | Dingwall C, Ernberg I, Gait MJ, Green SM, Heaphy S, Karn J, Lowe AD, Singh M, Skinner MA. HIV-1 tat protein stimulates transcription by binding to a U-rich bulge in the stem of the TAR RNA structure. EMBO J. 1990;9:4145-4153. [PubMed] |
| 176. | Kim J, Kim H. Recruitment and biological consequences of histone modification of H3K27me3 and H3K9me3. ILAR J. 2012;53:232-239. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 68] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 177. | Friedman J, Cho WK, Chu CK, Keedy KS, Archin NM, Margolis DM, Karn J. Epigenetic silencing of HIV-1 by the histone H3 lysine 27 methyltransferase enhancer of Zeste 2. J Virol. 2011;85:9078-9089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 234] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 178. | Le Douce V, Colin L, Redel L, Cherrier T, Herbein G, Aunis D, Rohr O, Van Lint C, Schwartz C. LSD1 cooperates with CTIP2 to promote HIV-1 transcriptional silencing. Nucleic Acids Res. 2012;40:1904-1915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 179. | du Chéné I, Basyuk E, Lin YL, Triboulet R, Knezevich A, Chable-Bessia C, Mettling C, Baillat V, Reynes J, Corbeau P. Suv39H1 and HP1gamma are responsible for chromatin-mediated HIV-1 transcriptional silencing and post-integration latency. EMBO J. 2007;26:424-435. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 238] [Cited by in RCA: 260] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 180. | Marban C, Suzanne S, Dequiedt F, de Walque S, Redel L, Van Lint C, Aunis D, Rohr O. Recruitment of chromatin-modifying enzymes by CTIP2 promotes HIV-1 transcriptional silencing. EMBO J. 2007;26:412-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 264] [Cited by in RCA: 299] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 181. | Imai K, Togami H, Okamoto T. Involvement of histone H3 lysine 9 (H3K9) methyltransferase G9a in the maintenance of HIV-1 latency and its reactivation by BIX01294. J Biol Chem. 2010;285:16538-16545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 210] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 182. | Kobayashi-Ishihara M, Yamagishi M, Hara T, Matsuda Y, Takahashi R, Miyake A, Nakano K, Yamochi T, Ishida T, Watanabe T. HIV-1-encoded antisense RNA suppresses viral replication for a prolonged period. Retrovirology. 2012;9:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 85] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 183. | Saayman S, Ackley A, Turner AM, Famiglietti M, Bosque A, Clemson M, Planelles V, Morris KV. An HIV-encoded antisense long noncoding RNA epigenetically regulates viral transcription. Mol Ther. 2014;22:1164-1175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 120] [Cited by in RCA: 142] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 184. | Chávez L, Kauder S, Verdin E. In vivo, in vitro, and in silico analysis of methylation of the HIV-1 provirus. Methods. 2011;53:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 185. | Brockdorff N. Noncoding RNA and Polycomb recruitment. RNA. 2013;19:429-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 236] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 186. | Thaler F, Mercurio C. Towards selective inhibition of histone deacetylase isoforms: what has been achieved, where we are and what will be next. ChemMedChem. 2014;9:523-526. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 187. | Lee NS, Rossi JJ. Control of HIV-1 replication by RNA interference. Virus Res. 2004;102:53-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 55] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 188. | Park WS, Hayafune M, Miyano-Kurosaki N, Takaku H. Specific HIV-1 env gene silencing by small interfering RNAs in human peripheral blood mononuclear cells. Gene Ther. 2003;10:2046-2050. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 189. | Lee NS, Dohjima T, Bauer G, Li H, Li MJ, Ehsani A, Salvaterra P, Rossi J. Expression of small interfering RNAs targeted against HIV-1 rev transcripts in human cells. Nat Biotechnol. 2002;20:500-505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 646] [Cited by in RCA: 671] [Article Influence: 29.2] [Reference Citation Analysis (0)] |
| 190. | Novina CD, Murray MF, Dykxhoorn DM, Beresford PJ, Riess J, Lee SK, Collman RG, Lieberman J, Shankar P, Sharp PA. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8:681-686. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 597] [Cited by in RCA: 586] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 191. | Martínez MA, Gutiérrez A, Armand-Ugón M, Blanco J, Parera M, Gómez J, Clotet B, Esté JA. Suppression of chemokine receptor expression by RNA interference allows for inhibition of HIV-1 replication. AIDS. 2002;16:2385-2390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 159] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 192. | Das AT, Brummelkamp TR, Westerhout EM, Vink M, Madiredjo M, Bernards R, Berkhout B. Human immunodeficiency virus type 1 escapes from RNA interference-mediated inhibition. J Virol. 2004;78:2601-2605. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 346] [Cited by in RCA: 350] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 193. | Westerhout EM, Ooms M, Vink M, Das AT, Berkhout B. HIV-1 can escape from RNA interference by evolving an alternative structure in its RNA genome. Nucleic Acids Res. 2005;33:796-804. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 314] [Cited by in RCA: 319] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 194. | von Eije KJ, ter Brake O, Berkhout B. Human immunodeficiency virus type 1 escape is restricted when conserved genome sequences are targeted by RNA interference. J Virol. 2008;82:2895-2903. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 98] [Cited by in RCA: 102] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 195. | ter Brake O, Konstantinova P, Ceylan M, Berkhout B. Silencing of HIV-1 with RNA interference: a multiple shRNA approach. Mol Ther. 2006;14:883-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 243] [Cited by in RCA: 240] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 196. | von Eije KJ, ter Brake O, Berkhout B. Stringent testing identifies highly potent and escape-proof anti-HIV short hairpin RNAs. J Gene Med. 2009;11:459-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 197. | Schopman NC, ter Brake O, Berkhout B. Anticipating and blocking HIV-1 escape by second generation antiviral shRNAs. Retrovirology. 2010;7:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 198. | Méndez-Ortega MC, Restrepo S, Rodríguez-R LM, Pérez I, Mendoza JC, Martínez AP, Sierra R, Rey-Benito GJ. An RNAi in silico approach to find an optimal shRNA cocktail against HIV-1. Virol J. 2010;7:369. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 199. | McIntyre GJ, Arndt AJ, Gillespie KM, Mak WM, Fanning GC. A comparison of multiple shRNA expression methods for combinatorial RNAi. Genet Vaccines Ther. 2011;9:9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 13] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 200. | Eekels JJ, Geerts D, Jeeninga RE, Berkhout B. Long-term inhibition of HIV-1 replication with RNA interference against cellular co-factors. Antiviral Res. 2011;89:43-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 201. | Suzuki K, Shijuuku T, Fukamachi T, Zaunders J, Guillemin G, Cooper D, Kelleher A. Prolonged transcriptional silencing and CpG methylation induced by siRNAs targeted to the HIV-1 promoter region. J RNAi Gene Silencing. 2005;1:66-78. [PubMed] |
| 202. | Suzuki K, Juelich T, Lim H, Ishida T, Watanebe T, Cooper DA, Rao S, Kelleher AD. Closed chromatin architecture is induced by an RNA duplex targeting the HIV-1 promoter region. J Biol Chem. 2008;283:23353-23363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 203. | Yamagishi M, Ishida T, Miyake A, Cooper DA, Kelleher AD, Suzuki K, Watanabe T. Retroviral delivery of promoter-targeted shRNA induces long-term silencing of HIV-1 transcription. Microbes Infect. 2009;11:500-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 204. | Suzuki K, Ishida T, Yamagishi M, Ahlenstiel C, Swaminathan S, Marks K, Murray D, McCartney EM, Beard MR, Alexander M. Transcriptional gene silencing of HIV-1 through promoter targeted RNA is highly specific. RNA Biol. 2011;8:1035-1046. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 205. | Ahlenstiel CL, Lim HG, Cooper DA, Ishida T, Kelleher AD, Suzuki K. Direct evidence of nuclear Argonaute distribution during transcriptional silencing links the actin cytoskeleton to nuclear RNAi machinery in human cells. Nucleic Acids Res. 2012;40:1579-1595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 206. | Mack KD, Jin X, Yu S, Wei R, Kapp L, Green C, Herndier B, Abbey NW, Elbaggari A, Liu Y. HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J Acquir Immune Defic Syndr. 2003;33:308-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 207. | Sloan RD, Wainberg MA. The role of unintegrated DNA in HIV infection. Retrovirology. 2011;8:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 100] [Cited by in RCA: 108] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 208. | Westerhout EM, ter Brake O, Berkhout B. The virion-associated incoming HIV-1 RNA genome is not targeted by RNA interference. Retrovirology. 2006;3:57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 209. | Hütter G, Nowak D, Mossner M, Ganepola S, Müssig A, Allers K, Schneider T, Hofmann J, Kücherer C, Blau O. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360:692-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1330] [Cited by in RCA: 1374] [Article Influence: 85.9] [Reference Citation Analysis (0)] |
| 210. | Carrington M, Dean M, Martin MP, O’Brien SJ. Genetics of HIV-1 infection: chemokine receptor CCR5 polymorphism and its consequences. Hum Mol Genet. 1999;8:1939-1945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 158] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 211. | Allers K, Hütter G, Hofmann J, Loddenkemper C, Rieger K, Thiel E, Schneider T. Evidence for the cure of HIV infection by CCR5Δ32/Δ32 stem cell transplantation. Blood. 2011;117:2791-2799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 525] [Cited by in RCA: 525] [Article Influence: 35.0] [Reference Citation Analysis (0)] |
| 212. | Naif HM. Pathogenesis of HIV Infection. Infect Dis Rep. 2013;5:e6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 70] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 213. | Ferrer P, Tello M, Montecinos L, Tordecilla R, Rodríguez C, Beltrán C, Guzmán MA, Ferrés M, Pérez CM, Afani A. Prevalence of R5 and X4 HIV variants in antiretroviral treatment experienced patients with virologic failure. J Clin Virol. 2014;60:290-294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 214. | Lim JK, Murphy PM. Chemokine control of West Nile virus infection. Exp Cell Res. 2011;317:569-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 57] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 215. | Sanchooli J, Sanadgol N, Kazemi Arababadi M, Kennedy D. CCR5 plays important roles in hepatitis B infection. Viral Immunol. 2014;27:2-6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 216. | Ghorban K, Dadmanesh M, Hassanshahi G, Momeni M, Zare-Bidaki M, Arababadi MK, Kennedy D. Is the CCR5 Δ 32 mutation associated with immune system-related diseases? Inflammation. 2013;36:633-642. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 217. | Burke BP, Boyd MP, Impey H, Breton LR, Bartlett JS, Symonds GP, Hütter G. CCR5 as a natural and modulated target for inhibition of HIV. Viruses. 2014;6:54-68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 218. | Walker JE, Chen RX, McGee J, Nacey C, Pollard RB, Abedi M, Bauer G, Nolta JA, Anderson JS. Generation of an HIV-1-resistant immune system with CD34(+) hematopoietic stem cells transduced with a triple-combination anti-HIV lentiviral vector. J Virol. 2012;86:5719-5729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 71] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 219. | Li MJ, Kim J, Li S, Zaia J, Yee JK, Anderson J, Akkina R, Rossi JJ. Long-term inhibition of HIV-1 infection in primary hematopoietic cells by lentiviral vector delivery of a triple combination of anti-HIV shRNA, anti-CCR5 ribozyme, and a nucleolar-localizing TAR decoy. Mol Ther. 2005;12:900-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 192] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 220. | DiGiusto DL, Krishnan A, Li L, Li H, Li S, Rao A, Mi S, Yam P, Stinson S, Kalos M. RNA-based gene therapy for HIV with lentiviral vector-modified CD34(+) cells in patients undergoing transplantation for AIDS-related lymphoma. Sci Transl Med. 2010;2:36ra43. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 324] [Cited by in RCA: 301] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 221. | Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, Spratt SK, Surosky RT, Giedlin MA, Nichol G. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:901-910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1057] [Cited by in RCA: 1039] [Article Influence: 94.5] [Reference Citation Analysis (0)] |
| 222. | Li L, Krymskaya L, Wang J, Henley J, Rao A, Cao LF, Tran CA, Torres-Coronado M, Gardner A, Gonzalez N. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21:1259-1269. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 151] [Cited by in RCA: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 223. | Ye L, Wang J, Beyer AI, Teque F, Cradick TJ, Qi Z, Chang JC, Bao G, Muench MO, Yu J. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Δ32 mutation confers resistance to HIV infection. Proc Natl Acad Sci USA. 2014;111:9591-9596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 241] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 224. | Hu W, Kaminski R, Yang F, Zhang Y, Cosentino L, Li F, Luo B, Alvarez-Carbonell D, Garcia-Mesa Y, Karn J. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci USA. 2014;111:11461-11466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 410] [Cited by in RCA: 406] [Article Influence: 36.9] [Reference Citation Analysis (0)] |
| 225. | Wayengera M. Proviral HIV-genome-wide and pol-gene specific zinc finger nucleases: usability for targeted HIV gene therapy. Theor Biol Med Model. 2011;8:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 226. | Morrison SJ, Kimble J. Asymmetric and symmetric stem-cell divisions in development and cancer. Nature. 2006;441:1068-1074. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 957] [Cited by in RCA: 937] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 227. | Bazrgar M, Gourabi H, Valojerdi MR, Yazdi PE, Baharvand H. Self-correction of chromosomal abnormalities in human preimplantation embryos and embryonic stem cells. Stem Cells Dev. 2013;22:2449-2456. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 228. | Guillemette B, Drogaris P, Lin HH, Armstrong H, Hiragami-Hamada K, Imhof A, Bonneil E, Thibault P, Verreault A, Festenstein RJ. H3 lysine 4 is acetylated at active gene promoters and is regulated by H3 lysine 4 methylation. PLoS Genet. 2011;7:e1001354. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 229. | Tang Z, Chen WY, Shimada M, Nguyen UT, Kim J, Sun XJ, Sengoku T, McGinty RK, Fernandez JP, Muir TW. SET1 and p300 act synergistically, through coupled histone modifications, in transcriptional activation by p53. Cell. 2013;154:297-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 138] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 230. | Keating ST, El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics. 2013;8:361-372. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 231. | Ali M, Hom RA, Blakeslee W, Ikenouye L, Kutateladze TG. Diverse functions of PHD fingers of the MLL/KMT2 subfamily. Biochim Biophys Acta. 2014;1843:366-371. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 65] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 232. | Abu-Farha M, Lambert JP, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol Cell Proteomics. 2008;7:560-572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 151] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 233. | Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119:941-953. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2977] [Cited by in RCA: 3206] [Article Influence: 160.3] [Reference Citation Analysis (0)] |
| 234. | Rasmussen PB, Staller P. The KDM5 family of histone demethylases as targets in oncology drug discovery. Epigenomics. 2014;6:277-286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 235. | Persson J, Ekwall K. Chd1 remodelers maintain open chromatin and regulate the epigenetics of differentiation. Exp Cell Res. 2010;316:1316-1323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 236. | Jones JM, Simkus C. The roles of the RAG1 and RAG2 “non-core” regions in V(D)J recombination and lymphocyte development. Arch Immunol Ther Exp (Warsz). 2009;57:105-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 237. | Lauberth SM, Nakayama T, Wu X, Ferris AL, Tang Z, Hughes SH, Roeder RG. H3K4me3 interactions with TAF3 regulate preinitiation complex assembly and selective gene activation. Cell. 2013;152:1021-1036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 279] [Cited by in RCA: 329] [Article Influence: 27.4] [Reference Citation Analysis (0)] |
| 238. | Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD, Patel DJ. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature. 2006;442:91-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 527] [Cited by in RCA: 590] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 239. | Lan F, Collins RE, De Cegli R, Alpatov R, Horton JR, Shi X, Gozani O, Cheng X, Shi Y. Recognition of unmethylated histone H3 lysine 4 links BHC80 to LSD1-mediated gene repression. Nature. 2007;448:718-722. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 354] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 240. | Tallen G, Riabowol K. Keep-ING balance: tumor suppression by epigenetic regulation. FEBS Lett. 2014;588:2728-2742. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 57] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 241. | Gu B, Sun P, Yuan Y, Moraes RC, Li A, Teng A, Agrawal A, Rhéaume C, Bilanchone V, Veltmaat JM. Pygo2 expands mammary progenitor cells by facilitating histone H3 K4 methylation. J Cell Biol. 2009;185:811-826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 95] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 242. | Gu B, Lee MG. Histone H3 lysine 4 methyltransferases and demethylases in self-renewal and differentiation of stem cells. Cell Biosci. 2013;3:39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 243. | Metzger E, Imhof A, Patel D, Kahl P, Hoffmeyer K, Friedrichs N, Müller JM, Greschik H, Kirfel J, Ji S. Phosphorylation of histone H3T6 by PKCbeta(I) controls demethylation at histone H3K4. Nature. 2010;464:792-796. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 217] [Cited by in RCA: 233] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 244. | Jin Q, Yu LR, Wang L, Zhang Z, Kasper LH, Lee JE, Wang C, Brindle PK, Dent SY, Ge K. Distinct roles of GCN5/PCAF-mediated H3K9ac and CBP/p300-mediated H3K18/27ac in nuclear receptor transactivation. EMBO J. 2011;30:249-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 698] [Cited by in RCA: 638] [Article Influence: 45.6] [Reference Citation Analysis (0)] |
| 245. | Michishita E, McCord RA, Berber E, Kioi M, Padilla-Nash H, Damian M, Cheung P, Kusumoto R, Kawahara TL, Barrett JC. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492-496. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 897] [Cited by in RCA: 838] [Article Influence: 49.3] [Reference Citation Analysis (0)] |
| 246. | Kanno T, Kanno Y, LeRoy G, Campos E, Sun HW, Brooks SR, Vahedi G, Heightman TD, Garcia BA, Reinberg D. BRD4 assists elongation of both coding and enhancer RNAs by interacting with acetylated histones. Nat Struct Mol Biol. 2014;21:1047-1057. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 229] [Cited by in RCA: 249] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 247. | Krishnan S, Horowitz S, Trievel RC. Structure and function of histone H3 lysine 9 methyltransferases and demethylases. Chembiochem. 2011;12:254-263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 248. | Mozzetta C, Pontis J, Fritsch L, Robin P, Portoso M, Proux C, Margueron R, Ait-Si-Ali S. The histone H3 lysine 9 methyltransferases G9a and GLP regulate polycomb repressive complex 2-mediated gene silencing. Mol Cell. 2014;53:277-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 203] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 249. | Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature. 2010;464:927-931. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 603] [Cited by in RCA: 600] [Article Influence: 40.0] [Reference Citation Analysis (0)] |
| 250. | Loh YH, Zhang W, Chen X, George J, Ng HH. Jmjd1a and Jmjd2c histone H3 Lys 9 demethylases regulate self-renewal in embryonic stem cells. Genes Dev. 2007;21:2545-2557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 251. | Brauchle M, Yao Z, Arora R, Thigale S, Clay I, Inverardi B, Fletcher J, Taslimi P, Acker MG, Gerrits B. Protein complex interactor analysis and differential activity of KDM3 subfamily members towards H3K9 methylation. PLoS One. 2013;8:e60549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 49] [Cited by in RCA: 67] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 252. | Whetstine JR, Nottke A, Lan F, Huarte M, Smolikov S, Chen Z, Spooner E, Li E, Zhang G, Colaiacovo M. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467-481. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 768] [Cited by in RCA: 788] [Article Influence: 41.5] [Reference Citation Analysis (0)] |
| 253. | Nishibuchi G, Machida S, Osakabe A, Murakoshi H, Hiragami-Hamada K, Nakagawa R, Fischle W, Nishimura Y, Kurumizaka H, Tagami H. N-terminal phosphorylation of HP1α increases its nucleosome-binding specificity. Nucleic Acids Res. 2014;42:12498-12511. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 65] [Cited by in RCA: 62] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 254. | Bua DJ, Kuo AJ, Cheung P, Liu CL, Migliori V, Espejo A, Casadio F, Bassi C, Amati B, Bedford MT. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 255. | Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010;29:3673-3687. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 103] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 256. | Zhang J, Gao Q, Li P, Liu X, Jia Y, Wu W, Li J, Dong S, Koseki H, Wong J. S phase-dependent interaction with DNMT1 dictates the role of UHRF1 but not UHRF2 in DNA methylation maintenance. Cell Res. 2011;21:1723-1739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 257. | Fischle W, Franz H, Jacobs SA, Allis CD, Khorasanizadeh S. Specificity of the chromodomain Y chromosome family of chromodomains for lysine-methylated ARK(S/T) motifs. J Biol Chem. 2008;283:19626-19635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 258. | Margueron R, Li G, Sarma K, Blais A, Zavadil J, Woodcock CL, Dynlacht BD, Reinberg D. Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms. Mol Cell. 2008;32:503-518. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 726] [Cited by in RCA: 672] [Article Influence: 39.5] [Reference Citation Analysis (0)] |
| 259. | Hong S, Cho YW, Yu LR, Yu H, Veenstra TD, Ge K. Identification of JmjC domain-containing UTX and JMJD3 as histone H3 lysine 27 demethylases. Proc Natl Acad Sci USA. 2007;104:18439-18444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 588] [Cited by in RCA: 545] [Article Influence: 30.3] [Reference Citation Analysis (0)] |
| 260. | Margueron R, Justin N, Ohno K, Sharpe ML, Son J, Drury WJ, Voigt P, Martin SR, Taylor WR, De Marco V. Role of the polycomb protein EED in the propagation of repressive histone marks. Nature. 2009;461:762-767. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 875] [Cited by in RCA: 914] [Article Influence: 57.1] [Reference Citation Analysis (0)] |
| 261. | Son J, Shen SS, Margueron R, Reinberg D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013;27:2663-2677. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 134] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 262. | Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol. 2012;13:115-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 640] [Cited by in RCA: 729] [Article Influence: 56.1] [Reference Citation Analysis (0)] |
| 263. | Yuan G, Ma B, Yuan W, Zhang Z, Chen P, Ding X, Feng L, Shen X, Chen S, Li G. Histone H2A ubiquitination inhibits the enzymatic activity of H3 lysine 36 methyltransferases. J Biol Chem. 2013;288:30832-30842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 264. | Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y, Yan X, Wei W, Pang Y, Cheng H. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet. 2014;46:287-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 197] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 265. | Martinez-Garcia E, Popovic R, Min DJ, Sweet SM, Thomas PM, Zamdborg L, Heffner A, Will C, Lamy L, Staudt LM. The MMSET histone methyl transferase switches global histone methylation and alters gene expression in t(4; 14) multiple myeloma cells. Blood. 2011;117:211-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 260] [Cited by in RCA: 282] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 266. | Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem. 2011;286:7983-7989. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 383] [Cited by in RCA: 416] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 267. | Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, Zhang Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811-816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1516] [Cited by in RCA: 1623] [Article Influence: 81.2] [Reference Citation Analysis (0)] |
| 268. | He J, Kallin EM, Tsukada Y, Zhang Y. The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat Struct Mol Biol. 2008;15:1169-1175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 262] [Cited by in RCA: 242] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 269. | Maltby VE, Martin BJ, Schulze JM, Johnson I, Hentrich T, Sharma A, Kobor MS, Howe L. Histone H3 lysine 36 methylation targets the Isw1b remodeling complex to chromatin. Mol Cell Biol. 2012;32:3479-3485. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 55] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 270. | Beck DB, Oda H, Shen SS, Reinberg D. PR-Set7 and H4K20me1: at the crossroads of genome integrity, cell cycle, chromosome condensation, and transcription. Genes Dev. 2012;26:325-337. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 223] [Cited by in RCA: 260] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 271. | Liu W, Tanasa B, Tyurina OV, Zhou TY, Gassmann R, Liu WT, Ohgi KA, Benner C, Garcia-Bassets I, Aggarwal AK. PHF8 mediates histone H4 lysine 20 demethylation events involved in cell cycle progression. Nature. 2010;466:508-512. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 362] [Cited by in RCA: 334] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 272. | Kalakonda N, Fischle W, Boccuni P, Gurvich N, Hoya-Arias R, Zhao X, Miyata Y, Macgrogan D, Zhang J, Sims JK. Histone H4 lysine 20 monomethylation promotes transcriptional repression by L3MBTL1. Oncogene. 2008;27:4293-4304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 124] [Cited by in RCA: 124] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 273. | Jørgensen S, Schotta G, Sørensen CS. Histone H4 lysine 20 methylation: key player in epigenetic regulation of genomic integrity. Nucleic Acids Res. 2013;41:2797-2806. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 253] [Cited by in RCA: 280] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 274. | Tsang LW, Hu N, Underhill DA. Comparative analyses of SUV420H1 isoforms and SUV420H2 reveal differences in their cellular localization and effects on myogenic differentiation. PLoS One. 2010;5:e14447. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 275. | Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ, Boutros M, Perrimon N, Rosenfeld MG, Glass CK. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell. 2012;48:28-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 157] [Cited by in RCA: 184] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 276. | Adams-Cioaba MA, Li Z, Tempel W, Guo Y, Bian C, Li Y, Lam R, Min J. Crystal structures of the Tudor domains of human PHF20 reveal novel structural variations on the Royal Family of proteins. FEBS Lett. 2012;586:859-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 277. | Min J, Allali-Hassani A, Nady N, Qi C, Ouyang H, Liu Y, MacKenzie F, Vedadi M, Arrowsmith CH. L3MBTL1 recognition of mono- and dimethylated histones. Nat Struct Mol Biol. 2007;14:1229-1230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 159] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 278. | Verdin E, Paras P, Van Lint C. Chromatin disruption in the promoter of human immunodeficiency virus type 1 during transcriptional activation. EMBO J. 1993;12:3249-3259. [PubMed] |
| 279. | Rohr O, Aunis D, Schaeffer E. COUP-TF and Sp1 interact and cooperate in the transcriptional activation of the human immunodeficiency virus type 1 long terminal repeat in human microglial cells. J Biol Chem. 1997;272:31149-31155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 79] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 280. | Jiang G, Espeseth A, Hazuda DJ, Margolis DM. c-Myc and Sp1 contribute to proviral latency by recruiting histone deacetylase 1 to the human immunodeficiency virus type 1 promoter. J Virol. 2007;81:10914-10923. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 166] [Cited by in RCA: 199] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 281. | Bernhard W, Barreto K, Raithatha S, Sadowski I. An upstream YY1 binding site on the HIV-1 LTR contributes to latent infection. PLoS One. 2013;8:e77052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 282. | Li C, Lai CF, Sigman DS, Gaynor RB. Cloning of a cellular factor, interleukin binding factor, that binds to NFAT-like motifs in the human immunodeficiency virus long terminal repeat. Proc Natl Acad Sci USA. 1991;88:7739-7743. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 84] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 283. | Mitra D, Sikder SK, Laurence J. Role of glucocorticoid receptor binding sites in the human immunodeficiency virus type 1 long terminal repeat in steroid-mediated suppression of HIV gene expression. Virology. 1995;214:512-521. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 34] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 284. | Kino T, Kopp JB, Chrousos GP. Glucocorticoids suppress human immunodeficiency virus type-1 long terminal repeat activity in a cell type-specific, glucocorticoid receptor-mediated fashion: direct protective effects at variance with clinical phenomenology. J Steroid Biochem Mol Biol. 2000;75:283-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 31] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 285. | Hanley TM, Viglianti GA. Nuclear receptor signaling inhibits HIV-1 replication in macrophages through multiple trans-repression mechanisms. J Virol. 2011;85:10834-10850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 286. | Duverger A, Wolschendorf F, Zhang M, Wagner F, Hatcher B, Jones J, Cron RQ, van der Sluis RM, Jeeninga RE, Berkhout B. An AP-1 binding site in the enhancer/core element of the HIV-1 promoter controls the ability of HIV-1 to establish latent infection. J Virol. 2013;87:2264-2277. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 287. | Malcolm T, Chen J, Chang C, Sadowski I. Induction of chromosomally integrated HIV-1 LTR requires RBF-2 (USF/TFII-I) and Ras/MAPK signaling. Virus Genes. 2007;35:215-223. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 43] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 288. | Dahabieh MS, Ooms M, Malcolm T, Simon V, Sadowski I. Identification and functional analysis of a second RBF-2 binding site within the HIV-1 promoter. Virology. 2011;418:57-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 289. | Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J. 2006;25:139-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 368] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 290. | Williams SA, Kwon H, Chen LF, Greene WC. Sustained induction of NF-kappa B is required for efficient expression of latent human immunodeficiency virus type 1. J Virol. 2007;81:6043-6056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 114] [Cited by in RCA: 119] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 291. | Romanchikova N, Ivanova V, Scheller C, Jankevics E, Jassoy C, Serfling E. NFAT transcription factors control HIV-1 expression through a binding site downstream of TAR region. Immunobiology. 2003;208:361-365. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 292. | Marban C, Redel L, Suzanne S, Van Lint C, Lecestre D, Chasserot-Golaz S, Leid M, Aunis D, Schaeffer E, Rohr O. COUP-TF interacting protein 2 represses the initial phase of HIV-1 gene transcription in human microglial cells. Nucleic Acids Res. 2005;33:2318-2331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 76] [Cited by in RCA: 89] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 293. | Rohr O, Lecestre D, Chasserot-Golaz S, Marban C, Avram D, Aunis D, Leid M, Schaeffer E. Recruitment of Tat to heterochromatin protein HP1 via interaction with CTIP2 inhibits human immunodeficiency virus type 1 replication in microglial cells. J Virol. 2003;77:5415-5427. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 294. | Malcolm T, Kam J, Pour PS, Sadowski I. Specific interaction of TFII-I with an upstream element on the HIV-1 LTR regulates induction of latent provirus. FEBS Lett. 2008;582:3903-3908. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 24] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 295. | Imai K, Okamoto T. Transcriptional repression of human immunodeficiency virus type 1 by AP-4. J Biol Chem. 2006;281:12495-12505. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 123] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 296. | Ou SH, Garcia-Martínez LF, Paulssen EJ, Gaynor RB. Role of flanking E box motifs in human immunodeficiency virus type 1 TATA element function. J Virol. 1994;68:7188-7199. [PubMed] |
| 297. | He G, Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Mol Cell Biol. 2002;22:2965-2973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 199] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 298. | Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. J Virol. 2000;74:6790-6799. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 273] [Cited by in RCA: 287] [Article Influence: 11.5] [Reference Citation Analysis (0)] |