Published online May 12, 2015. doi: 10.5501/wjv.v4.i2.124
Peer-review started: November 10, 2014
First decision: December 26, 2014
Revised: January 20, 2015
Accepted: February 10, 2015
Article in press: February 12, 2015
Published online: May 12, 2015
Processing time: 172 Days and 15.7 Hours
Host and viral factors deeply influence the human immunodeficiency virus (HIV) disease progression. Among them human leukocyte antigen (HLA) locus plays a key role at different levels. In fact, genes of the HLA locus have shown the peculiar capability to modulate both innate and adaptive immune responses. In particular, HLA class I molecules are recognized by CD8+ T-cells and natural killers (NK) cells towards the interaction with T cell receptor (TCR) and Killer Immunoglobulin Receptor (KIR) 3DL1 respectively. Polymorphisms within the different HLA alleles generate structural changes in HLA class I peptide-binding pockets. Amino acid changes in the peptide-binding pocket lead to the presentation of a different set of peptides to T and NK cells. This review summarizes the role of HLA in HIV progression toward acquired immunodeficiency disease syndrome and its receptors. Recently, many studies have been focused on determining the HLA binding-peptides. The novel use of immune-informatics tools, from the prediction of the HLA-bound peptides to the modification of the HLA-receptor complexes, is considered. A better knowledge of HLA peptide presentation and recognition are allowing new strategies for immune response manipulation to be applied against HIV virus.
Core tip: Human immunodeficiency virus (HIV) disease progression depends on several host factors. Among them human leukocyte antigen (HLA) locus has a main role due to the peculiar capability to modulate both innate and adaptive immune response. In this review, the role of HLA molecules and its receptors in HIV progression toward acquired immunodeficiency disease syndrome is summarized. A better knowledge about HLA-peptide presentation and recognition by immune cells will open new applications in HIV vaccine and diagnostics design.
- Citation: Grifoni A, Montesano C, Colizzi V, Amicosante M. Key role of human leukocyte antigen in modulating human immunodeficiency virus progression: An overview of the possible applications. World J Virology 2015; 4(2): 124-133
- URL: https://www.wjgnet.com/2220-3249/full/v4/i2/124.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i2.124
Different host’s genetic factors have been associated both with rapid and slow progression to acquired immunodeficiency disease syndrome (AIDS). This suggests that the efficient control of human immunodeficiency virus (HIV) -1 infection lays on different variants of immune response associated genes. In this context, genetic association studies have been strongly limited by different factors. HIV-1 is a quasi-species virus with large variability among the population even if small geographical areas are examined. In this review, the strong contribution of human leukocyte antigen (HLA) locus in HIV progression is highlighted. The use of immune-informatics is capable to efficiently predict the HLA binding peptides, adding important information in this context. Overall, this might lead to the design of preventive vaccine and immunotherapies capable to improve the HIV immune response.
HIV immune response depends on both innate and adaptive compartment of the immune system. The primary HIV infection typically occurs in the mucosa. At this level, resident memory CD4+ T cells are infected together with dendritic cells, granulocytes, natural killer (NK) cells and macrophages[1]. Subsequently, infected cells and virus particles bounded by dendritic cells and B lymphocytes reach the lymph nodes. Within the lymph nodes, HIV-1 infects also the effector-memory and the activated CD4+ T cells.
These processes are responsible for the increase of viral spread, viremia and decrease in the number of CD4+ T cells[1]. Early events, which occur directly after HIV infection, determine the course of HIV disease progression. The reduction of viral replication often occurs before the development of the adaptive immune response against HIV, suggesting that the innate immune system has an essential role in controlling the infection[2,3].
Studies on primary HIV infection before the seroconversion show the presence of HIV-specific adaptive immune response exert by CD8+ T lymphocytes (CTL)[4-6]. CTL immune responses play a central role in the control of viral replication as it has been observed in Long Term Non-Progressor (LTNP) patients. Different mechanisms for viral inhibition mediated by CTL immune response have been observed.
HIV infected cells are recognized by the TCR of HIV-specific CTLs when viral peptides are presented at the cell surface in the context of HLA class I molecules. This recognition leads to CTL cytotoxic immune response[7].
Humoral immune response has a secondary role in the control of HIV infection. Although neutralizing antibodies reduce the virus particles and therefore the viral spread. However, serum of the infected patients does not reduce the viral infectivity in vitro and the efficacy of gp120 neutralizing antibodies is reduced. This is due to the fact that gp120, HIV glycoprotein responsible for the viral entry, has a high mutation frequency which leads to conformational changes impairing the antibody binding[7].
Due to the lack of capability of the immune response to eradicate HIV, the infection becomes chronic and the virus is integrated in a latent form in the human genome. Despite the return of circulating CD4+T cells to normal levels, massive immune activation and accelerated cell turnover takes place. The ultimate consequence of immune activation is the depletion of CD4+ T cells. In absence of T helper response the immune system is not able to control other infections, therefore opportunistic infections occur and lead to AIDS[1].
In general, HIV protective immune response is associated with recognition and activation of the cytolytic function exerted mostly by NK and CD8+ T cells. Thus, the contribution of HLA molecules and its ligands play a key role in controlling HIV disease progression[8].
The progression of HIV infection has different phases. In the primary infection, HIV infects mainly macrophages and dendritic cells by using the co-receptor C-C chemokine receptor 5 (CCR5) together with the CD4 molecule. Virus replication in the lymph nodes leads to the viremic peak characteristic of acute infection[4,5]. The viremia increases the viral spread in the other lymph nodes of the entire organism. The immune system mounts a response to control the viremia, which decreases towards a stationary phase named “set point”. In most of the cases, the immune response is not capable to eradicate the infection. Therefore, an equilibrium between host and virus occurs and the viral DNA is integrated in a latent form that could not be detected by the immune system[6].
In the late phase of the infection, the constant viral replication induces a tropism shift. The virus prefers C-X-C chemokine receptor type 4 co-receptor (CXCR4) and infects mainly the CD4+ T-cells. The CD4+ T-cells depletion (< 200 cell/mm3) and the increase of the viral load lead to an impairment of the entire immune system. Therefore, opportunistic infections occur, leading to AIDS and often to death[7]. Clinical latency period has a large variability in the HIV-infected subjects with different disease progression rates in absence of antiretroviral therapy.
Most of the infected individuals (70%-80%) are defined as slow progressors (SP). SP are characterized by increasing of viral load and CD4+ T-cell count decline towards AIDS within 6-10 years of HIV infection. A smaller percentage of individuals (10%-15%), defined as fast progressors, have a fast CD4 count decline and develop AIDS within few years after infection. The LTNP represent about 5% of the infected cases and do not have significant changes in CD4 count, viral load or clinical symptoms for over 10 years[7-9]. Among them, a subgroup named elite controllers (EC) is characterized by stable CD4+ T-cell count, undetectable viremia and no clinical symptoms overtime[10].
Overall, the strong individual variability to HIV infection highlights the importance of the host factors in delaying HIV progression toward AIDS. Different host factors have been widely associated with HIV progression and can be classically divided into two different groups: one related to a reduction in the viral entry capability and the other one with the interference with the viral replication process.
Reduction of viral entry has been associated with different receptors, co-receptors and ligands. Among them the CCR5Δ32 in combination with higher C-C Chemokine ligand 3-like 1 (CCL3L1) copy number and RANTES or Stromal cell–derived factor 1 chemokine variants have been extensively studied[9,11-14]. Regarding the viral replication processes, a pioneer work of Brass led to the identification of all the possible host endogenous proteins related with HIV infection[15].
Among them Zinc Ribbon Domain-containing 1 (ZNRD1), HLA Complex P5, (HCP5) Apoipoprotein B mRNA editing enzyme catalytic polypeptide-like 3G (APOBEC3G) genes have been extensively studied in association with delayed HIV progression. However, further studies regarding other possible interacting proteins still need to be addressed[16-18].
More recently, a contribution of micro-RNA has been described in HIV context leading towards interesting alternative approaches[19].
In addition, other immune related mechanisms have been associated with HIV control by immune response. This is the case of TNF-α and Ig enhancer HS1, 2 last but not least in showing a role in controlling HIV progression. Although, these factors barely play a role in delaying HIV progression compared with other host factors[20,21].
Beside the constant discovery of novel host variants, multiple issues such as population dependency might increase the difficulty to perform an association with HIV progression. For these reasons, HLA locus remains the unique factor clearly associated with HIV progression among the human population. However, different HLA alleles play a main role in HIV disease progression depending on the population considered.
Moreover, the HLA locus is the only one capable to modulate both innate and adaptive immune responses against viral infections respect to other immune related genes. Therefore, HLA locus might be used not only for diagnostic purpose, but also for drug and vaccine design approaches.
The HLA gene products are highly polymorphic molecules, characterized by co-dominant expression and polygeny. The combination of polygenicity and polymorphism has two important consequences. First, it ensures that each individual will be able to present a broad range of peptides. Second, the population will be consisted of individuals presenting different peptide’s repertoires[22].
It is possible to distinguish the HLA molecules in two different classes: HLA class I and HLA class II[23].
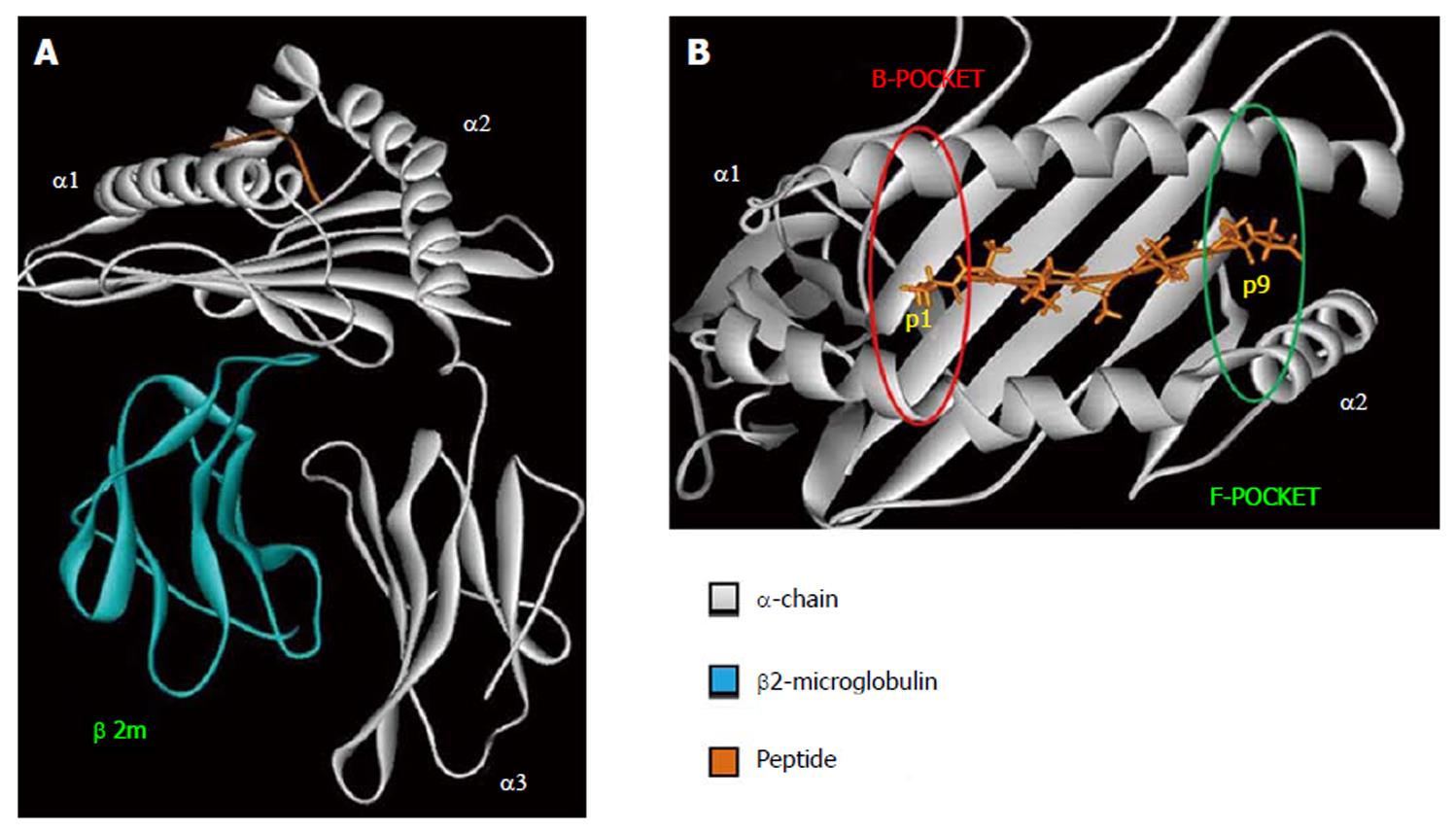
HLA class I is expressed on all nucleated cells and are recognized by CD8+ T-cells[23]. The overall structure of HLA class I molecule is shown in Figure 1A. The β2-microglobulin is a monomorphic polypeptidic chain and its main role is to keep the tridimensional structure of HLA class I molecules. The α-chain is responsible for the peptide binding and interacts with TCR, CD8 and innate immune receptors. The binding of the peptide as well as the TCR interaction are mediated by α1 and α2 domains. Both of them present two main interaction pockets (B and F), which directly interact with the bound peptide (Figure 1B). HLA class I molecules bind peptides between 8–12 amino acids long which are derived from proteolysed endogenous protein fragments[23].
HLA class II is expressed only on antigen presenting cells and are recognized by CD4+ T-cells[23].
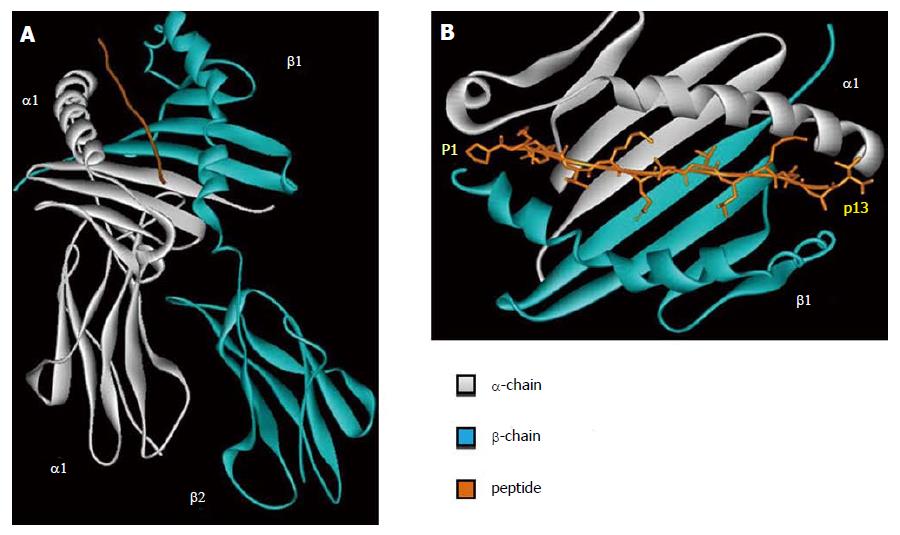
HLA class II molecule is composed by two polypeptidic chains (α and β) with a similar structure and belong to Ig superfamily. Both α and β chains participate in the peptide binding (Figure 2A). The two chains are bound in a non-covalent manner and can be further divided in two different domains. The first domain of each chain (α1 and β1) is responsible for the peptide binding and TCR interaction. The second domain of each chain (α2 and β2) has an important role in the HLA class II structure and in the interaction with CD4 molecules.
The two HLA molecules have a distinct pattern of expression and cellular interaction. HLA class I molecules are expressed by all nucleated cells and recognized by CTL. HLA class II molecules are selectively expressed on antigen presenting cells such as macrophages, monocytes, B lymphocytes and dendritic cells. HLA class II molecules are recognized by T helper lymphocytes[24]. The main role of HLA molecules is to present the antigen to different immunological receptors. In first approximation, HLA class I molecule presents peptides derived from endogenous/cytosolic proteins, while HLA class II presents peptides derived from exogenous proteins[25].
In addition, HLA class I molecules play an important role in the activation of the innate immune response. In fact, HLA class I molecules interact also with innate immune receptor expressed by NK cells[26]. The wide inter- and intra-population diversity in HLA locus and the presence of other immune associated genes increases the difficulty to select the genetic variant(s) responsible for the disease susceptibility. However, HLA alleles’ association with particular immunological profile has been consistently assessed for different chronic viral infections including HIV.
In this context, heterozygosis for HLA class I molecules has been associated with HIV delayed disease progression and lower mortality in HIV infected patients[27,28]. In addition, various HLA alleles have been associated with an increase or decrease risk of HIV vertical and horizontal transmission and hypersensitivity to anti-HIV therapy[29,30].
HLA/HIV association studies are useful to evaluate the host-pathogen interaction. HLA is important not only for the adaptive immune response but also for innate immune response. Polymorphisms within the different HLA class I alleles generate structural changes in peptide-binding pockets. Amino acid changes in the peptide-binding pockets lead to the presentation of a different set of peptides to CTLs[31-33].
The ability of particular HLA alleles to induce a viral selection could predict the HIV viral load. This could provide an “a priori” information about the disease progression[34]. Evaluations of HLA supertypes, group of alleles that share specific peptide-binding preferences, simplify the association studies with different disease progression.
The study of EC sheds light on the contribution of Human Leukocyte Antigen B (HLA-B)*57:01 (Supertype B*58) allele with HIV delayed disease progression. This allele is able to recognize a conserved epitope of HIV Gag protein, leading to a higher CD4+ T-cell count and lower viral load in absence of Highly Active Antiretroviral Therapy therapy (HAART). HLA-B*57:01 is also characterized by the presence of unique valine at position 97 that contributes to the formation of the C-pocket in the peptide-binding cleft[31,33].
When a subject that do not carry HLA-B*57:01 allele is infected with a viral strain derived from a B*58 patient, it resembles the same CD4+ T-cell count and viremia of the B*58 patient. This observation suggests that HLA exerts a strong restriction on the viral replication and the viral mutant selected have a lower fitness[35].
Other studies have associated HLA-B*27 and HLA-B*58 with a low viral load and higher CD4+ T-cell count. In this context, the selectivity exerted by CTL after antigen recognition by HLA class I molecule is responsible for delaying the HIV progression[28,31,33,36-38].
Several HLA-B alleles have been associated with HIV rapid disease progression. Among them HLA-B*35 supertype contributes to a reduction of CTL peptide recognition and therefore leads to a non-efficient viral control[28,37]. Further, supertype B*7 has been associated with high viral load, decrease CTL response and consequently rapid HIV progression towards AIDS[28,31].
Multiple issues such as the viral strain variability within the subjects and the different genetic background of the population have limited the association studies related with HIV progression.
In this context, we performed a study in a defined cohort of children infected during a hospital outbreak with a monophyletic strain of HIV-1[39]. The role of HLA amino acid polymorphisms determining specific characteristics of the HLA peptide-binding pocket has been assessed. In particular, HLA-B peptide binding pockets present a specific set of epitopes against which the subject can mount a HIV-specific immune response. According to previous observations, these findings might represent the basis of the HIV disease progression[40-44].
As expected from previous immunogenetic studies, a large number of residues found in association with LTNP or progression to AIDS have been located in the HLA-B locus[42-46]. Recently, we have further supported this notion with in silico identification of the HIV gag protein epitopes. The study has been performed on the same outbreak cohort using HIV-1 viral sequences and HLA alleles. Peptides deriving from the HIV-1 sequences and recognized by the HLA allele combinations of the study subjects have been further analyzed.
Non-progressors recognized a higher number of epitopes compared to progressors in any HLA locus analyzed[47]. This is in agreement with previous observations showing an important contribution of CTL immune response in controlling the HIV disease progression. In a nutshell, HLA class I molecules and the recognition of large set of CTL epitopes are the key factors for delaying HIV progression[48-50].
CTL also determines escape mutants of the virus in different genes of HIV-1 such as Protease, Reverse Transcriptase (RT), Vpr and Nef[38,51]. Different HLA alleles, such as HLA-B*580[52], efficiently cross-recognize HIV-1 CD8+ T-cell epitopes leading to delayed progression[53].
Recently, many studies have been focused on determining the HLA binding-peptides. The approaches are from direct measurement to the development of different Major Histocompatibility Complex (MHC) class I binding prediction systems[54-56]. Different online databases are capable to extract epitopes obtained from experimental and in silico studies giving also the opportunity to predict HLA binding epitopes using any target protein sequence[57-60]. The choice of the prediction system is very important and often the combination of more than one prediction system has shown the best performance[56,61-63].
Once obtained the predicted epitope, it is always very useful to perform a comparison with literature data. Thanks to the large in vitro characterization of HIV epitopes, it has been determined that most of the in silico predicted epitopes are also described within the literature. This supports the efficiency of prediction methods related with the epitope discovery[59,60].
Overall, HIV-specific T-cell response, and in particular CTL, plays a key role in controlling HIV infection[40,41]. T-cell response depends on HLA molecules. Thus, the individual’s variations in the HLA class I and II alleles has a profound effect on the outcome of infection and disease progression toward AIDS[40,42].
HLA-B polymorphic variants 80I, 81A, 82L, 83R have been associated with LTNP[46]. These positions interact with the peptide in the F-pocket of HLA-B[46,64]. The LTNP associated pattern 80I, 81A, 82R, 83L is typical for HLA-B supertypes B58 and B27 which are already found associated with a slow progression to AIDS[42]. The same amino acid positions are involved in the formation of the structurally related HLA serotyping epitope Bw4 and Bw6. When HLA-B alleles are classified accordingly with carrying Bw4/Bw6 epitope, we have shown a strong contribution of Bw4 homozygosis in delaying HIV progression[46,65]. These results are in agreement with previous associations between Bw4 homozygosis and the control of HIV viremia[66].
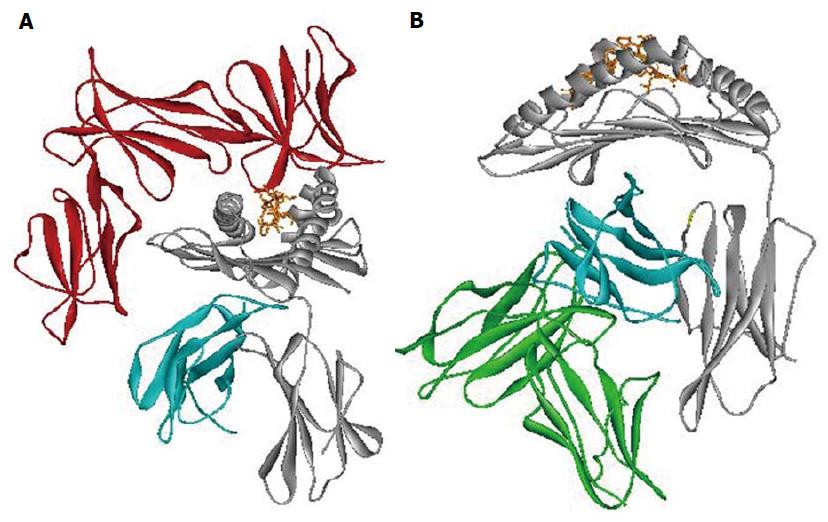
The importance of epitope Bw4 is due to different aspects. First, it has a direct interaction with the HLA bound peptide involved with CD8+ T cell recognition. Second, it is also a ligand for Killer Immunoglobulin Receptor 3DL1 (KIR3DL1), an NK’s inhibitory receptor (Figure 3A)[67,68].
This evidence suggests a strong contribution of the innate immune response in controlling HIV progression and confirming the key role played by HLA-B molecules[2]. Recent studies evaluated the different contribution of KIR3DL1/HLA-B allele’s interaction in modulating the innate immune system[68-71]. The presence of HLA-B Bw4 epitope leads to a stronger interaction with all the different KIR3DL1 alleles. This is particularly evident within the HLA-B alleles belonging to the same supertype, in agreement with previous data[72-75].
Different studies evaluate the contribution of Leukocyte Immunoglobulin-Like Receptor subfamily B member 1 (LILRB1) interaction with HLA class I in the context of several infections (Figure 3B)[76-78]. Among HLA class I polymorphisms, we associated the HLA-B alpha 3 domain amino acid position 194 with different HIV progression[46,65]. Amino acid position 194 of HLA-B has been found to take a part in the interaction with LILRB1 receptor (ILT2/LIR1/CD85j) when the Val variant is present[69,78,79]. Moreover, Val 194 was in association with LTNP[46,65]. Change in the strength of interaction between HLA-B alleles carrying Ile 194 and LILRB1 receptor might lead to rapid HIV progression. Previous data suggests that the expression of LILRB1 receptor on the cell surface remains unchanged in subjects with different HIV progression[80]. However, the presence of different amino acids at the polymorphic position 194 of HLA-B might modify the interaction with LILRB1. This might influence the LILRB1 strength of binding, as already reported for the LIR1-HLA-A interaction[77]. These results show the influence of HLA allelic variation and conformation on LILR binding capability. These findings are according to recent studies particular in the HIV context[77,78].
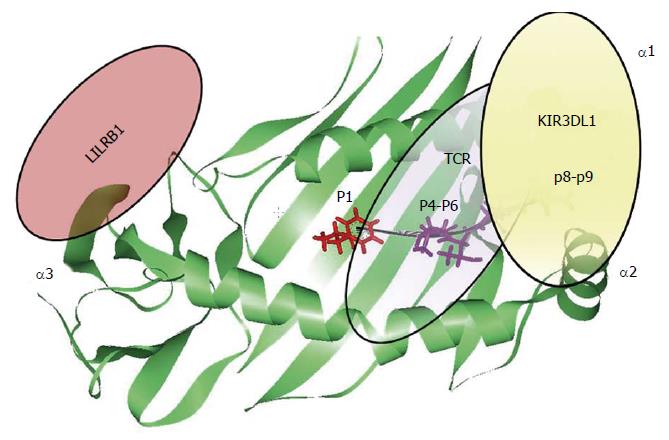
The contribution of the HLA-bound peptide seems to be the key point able to disrupt HLA interaction with the different immune receptors (Figure 4). In the context of HLA-B/KIR3DL1 interaction, the HLA-bound peptide position P8 is the main one that is able to disrupt KIR3DL1 binding. This has been previously observed in KIR3DL1 interaction with HLA-B*27:05 and HLA-B*57:01 alleles due to the conserved amino acid residue Glu282 of KIR3DL1 receptor[68,81-85]. The strong influence of the HLA bound peptide in the modulation of the innate immune response, point out similarity between T-cell and NK cell immune response. Individual selection pressures exerted on HLA class I by T-cell and NK-cell might cause a competition between the two different immune responses. Therefore, depending on the HLA class I allelic variant and the antigenic peptide loaded on HLA molecule we might observe a beneficial NK or T-cell response with detrimental consequences for the other one[86].
Altogether, the observations suggest that each peptide binding pocket position of the HLA class I molecule is capable of modulating innate and adaptive immune receptors leading to different immune responses (Figure 4).
Notably, identification of T-cell epitopes is actually made with the strategy of the reverse vaccinology. This strategy is based on HLA binding specificity and takes in consideration only the interaction with adaptive immune receptor. Future studies should be focused on the prediction of binding epitopes with wider characteristics. Peptides should be capable not only to be recognized by adaptive immune receptor, but also to modulate the innate immune receptor. These peptide characteristics could allow better fitting strategies for vaccination and diagnostics.
In conclusion, HLA molecules play a key role in modulating both adaptive and innate immune responses. The protective cytotoxic immune response is modulated by the interactions with TCR as well as other innate receptors[31,33]. The modulation of innate immune responses depends also on the peptide-binding capability of HLA-B and on the interaction between HLA-B and NK’s inhibitory receptors such as KIR3DL1 and LILRB1. The observed fine tune regulation might play a key role in the progression of HIV infection. The application of immune-informatics to immunogenic studies might shed new lights on the mechanisms behind the association of HLA genetic susceptibility to viral infections. This represents a powerful tool for novel design of vaccine and diagnostics, ensuring wider population coverage with the inclusion of genetically susceptible subjects.
P- Reviewer: Chen YD, Ghiringhelli PD, Qiu HJ S- Editor: Tian YL L- Editor: A E- Editor: Jiao XK
| 1. | Mogensen TH, Melchjorsen J, Larsen CS, Paludan SR. Innate immune recognition and activation during HIV infection. Retrovirology. 2010;7:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 2. | Carrington M, Alter G. Innate immune control of HIV. Cold Spring Harb Perspect Med. 2012;2:a007070. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 3. | Alter G, Altfeld M. NK cells in HIV-1 infection: evidence for their role in the control of HIV-1 infection. J Intern Med. 2009;265:29-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 147] [Cited by in RCA: 143] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 4. | Mothe B, Llano A, Ibarrondo J, Zamarreño J, Schiaulini M, Miranda C, Ruiz-Riol M, Berger CT, Herrero MJ, Palou E. CTL responses of high functional avidity and broad variant cross-reactivity are associated with HIV control. PLoS One. 2012;7:e29717. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 112] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 5. | Draenert R, Goebel FD. What’s new in HIV/AIDS. Protective immunity in HIV infection: where do we stand? Infection. 2004;32:250-252. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 6. | Leslie AJ, Pfafferott KJ, Chetty P, Draenert R, Addo MM, Feeney M, Tang Y, Holmes EC, Allen T, Prado JG. HIV evolution: CTL escape mutation and reversion after transmission. Nat Med. 2004;10:282-289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 680] [Cited by in RCA: 684] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 7. | Sudharshan S, Biswas J. Introduction and immunopathogenesis of acquired immune deficiency syndrome. Indian J Ophthalmol. 2008;56:357-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Goulder PJ, Walker BD. HIV and HLA class I: an evolving relationship. Immunity. 2012;37:426-440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 252] [Article Influence: 19.4] [Reference Citation Analysis (0)] |
| 9. | Lama J, Planelles V. Host factors influencing susceptibility to HIV infection and AIDS progression. Retrovirology. 2007;4:52. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 99] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 10. | Julg B, Pereyra F, Buzón MJ, Piechocka-Trocha A, Clark MJ, Baker BM, Lian J, Miura T, Martinez-Picado J, Addo MM. Infrequent recovery of HIV from but robust exogenous infection of activated CD4(+) T cells in HIV elite controllers. Clin Infect Dis. 2010;51:233-238. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 96] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 11. | Huik K, Sadam M, Karki T, Avi R, Krispin T, Paap P, Rüütel K, Uusküla A, Talu A, Abel-Ollo K. CCL3L1 copy number is a strong genetic determinant of HIV seropositivity in Caucasian intravenous drug users. J Infect Dis. 2010;201:730-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 12. | Armand-Ugón M, Moncunill G, Gonzalez E, Mena M, Ballana E, Clotet B, Esté JA. Different selection patterns of resistance and cross-resistance to HIV-1 agents targeting CCR5. J Antimicrob Chemother. 2010;65:417-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 13. | Bhattacharya T, Stanton J, Kim EY, Kunstman KJ, Phair JP, Jacobson LP, Wolinsky SM. CCL3L1 and HIV/AIDS susceptibility. Nat Med. 2009;15:1112-1115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 49] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 14. | Kulkarni H, Agan BK, Marconi VC, O’Connell RJ, Camargo JF, He W, Delmar J, Phelps KR, Crawford G, Clark RA. CCL3L1-CCR5 genotype improves the assessment of AIDS Risk in HIV-1-infected individuals. PLoS One. 2008;3:e3165. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 15. | Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921-926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1207] [Cited by in RCA: 1132] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 16. | Ballana E, Senserrich J, Pauls E, Faner R, Mercader JM, Uyttebroeck F, Palou E, Mena MP, Grau E, Clotet B. ZNRD1 (zinc ribbon domain-containing 1) is a host cellular factor that influences HIV-1 replication and disease progression. Clin Infect Dis. 2010;50:1022-1032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 17. | van Manen D, Kootstra NA, Boeser-Nunnink B, Handulle MA, van’t Wout AB, Schuitemaker H. Association of HLA-C and HCP5 gene regions with the clinical course of HIV-1 infection. AIDS. 2009;23:19-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 18. | Singh KK, Wang Y, Gray KP, Farhad M, Brummel S, Fenton T, Trout R, Spector SA. Genetic variants in the host restriction factor APOBEC3G are associated with HIV-1-related disease progression and central nervous system impairment in children. J Acquir Immune Defic Syndr. 2013;62:197-203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 19. | Munshi SU, Panda H, Holla P, Rewari BB, Jameel S. MicroRNA-150 is a potential biomarker of HIV/AIDS disease progression and therapy. PLoS One. 2014;9:e95920. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 63] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 20. | Montesano C, Giambra V, Frezza D, Palma P, Serone E, Gattinara GC, Mattei M, Mancino G, Colizzi V, Amicosante M. HS1,2 Ig enhancer alleles association to AIDS progression in a pediatric cohort infected with a monophyletic HIV-strain. Biomed Res Int. 2014;2014:637523. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 2] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 21. | Veloso S, Olona M, García F, Domingo P, Alonso-Villaverde C, Broch M, Peraire J, Viladés C, Plana M, Pedrol E. Effect of TNF-alpha genetic variants and CCR5 Delta 32 on the vulnerability to HIV-1 infection and disease progression in Caucasian Spaniards. BMC Med Genet. 2010;11:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 21] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 22. | de Verteuil D, Granados DP, Thibault P, Perreault C. Origin and plasticity of MHC I-associated self peptides. Autoimmun Rev. 2012;11:627-635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 43] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 23. | Binkowski TA, Marino SR, Joachimiak A. Predicting HLA class I non-permissive amino acid residues substitutions. PLoS One. 2012;7:e41710. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Jones EY. MHC class I and class II structures. Curr Opin Immunol. 1997;9:75-79. [PubMed] |
| 25. | Wubbolts R, Neefjes J. Intracellular transport and peptide loading of MHC class II molecules: regulation by chaperones and motors. Immunol Rev. 1999;172:189-208. [PubMed] |
| 26. | Carrington M, Martin MP, van Bergen J. KIR-HLA intercourse in HIV disease. Trends Microbiol. 2008;16:620-627. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 104] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 27. | Burgner D, Jamieson SE, Blackwell JM. Genetic susceptibility to infectious diseases: big is beautiful, but will bigger be even better? Lancet Infect Dis. 2006;6:653-663. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 174] [Cited by in RCA: 154] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 28. | Kaur G, Mehra N. Genetic determinants of HIV-1 infection and progression to AIDS: susceptibility to HIV infection. Tissue Antigens. 2009;73:289-301. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 35] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 29. | Mallal S, Nolan D, Witt C, Masel G, Martin AM, Moore C, Sayer D, Castley A, Mamotte C, Maxwell D. Association between presence of HLA-B*5701, HLA-DR7, and HLA-DQ3 and hypersensitivity to HIV-1 reverse-transcriptase inhibitor abacavir. Lancet. 2002;359:727-732. [PubMed] |
| 30. | Mallal S, Phillips E, Carosi G, Molina JM, Workman C, Tomazic J, Jägel-Guedes E, Rugina S, Kozyrev O, Cid JF. HLA-B*5701 screening for hypersensitivity to abacavir. N Engl J Med. 2008;358:568-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1336] [Cited by in RCA: 1250] [Article Influence: 73.5] [Reference Citation Analysis (0)] |
| 31. | Stephens HA. HIV-1 diversity versus HLA class I polymorphism. Trends Immunol. 2005;26:41-47. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 32. | Sidney J, Peters B, Frahm N, Brander C, Sette A. HLA class I supertypes: a revised and updated classification. BMC Immunol. 2008;9:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 584] [Cited by in RCA: 556] [Article Influence: 32.7] [Reference Citation Analysis (0)] |
| 33. | Stephens HA. Immunogenetic surveillance of HIV/AIDS. Infect Genet Evol. 2012;12:1481-1491. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 10] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 34. | Brumme ZL, Tao I, Szeto S, Brumme CJ, Carlson JM, Chan D, Kadie C, Frahm N, Brander C, Walker B. Human leukocyte antigen-specific polymorphisms in HIV-1 Gag and their association with viral load in chronic untreated infection. AIDS. 2008;22:1277-1286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 35. | Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med. 2006;203:1357-1369. [PubMed] |
| 36. | Bansal A, Yue L, Conway J, Yusim K, Tang J, Kappes J, Kaslow RA, Wilson CM, Goepfert PA. Immunological control of chronic HIV-1 infection: HLA-mediated immune function and viral evolution in adolescents. AIDS. 2007;21:2387-2397. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 33] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 37. | Blackwell JM, Jamieson SE, Burgner D. HLA and infectious diseases. Clin Microbiol Rev. 2009;22:370-385, Table of Contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 264] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 38. | Duda A, Lee-Turner L, Fox J, Robinson N, Dustan S, Kaye S, Fryer H, Carrington M, McClure M, McLean AR. HLA-associated clinical progression correlates with epitope reversion rates in early human immunodeficiency virus infection. J Virol. 2009;83:1228-1239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 39. | de Oliveira T, Pybus OG, Rambaut A, Salemi M, Cassol S, Ciccozzi M, Rezza G, Gattinara GC, D’Arrigo R, Amicosante M. Molecular epidemiology: HIV-1 and HCV sequences from Libyan outbreak. Nature. 2006;444:836-837. [PubMed] |
| 40. | McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature. 2001;410:980-987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 440] [Article Influence: 18.3] [Reference Citation Analysis (0)] |
| 41. | Kaur G, Mehra N. Genetic determinants of HIV-1 infection and progression to AIDS: immune response genes. Tissue Antigens. 2009;74:373-385. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 39] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Trachtenberg EA. A Review of the Role of the Human Leukocyte Antigen (HLA) System as a Host Immunogenetic Factor Influencing HIV Transmission and Progression to AIDS. Korber BT, Brander C, Haynes BF, Koup R, Kuiken C, Moore JP, Walker BD, Watkins D, editors. Los Alamos, NM: Theoretical Biology and Biophysics Group, Los Alamos National Laboratory 2001; 43-60. |
| 43. | Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, Walker BD, Ripke S, Brumme CJ, Pulit SL, Carrington M, Kadie CM, Carlson JM, Heckerman D, Graham RR, Plenge RM, Deeks SG, Gianniny L, Crawford G, Sullivan J, Gonzalez E, Davies L, Camargo A, Moore JM, Beattie N, Gupta S, Crenshaw A, Burtt NP, Guiducci C, Gupta N, Gao X, Qi Y, Yuki Y, Piechocka-Trocha A, Cutrell E, Rosenberg R, Moss KL, Lemay P, O’Leary J, Schaefer T, Verma P, Toth I, Block B, Baker B, Rothchild A, Lian J, Proudfoot J, Alvino DM, Vine S, Addo MM, Allen TM, Altfeld M, Henn MR, Le Gall S, Streeck H, Haas DW, Kuritzkes DR, Robbins GK, Shafer RW, Gulick RM, Shikuma CM, Haubrich R, Riddler S, Sax PE, Daar ES, Ribaudo HJ, Agan B, Agarwal S, Ahern RL, Allen BL, Altidor S, Altschuler EL, Ambardar S, Anastos K, Anderson B, Anderson V, Andrady U, Antoniskis D, Bangsberg D, Barbaro D, Barrie W, Bartczak J, Barton S, Basden P, Basgoz N, Bazner S, Bellos NC, Benson AM, Berger J, Bernard NF, Bernard AM, Birch C, Bodner SJ, Bolan RK, Boudreaux ET, Bradley M, Braun JF, Brndjar JE, Brown SJ, Brown K, Brown ST, Burack J, Bush LM, Cafaro V, Campbell O, Campbell J, Carlson RH, Carmichael JK, Casey KK, Cavacuiti C, Celestin G, Chambers ST, Chez N, Chirch LM, Cimoch PJ, Cohen D, Cohn LE, Conway B, Cooper DA, Cornelson B, Cox DT, Cristofano MV, Cuchural G Jr, Czartoski JL, Dahman JM, Daly JS, Davis BT, Davis K, Davod SM, DeJesus E, Dietz CA, Dunham E, Dunn ME, Ellerin TB, Eron JJ, Fangman JJ, Farel CE, Ferlazzo H, Fidler S, Fleenor-Ford A, Frankel R, Freedberg KA, French NK, Fuchs JD, Fuller JD, Gaberman J, Gallant JE, Gandhi RT, Garcia E, Garmon D, Gathe JC Jr, Gaultier CR, Gebre W, Gilman FD, Gilson I, Goepfert PA, Gottlieb MS, Goulston C, Groger RK, Gurley TD, Haber S, Hardwicke R, Hardy WD, Harrigan PR, Hawkins TN, Heath S, Hecht FM, Henry WK, Hladek M, Hoffman RP, Horton JM, Hsu RK, Huhn GD, Hunt P, Hupert MJ, Illeman ML, Jaeger H, Jellinger RM, John M, Johnson JA, Johnson KL, Johnson H, Johnson K, Joly J, Jordan WC, Kauffman CA, Khanlou H, Killian RK, Kim AY, Kim DD, Kinder CA, Kirchner JT, Kogelman L, Kojic EM, Korthuis PT, Kurisu W, Kwon DS, LaMar M, Lampiris H, Lanzafame M, Lederman MM, Lee DM, Lee JM, Lee MJ, Lee ET, Lemoine J, Levy JA, Llibre JM, Liguori MA, Little SJ, Liu AY, Lopez AJ, Loutfy MR, Loy D, Mohammed DY, Man A, Mansour MK, Marconi VC, Markowitz M, Marques R, Martin JN, Martin HL Jr, Mayer KH, McElrath MJ, McGhee TA, McGovern BH, McGowan K, McIntyre D, Mcleod GX, Menezes P, Mesa G, Metroka CE, Meyer-Olson D, Miller AO, Montgomery K, Mounzer KC, Nagami EH, Nagin I, Nahass RG, Nelson MO, Nielsen C, Norene DL, O’Connor DH, Ojikutu BO, Okulicz J, Oladehin OO, Oldfield EC 3rd, Olender SA, Ostrowski M, Owen WF Jr, Pae E, Parsonnet J, Pavlatos AM, Perlmutter AM, Pierce MN, Pincus JM, Pisani L, Price LJ, Proia L, Prokesch RC, Pujet HC, Ramgopal M, Rathod A, Rausch M, Ravishankar J, Rhame FS, Richards CS, Richman DD, Rodes B, Rodriguez M, Rose RC 3rd, Rosenberg ES, Rosenthal D, Ross PE, Rubin DS, Rumbaugh E, Saenz L, Salvaggio MR, Sanchez WC, Sanjana VM, Santiago S, Schmidt W, Schuitemaker H, Sestak PM, Shalit P, Shay W, Shirvani VN, Silebi VI, Sizemore JM Jr, Skolnik PR, Sokol-Anderson M, Sosman JM, Stabile P, Stapleton JT, Starrett S, Stein F, Stellbrink HJ, Sterman FL, Stone VE, Stone DR, Tambussi G, Taplitz RA, Tedaldi EM, Telenti A, Theisen W, Torres R, Tosiello L, Tremblay C, Tribble MA, Trinh PD, Tsao A, Ueda P, Vaccaro A, Valadas E, Vanig TJ, Vecino I, Vega VM, Veikley W, Wade BH, Walworth C, Wanidworanun C, Ward DJ, Warner DA, Weber RD, Webster D, Weis S, Wheeler DA, White DJ, Wilkins E, Winston A, Wlodaver CG, van’t Wout A, Wright DP, Yang OO, Yurdin DL, Zabukovic BW, Zachary KC, Zeeman B, Zhao M; International HIV Controllers Study. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551-1557. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 989] [Cited by in RCA: 958] [Article Influence: 63.9] [Reference Citation Analysis (0)] |
| 44. | Amicosante M, Gioia C, Montesano C, Casetti R, Topino S, D’Offizi G, Cappelli G, Ippolito G, Colizzi V, Poccia F. Computer-based design of an HLA-haplotype and HIV-clade independent cytotoxic T-lymphocyte assay for monitoring HIV-specific immunity. Mol Med. 2002;8:798-807. [PubMed] |
| 45. | Hertz T, Nolan D, James I, John M, Gaudieri S, Phillips E, Huang JC, Riadi G, Mallal S, Jojic N. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol. 2011;85:1310-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 46. | Montesano C, Bonanno CT, Grifoni A, Di Sano C, Palma P, Castelli-Gattinara G, Mattei M, Mancino G, Salerno A, Colizzi V. Impact of Human Leukocyte Antigen Polymorphisms in Human Immunodeficiency Virus Progression in a Paediatric Cohort Infected with a Mono-phyletic Human Immunodeficiency Virus-1 Strain. J AIDS Clin Res. 2014;In press. |
| 47. | Grifoni A, Montesano C, Palma P, Giovannetti M, Castelli-Gattinara G, Ciccozzi M, Mattei M, Mancino G, Salerno A, Colizzi V. Role of individual’s T-cell immunome in controlling HIV-1 progression. Immunology. 2014;143:631-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 48. | Zhai S, Zhuang Y, Song Y, Li S, Huang D, Kang W, Li X, Liao Q, Liu Y, Zhao Z. HIV-1-specific cytotoxic T lymphocyte (CTL) responses against immunodominant optimal epitopes slow the progression of AIDS in China. Curr HIV Res. 2008;6:335-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 49. | Chouquet C, Autran B, Gomard E, Bouley JM, Calvez V, Katlama C, Costagliola D, Rivière Y. Correlation between breadth of memory HIV-specific cytotoxic T cells, viral load and disease progression in HIV infection. AIDS. 2002;16:2399-2407. [PubMed] |
| 50. | Schmid BV, Keşmir C, de Boer RJ. The distribution of CTL epitopes in HIV-1 appears to be random, and similar to that of other proteomes. BMC Evol Biol. 2009;9:184. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 51. | Brumme ZL, Brumme CJ, Heckerman D, Korber BT, Daniels M, Carlson J, Kadie C, Bhattacharya T, Chui C, Szinger J. Evidence of differential HLA class I-mediated viral evolution in functional and accessory/regulatory genes of HIV-1. PLoS Pathog. 2007;3:e94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 126] [Cited by in RCA: 141] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 52. | Ngumbela KC, Day CL, Mncube Z, Nair K, Ramduth D, Thobakgale C, Moodley E, Reddy S, de Pierres C, Mkhwanazi N. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res Hum Retroviruses. 2008;24:72-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 53. | Turnbull EL, Lopes AR, Jones NA, Cornforth D, Newton P, Aldam D, Pellegrino P, Turner J, Williams I, Wilson CM. HIV-1 epitope-specific CD8+ T cell responses strongly associated with delayed disease progression cross-recognize epitope variants efficiently. J Immunol. 2006;176:6130-6146. [PubMed] |
| 54. | Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Keşmir C, Peters B. Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol. 2013;9:e1003266. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 410] [Cited by in RCA: 611] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 55. | HIV Databases. Division of AIDS of the National Institute of Allergy and Infectious Diseases. Available from: http: //www.hiv.lanl.gov/. |
| 56. | Salimi N, Fleri W, Peters B, Sette A. The immune epitope database: a historical retrospective of the first decade. Immunology. 2012;137:117-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 57. | De Groot AS, Jesdale B, Martin W, Saint Aubin C, Sbai H, Bosma A, Lieberman J, Skowron G, Mansourati F, Mayer KH. Mapping cross-clade HIV-1 vaccine epitopes using a bioinformatics approach. Vaccine. 2003;21:4486-4504. [PubMed] |
| 58. | Erup Larsen M, Kloverpris H, Stryhn A, Koofhethile CK, Sims S, Ndung’u T, Goulder P, Buus S, Nielsen M. HLArestrictor--a tool for patient-specific predictions of HLA restriction elements and optimal epitopes within peptides. Immunogenetics. 2011;63:43-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 59. | Kumar N, Mohanty D. Structure-based identification of MHC binding peptides: Benchmarking of prediction accuracy. Mol Biosyst. 2010;6:2508-2520. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 60. | Lundegaard C, Lund O, Buus S, Nielsen M. Major histo compatibility complex class I binding predictions as a tool in epitope discovery. Immunology. 2010;130:309-318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 99] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 61. | Peters B, Sette A. Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinformatics. 2005;6:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 377] [Cited by in RCA: 403] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 62. | Bui HH, Sidney J, Peters B, Sathiamurthy M, Sinichi A, Purton KA, Mothé BR, Chisari FV, Watkins DI, Sette A. Automated generation and evaluation of specific MHC binding predictive tools: ARB matrix applications. Immunogenetics. 2005;57:304-314. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 203] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 63. | Nielsen M, Lundegaard C, Worning P, Lauemøller SL, Lamberth K, Buus S, Brunak S, Lund O. Reliable prediction of T-cell epitopes using neural networks with novel sequence representations. Protein Sci. 2003;12:1007-1017. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 806] [Cited by in RCA: 833] [Article Influence: 37.9] [Reference Citation Analysis (0)] |
| 64. | Madden DR. The three-dimensional structure of peptide-MHC complexes. Annu Rev Immunol. 1995;13:587-622. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 627] [Cited by in RCA: 612] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 65. | Grifoni A, Montesano C, Palma P, Salerno A, Colizzi V, Amicosante M. Role of HLA-B α-3 domain amino acid position 194 in HIV disease progression. Mol Immunol. 2013;53:410-413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 66. | Flores-Villanueva PO, Yunis EJ, Delgado JC, Vittinghoff E, Buchbinder S, Leung JY, Uglialoro AM, Clavijo OP, Rosenberg ES, Kalams SA. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc Natl Acad Sci USA. 2001;98:5140-5145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 191] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 67. | Thananchai H, Gillespie G, Martin MP, Bashirova A, Yawata N, Yawata M, Easterbrook P, McVicar DW, Maenaka K, Parham P. Cutting Edge: Allele-specific and peptide-dependent interactions between KIR3DL1 and HLA-A and HLA-B. J Immunol. 2007;178:33-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 184] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 68. | Vivian JP, Duncan RC, Berry R, O’Connor GM, Reid HH, Beddoe T, Gras S, Saunders PM, Olshina MA, Widjaja JM. Killer cell immunoglobulin-like receptor 3DL1-mediated recognition of human leukocyte antigen B. Nature. 2011;479:401-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 161] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 69. | Sanjanwala B, Draghi M, Norman PJ, Guethlein LA, Parham P. Polymorphic sites away from the Bw4 epitope that affect interaction of Bw4+ HLA-B with KIR3DL1. J Immunol. 2008;181:6293-6300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 70. | Carr WH, Pando MJ, Parham P. KIR3DL1 polymorphisms that affect NK cell inhibition by HLA-Bw4 ligand. J Immunol. 2005;175:5222-5229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 174] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 71. | O’Connor GM, Guinan KJ, Cunningham RT, Middleton D, Parham P, Gardiner CM. Functional polymorphism of the KIR3DL1/S1 receptor on human NK cells. J Immunol. 2007;178:235-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 139] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 72. | Gumperz JE, Valiante NM, Parham P, Lanier LL, Tyan D. Heterogeneous phenotypes of expression of the NKB1 natural killer cell class I receptor among individuals of different human histocompatibility leukocyte antigens types appear genetically regulated, but not linked to major histocompatibililty complex haplotype. J Exp Med. 1996;183:1817-1827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 121] [Cited by in RCA: 124] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 73. | Litwin V, Gumperz J, Parham P, Phillips JH, Lanier LL. NKB1: a natural killer cell receptor involved in the recognition of polymorphic HLA-B molecules. J Exp Med. 1994;180:537-543. [PubMed] |
| 74. | Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Variable NK cell receptors exemplified by human KIR3DL1/S1. J Immunol. 2011;187:11-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 75. | Grifoni A, Montesano C, Patronov A, Colizzi V, Amicosante M. Immunoinformatic docking approach for the analysis of KIR3DL1/HLA-B interaction. Biomed Res Int. 2013;2013:283805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 76. | Chapman TL, Heikeman AP, Bjorkman PJ. The inhibitory receptor LIR-1 uses a common binding interaction to recognize class I MHC molecules and the viral homolog UL18. Immunity. 1999;11:603-613. [PubMed] |
| 77. | Jones DC, Kosmoliaptsis V, Apps R, Lapaque N, Smith I, Kono A, Chang C, Boyle LH, Taylor CJ, Trowsdale J. HLA class I allelic sequence and conformation regulate leukocyte Ig-like receptor binding. J Immunol. 2011;186:2990-2997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 111] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 78. | Lichterfeld M, Yu XG. The emerging role of leukocyte immunoglobulin-like receptors (LILRs) in HIV-1 infection. J Leukoc Biol. 2012;91:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 79. | Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4:913-919. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 207] [Cited by in RCA: 214] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 80. | Mavilio D, Benjamin J, Daucher M, Lombardo G, Kottilil S, Planta MA, Marcenaro E, Bottino C, Moretta L, Moretta A. Natural killer cells in HIV-1 infection: dichotomous effects of viremia on inhibitory and activating receptors and their functional correlates. Proc Natl Acad Sci USA. 2003;100:15011-15016. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 298] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 81. | Peruzzi M, Parker KC, Long EO, Malnati MS. Peptide sequence requirements for the recognition of HLA-B*2705 by specific natural killer cells. J Immunol. 1996;157:3350-3356. [PubMed] |
| 82. | Peruzzi M, Wagtmann N, Long EO. A p70 killer cell inhibitory receptor specific for several HLA-B allotypes discriminates among peptides bound to HLA-B*2705. J Exp Med. 1996;184:1585-1590. [PubMed] |
| 83. | Stewart-Jones GB, di Gleria K, Kollnberger S, McMichael AJ, Jones EY, Bowness P. Crystal structures and KIR3DL1 recognition of three immunodominant viral peptides complexed to HLA-B*2705. Eur J Immunol. 2005;35:341-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 93] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 84. | Kollnberger S, Chan A, Sun MY, Chen LY, Wright C, di Gleria K, McMichael A, Bowness P. Interaction of HLA-B27 homodimers with KIR3DL1 and KIR3DL2, unlike HLA-B27 heterotrimers, is independent of the sequence of bound peptide. Eur J Immunol. 2007;37:1313-1322. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 90] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 85. | O’Connor GM, Vivian JP, Widjaja JM, Bridgeman JS, Gostick E, Lafont BA, Anderson SK, Price DA, Brooks AG, Rossjohn J. Mutational and structural analysis of KIR3DL1 reveals a lineage-defining allotypic dimorphism that impacts both HLA and peptide sensitivity. J Immunol. 2014;192:2875-2884. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 86. | Parham P, Norman PJ, Abi-Rached L, Guethlein LA. Human-specific evolution of killer cell immunoglobulin-like receptor recognition of major histocompatibility complex class I molecules. Philos Trans R Soc Lond B Biol Sci. 2012;367:800-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 142] [Cited by in RCA: 158] [Article Influence: 12.2] [Reference Citation Analysis (0)] |