Published online Aug 12, 2013. doi: 10.5501/wjv.v2.i3.123
Revised: July 26, 2013
Accepted: August 8, 2013
Published online: August 12, 2013
Processing time: 111 Days and 18 Hours
AIM: To search for the presence of cis elements in hepatitis D virus (HDV) genomic and antigenomic RNA capable of promoting nuclear export.
METHODS: We made use of a well characterized chloramphenicol acetyl-transferase reporter system based on plasmid pDM138. Twenty cDNA fragments corresponding to different HDV genomic and antigenomic RNA sequences were inserted in plasmid pDM138, and used in transfection experiments in Huh7 cells. The relative amounts of HDV RNA in nuclear and cytoplasmic fractions were then determined by real-time polymerase chain reaction and Northern blotting. The secondary structure of the RNA sequences that displayed nuclear export ability was further predicted using a web interface. Finally, the sensitivity to leptomycin B was assessed in order to investigate possible cellular pathways involved in HDV RNA nuclear export.
RESULTS: Analysis of genomic RNA sequences did not allow identifying an unequivocal nuclear export element. However, two regions were found to promote the export of reporter mRNAs with efficiency higher than the negative controls albeit lower than the positive control. These regions correspond to nucleotides 266-489 and 584-920, respectively. In addition, when analyzing antigenomic RNA sequences a nuclear export element was found in positions 214-417. Export mediated by the nuclear export element of HDV antigenomic RNA is sensitive to leptomycin B suggesting a possible role of CRM1 in this transport pathway.
CONCLUSION: A cis-acting nuclear export element is present in nucleotides 214-417 of HDV antigenomic RNA.
Core tip: Hepatitis D virus (HDV) replicates in the nucleus and export of HDV RNPs to the cytoplasm is thought to be mediated by cis-elements present in virus RNA. We used a chloramphenicol acetyl-transferase reporter system in an attempt to identify the RNA sequences that mediate export to the cytoplasm. Several cDNA constructs coding for different HDV RNA (genomic and antigenomic) sequences were tested. Our results show that a cis-acting nuclear export element is present in positions 214-417 of antigenomic RNA. Two regions in genomic RNA were found to promote nuclear export with efficiency higher than the negative control although lower that the positive control.
- Citation: Freitas N, Cunha C. Searching for nuclear export elements in hepatitis D virus RNA. World J Virol 2013; 2(3): 123-135
- URL: https://www.wjgnet.com/2220-3249/full/v2/i3/123.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i3.123
Hepatitis D virus (HDV) is the only member of the Deltavirus genus and is considered to be a satellite virus of the hepatitis B virus (HBV)[1]. When compared with HBV alone, infection of human hepatocytes with both viruses increases liver damage and the risk of cirrhosis and fulminant disease[2,3]. The two viruses are associated due to the fact that the outer envelope of HDV consists of HBV surface antigens (HBsAgs) which are necessary for virus packaging and propagation of infection[4].
The HDV genome consists of a circular, closed ssRNA molecule of approximately 1.7 kb and negative polarity. It is estimated that about 70% of this RNA molecule is internally base-paired resulting in the formation of a rod-like structure similar to plant viroids[5]. There is still a considerable lack of information, and even some controversy, concerning the mechanisms and host factors involved in HDV RNA replication. It seems to be generally accepted that replication occurs through a double rolling-circle mechanism involving the participation of at least host RNA polymerase II, and resulting in the synthesis of multimeric antigenomic molecules[6,7]. Subsequently, these multimeric antigenomic molecules are self-cleaved and ligated at precise monomeric intervals by the HDV RNA ribozyme activity[8]. The monomeric antigenomes serve as templates for a second round of replication, by a similar mechanism, thus resulting in the synthesis of monomeric genomic RNA molecules. The HDV genome contains a single ORF that codes for a 24 kDa protein, the so-called small delta antigen (S-HDAg)[9]. As a consequence of an editing mechanism that converts an amber stop codon UAG into a tryptophan codon UGG in the antigenome, the ORF is extended by 19 additional aminoacids[10]. As a result, a 27 kDa protein, the large delta antigen (L-HDAg) is produced. These two proteins are thought to play different roles in the HDV replication cycle. S-HDAg is necessary for accumulation of virus RNA[11] and positively regulates ribozyme activity[12], and L-HDAg inhibits replication and interacts with HBsAgs to promote virus packaging[13,14]. HDV packaging occurs in the cytoplasm where the newly synthesized RNPs meet the HBsAgs to assemble mature virions. It has been previously shown that HDV RNPs shuttle continuously between the nucleus and the cytoplasm[15]. While nuclear import of virus RNPs is mediated by a nuclear localization signal in HDAgs[16], the export to the cytoplasm is believed to be mediated by a cis element present in the RNA molecule. This is supported by the fact that export of both genomic and antigenomic HDV RNAs was found to be independent of the presence of HDAgs[15]. Furthermore, Macnaughton and Lai reported that both genomic and antigenomic RNAs (gRNA and agRNA, respectively) are exported with similar efficiency at early times during replication[17]. Although cells expressing L-HDAg, HBsAgs, and agRNA were found to secret virus-like particles containing HDV agRNA[18], it is widely accepted that only gRNA molecules are packaged into newly synthesized virions. This observation led to the hypothesis that packaging is restricted to gRNA molecules due to the nuclear retention, and eventual further degradation, of HDV antigenomes. However, to our knowledge, no experimental evidences were obtained supporting this idea.
Simple retroviruses such as simian type D retroviruses have evolved mechanisms of RNA export based on the direct interaction of a cis-acting transport element [constitutive transport element (CTE)] with cellular transport receptors. The TAP protein, the human homologue of yeast Mex67p, is one of best studied host factors shown to interact with the CTE to promote nuclear export of unspliced simian retrovirus type D mRNAs[19]. TAP was also identified as one of the proteins responsible for export of cellular mRNAs[20]. On the other hand, complex retroviruses were shown to use a different pathway for export of intron-containing mRNAs. This pathway involves the participation of the cellular protein CRM1[21]. In the case of human immunodeficiency virus-1 (HIV-1), the association of CRM1 with intron-containing mRNAs is mediated by the virus protein Rev which recognizes a specific sequence named rev-responsive element (RRE)[22]. Additionally, the HBV posttranscriptional regulatory element (PRE), which was reported to play a crucial role in export of virus mRNAs to the cytoplasm, seems to use a distinct, not yet identified nuclear export pathway[23].
In an attempt to clarify whether HDV gRNA and agRNA contain cis elements capable of promoting the export to the cytoplasm, we made use of a chloranphenicol acetyl-transferase (CAT) reporter system, in transfection experiments, to identify and characterize putative nuclear export elements.
HuH-7 cells were cultured in RPMI 1640 medium (Sigma) supplemented with 10%FBS (Invitrogen). Cells were grown as monolayers at 37 °C, in a humidified atmosphere containing 5%CO2. Transfection assays were performed using the Fugene6 Transfection Reagent (Roche) and 1 μg plasmid DNA per 35-mm well, according to the manufacturer’s instructions. To control for transfection effiency in CAT assays, 20 ng of plasmid pSV-β-galactosidase (Promega) were cotransfected with 100 ng of reporter CAT constructs, and 1.88 μg pUC19. Cells were analysed 24 h post-transfection. In some experiments, 10 nmol/L leptomycin B was added to the medium 18 h after transfection and cells were subsequently incubated for 6 h before analysis as earlier described[24].
Plasmid pDM138[25,26] was a kind gift of Tristram Parslow (Emory University School of Medicine, Atlanta, United States). Plasmids pDM138-PRE(+) and pDM138-PRE(-) were generously provided by Benedict Yen (University of California, San Francisco, United States). Plasmids pDM138-PRE(+) and pDM138-PRE(-) contain a DNA fragment of approximately 570 bp that codes for the HBV post-transcriptional regulatory element (PRE). This fragment was inserted in the unique ClaI site of pDM138 in both sense and antisense orientations, originating plasmids pDM138-PRE(+) and pDM138-PRE(-), respectively[27].
Twenty vectors containing cDNA inserts corresponding to 10 different regions of the HDV agRNA cloned, in both orientations, in the unique ClaI site of pDM138 were generated by polymerase chain reaction (PCR), using the primers listed in Table 1, and plasmid pDL481[18] as template. This plasmid was designed to code for full-length HDV antigenomic RNA and was a kind gift of John Taylor (Fox Chase Cancer Center, Philadelphia, United States). The primers were designed in order to include a ClaI site in the 5’ end. The obtained PCR fragments were purified using the GFX PCR and Gel Band kit (GE Healthcare) and ligated with ClaI digested pDM138 using the Rapid DNA Ligation kit (Roche) according with the instructions of the manufacturer. The correct insertion in sense or antisense orientations of the fragments was first monitored by restriction endonuclease analysis with BanII, EcoRI, BanII and NheI, PstI, XhoI, BglII, and BamHI (Fermentas) followed by DNA sequencing.
| HDV cDNA fragments | Forward primer 5'→3' | Reverse primer 5'→3' | Fragment length (bp) | Genomelocation(Sense) | Antigenomelocation(Antisense) |
| A1 | CGCATCGATACTCCCTGCAGATTGGGGA | CGCATCGATATTCACCGACAAGGAGAGGC | 243 | 360-602 | 1078-1320 |
| A2 | CGCATCGATGCCTCTCCTTGTCGGTGAAT | CGCATCGATGAGACCTCCGGAAGACAAAGA | 204 | 176-379 | 1301-1504 |
| A3 | CGCATCGATTTCCGGAGGTCTCTCTCGAGT | CGCATCGATTCTCCTCGCTCGGAACTTG | 214 | 1654-679/188 | 1492-1679/26 |
| A4 | CGCATCGATTTCCTCGGTCAACCTCCTGA | CGCATCGATATAAGGATGGAGAGGGGGCT | 224 | 266-489 | 1191-1414 |
| A5 | CGCATCGATCCCCTCTCCATCCTTATCCT | CGCATCGATAGGGAGAGAAGAGATCCTCGA | 172 | 114-285 | 1395-1566 |
| A6 | CGCATCGATCGAATGGGACCCACAAATCT | CGCATCGATTCCCCAATCTGCAGGGAGT | 337 | 584-920 | 760-1096 |
| A7 | CGCATCGATCCCAATCCCAGATCTGGAGA | CGCATCGATTTTGCTTTCCTCCTCGCTTC | 204 | 1263-1466 | 214-417 |
| A8 | CGCATCGATAAAGAAAGCAACGGGGCTAG | CGCATCGATGGGAGTCGGAATCGAGCAT | 199 | 1067-1265 | 415-613 |
| A9 | CGCATCGATATGCTCGATTCCGACTCCC | CGCATCGATCCTAGAGAGATTTGTGGGTCCC | 191 | 895-1085 | 595-785 |
| A10 | CGCATCGATCAAGTTCCGAGCGAGGAGAC | CGCATCGATTCTCCAGATCTGGGATTGGG | 227 | 1446-1672 | 8-234 |
Additionally, we constructed eight pDM138 derived vectors containing different portions of the cDNA complementary to the HDV agRNA sequence comprised between nt 214 and 417. The strategy was similar to the one described above and the primers used in PCR reactions are listed in Table 2.
| HDV cDNA fragments | Forward primer 5'→3' | Reverse primer 5'→3' | Fragment length (bp) |
| 269-417 | CGCATCGATGGGAGGAATCCACTCGGAGA | CGCATCGATTTTGCTTTCCTCCTCGCTTC | 149 |
| 214-379 | CGCATCGATCCCAATCCCAGATCTGGAGA | CGCATCGATGCATCTCCTCCTATCGCTATGG | 166 |
| 214-403 | CGCATCGATCCCAATCCCAGATCTGGAGA | CGCATCGATGCTTCGGTCTCCCCCTACTC | 190 |
| 244-417 | CGCATCGATCCCGAAGGGTTGAGTAGCAC | CGCATCGATTTTGCTTTCCTCCTCGCTTC | 174 |
| 244-403 | CGCATCGATCCCGAAGGGTTGAGTAGCAC | CGCATCGATGCTTCGGTCTCCCCCTACTC | 160 |
| 314-417 | CGCATCGATACCCCTTCAGCGAACAAGAG | CGCATCGATTTTGCTTTCCTCCTCGCTTC | 104 |
| 214-322 | CGCATCGATCCCAATCCCAGATCTGGAGA | CGCATCGATTGAAGGGGTCCTCGGAGGT | 109 |
| 269-379 | CGCATCGATGGGAGGAATCCACTCGGAGA | CGCATCGATGCATCTCCTCCTATCGCTATGG | 111 |
Plasmid pDL481ΔNEE, containing full-length HDV agRNA from which the sequence corresponding to the putative nuclear export element (NEE) was removed (nt 2473-2696), was constructed as follows: first we digested plasmid pDL481 with ApaI (Invitrogen) which cuts at positions 2696 and 3208. The two resulting 5762 bp and 512 bp fragments were separated by electrophoresis, and the 5762 bp fragment was recovered and purified using the GFX PCR DNA kit (GE Healthcare). Subsequently, this fragment was further digested with NheI (GE Healthcare). Two fragments were obtained with 5539 bp and 223 bp, respectively. The 5539 bp fragment was purified as above. The next step consisted of the amplification of the 2696-3208 nt region of plasmid pDL481. To do this, we used the following primers: Fwd 5’GGGCCCGCTTAGCGCCCCTTTTTCTTCCACCTT 3’ in which a ApaI and a NheI restriction sites were included in the 5’ end, and Rev 5’ GGGCCCACCGGTGCCCCCTCTCCATCCTTAT 3’ in which a ApaI (underlined) and a AgeI (grey box) restriction sites were also added at the 5’ end. The amplified 512 bp fragment was purified as above, and the two 512 bp and 5539 bp fragments were then ligated using the Rapid DNA Ligation kit (Roche) according to the specifications of the manufacturer. The correct construction of the recombinant vector was tested by digestion with XhoI followed by DNA sequencing.
Plasmid pDL481Δδ was constructed by removing a 189 bp sequence, comprised between nucleotides 3208 and 3397 in pDL481. This sequence is complementary to the putative NEE in the HDV antigenome. To do this we first digested plasmid pDL481 with BpiI (Fermentas) which cuts at positions 3098 and 3397 generating two 5975 bp and 299 bp fragments, respectively. Next, the two fragments were purified and incubated with 5 U Klenow enzyme (Fermentas), 0.05 mmol/L dNTPs, and Klenow buffer (Fermentas), for 10 min at 37 °C to generate blunt ends. The resulting blunt-ended fragments were digested with ApaI (Fermentas) and 4 fragments were obtained with 110, 189, 402 and 5573 bp, respectively. The 5573 bp fragment was purified as above and used in ligation reactions with the 512 bp fragment of pDL481 (nt 2696-3208) which was amplified by PCR as described before. Prior to ligation, compatible ends were generated in the 512 bp amplified DNA fragment. To do this, we first digested this fragment with AgeI. Following incubation with Klenow and dNTPs to generate blunt ends, as above described, this fragment was next digested with ApaI. After digestion, the DNA was purified using the GFX PCR DNA kit (GE Healthcare), and subsequently used in ligation reactions with the 5573 bp fragment. Ligations were performed using the Rapid DNA Ligation kit (Roche) following the instructions of the manufacturer. The correct construction of the recombinant vector was monitored by restriction endonuclease analysis with XhoI followed by DNA sequencing.
Plasmid pDL481ΔNEEδ, from which the putative NEE and the corresponding complementary sequence in the HDV antigenome were deleted, was generated by removing a 223 bp sequence between positions 2473 and 2696 and a 189 bp sequence comprised between nucleotides 3208 and 3397 in plasmid pDL481. The first approach consisted of digesting plasmid pDL481 with BpiI followed by generation of blunt ends with Klenow enzyme, as above described for plasmid pDL481Δδ. Next, we digested the two resulting fragments with NheI. Three fragments of 299, 625 and 5350 bp, respectively, were obtained and separated by agarose gel electrophoresis. The 5350 bp fragment was purified from the gel using the GFX PCR DNA kit (GE Healthcare) and used in subsequent ligation reactions. Before ligation with the 512 bp fragment of pDL481 (nt 2696-3208) obtained by PCR, compatible ends were generated. This was performed by digesting the 512 bp fragment with AgeI. After filling the resulting cohesive ends with dNTPs, as above described, the obtained blunt fragment was further digested with NheI. After purification, this DNA fragment was finally ligated with the previously obtained 5350 bp DNA fragment, as described. Finally, we tested the correct construction of the recombinant plasmid by digestion with XhoI followed by DNA sequencing.
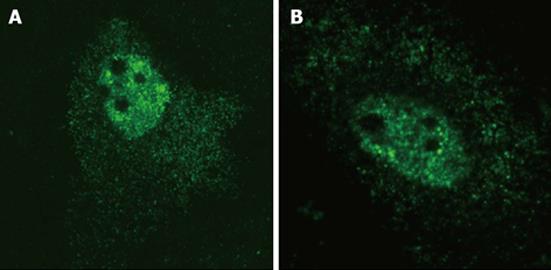
In situ hybridization was performed on pDL481 and pDL542 transfected HuH-7 cells essentially as described[15]. After transfection, cells were incubated at 37 °C for 24 h. All fixation, permeabilization, and denaturation steps were exactly as described[15]. Plasmid pSVL(D3) was labeled by nick-translation with digoxigenin-11-dUTP and used as a probe. This plasmid contains a trimer of full-length HDV cDNA cloned in pSVL (GE Healthcare). Hybridization was performed overnight at 37 °C and the probe was detected using a monoclonal anti-digoxigenin antibody conjugated with FITC (Roche) and a secondary anti-FITC antibody conjugated with Alexa-488 (Jackson ImmunoResearch Laboratories). Samples were analyzed under a Zeiss META LSM 510 microscope calibrated with multicolor fluorescent beads (Molecular probes). Green fluorescence was detected using a 488 nm Argon laser.
For Northern blotting, cytoplasmic mRNA was extracted from HuH-7 cells using the Oligotex Direct mRNA Mini kit (Qiagen). For each obtained sample, 10 μg mRNA was separated by formaldehyde agarose gel electrophoresis and transferred to Nylon membranes (Hybond-N, GE Healthcare) using standard protocols[28]. Hybridization was performed using a digoxigenin-11-dUTP (dig-11-dUTP) labeled DNA probe. Plasmid pDM138 was used as template to amplify and label, by asymmetric PCR, a 481 bp region in the ORF of the CAT protein (nucleotide position 109-590). The primers used in PCR reactions were: Fwd 5’ GTTCAGCTGGATATTACGGCC 3’ and Rev 5’ TCACAGACGGCATGATGAAC 3’. Typically, reaction mixtures contained 2 mmol/L MgCl2, 0.2 mmol/L dATP, dCTP and dGTP, 0.13 mmol/L dTTP, 0.07 mmol/L dig-11-dUTP (Roche), 0.1 μmol/L forward primer, 1 μmol/L reverse primer, 10 ng template DNA, 2.5 U Taq DNA polymerase (Fermentas), in PCR buffer for a final volume of 50 μL. After amplification and labeling, probes were purified using the GFX PCR DNA kit (GE Healthcare), and used for hybridization.
Hybridization was performed according to standard protocols[28] and the hybridized probe was detected with a monoclonal anti-digoxigenin antibody conjugated with peroxidase (Roche). Membrane development was achieved with the Lumi-lightPLUS Western Blotting Kit, Mouse/Rabbit (Roche) under the conditions indicated by the manufacturer.
Nuclear and cytoplasmic HuH-7 cell fractions were obtained according to a previously described method[29], and used for isolation of RNA with the NucleoSpin® RNA/protein kit (Macherey-Nagel) following the manufacturer’s specifications. The RNA samples were then treated with DNase I using the DNA-free™ kit (Ambion), also according to the instructions of the manufacturer, and used as templates for synthesis of cDNA. cDNA synthesis reactions typically contained approximately 5 μg total RNA, 0.2 μg random primers, 2 mmol/L dNTPs, 200 U Revert Aid™ M-MuLV Reverse Transcriptase (Fermentas), and 20 U RNase inhibitor (Fermentas) in a final volume of 20 μL. Reactions were performed at 42 °C, for 1 h, and the obtained cDNA was finally purified using the GFX PCR DNA and Gel Band purification kit (GE Healthcare).
Real-time PCR experiments were performed essentially as described[30]. The qPCR Core kit for SYBR® Green I (Eurogentec) was used following the specifications of the manufacturer. Reaction mixtures typically contained 3.5 mmol/L MgCl2, 200 μmol/L each dNTP, 300 nmol/L each primer, 0.025 U/μL HotGoldStar enzyme, and reaction buffer in a final volume of 20 μL. Reactions were performed in 96-well plates with optical caps in a GeneAmp® 5700 Sequence Detector System (all from Applied Biosystems). The PCR program used for amplification was: 10 min at 95 °C, 40 cycles with 15 s at 95 °C and 1 min at 60 °C. Each sample was assayed in triplicate and analysed with the GeneAmp® 5700 SDS v1.1 software and Microsoft Excel.
The relative quantification of RNA was performed according to the 2-ΔΔCt method earlier described[31]. The β-2-microglobulin gene (β2MG; GenBank accession number NM_004048) was used as reference gene to which all the samples were compared with. The program Primer Express™ (Applied Biosystems) and the bioinformatics tool Oligonucleotide Properties Calculator (http://www.basic.northwestern.edu/biotools/oligocalc.html) were used to design primers for the reference gene and target HDV cDNA sequence (GenBank accession number M21012). Melting temperature, GC content, length, and secondary structure were taken in consideration for primer design. The cDNA sequences were obtained from GenBank database from NCBI. The primers used in these experiments were, respectively: HDV Fwd 5’ CAGAGATTCTCCGGCGTTGT 3’, Rev 5’ CGGTAAAGAGCATTGGAACG 3’; β2MG Fwd 5’ GGCTATCCAGCGTACTCCAA 3’, Rev 5’ TCACACGGCAGGCATACTC 3’.
For western blot, protein extracts were prepared using with the NucleoSpin® RNA/protein kit (Macherey-Nagel) according to the manufacturer’s instructions and dissolved in sample buffer. Proteins were separated by electrophoresis on 12%SDS-polyacrylamide gels, and subsequently electroblotted onto nitrocellulose membranes (Schleicher and Schuell) as previously described[32]. Membranes were blocked with 5% low fat milk powder in PBS, and incubated for 1 h with 1 μg/mL of a primary mouse monoclonal antibody anti-GAPDH (Ambion). After washing with 2% low fat milk powder in PBS, membranes were further incubated with a secondary anti-mouse IgG antibody conjugated with horseradish peroxidase (BioRad). After washing, membranes were rinsed with PBS and subsequently developed using the ECL™ Western blotting analysis system (GE Healthcare).
Determination of CAT expression was performed using a commercial enzyme-linked immunosorbent assay (ELISA) kit (Roche). Briefly, 24 h post-transfection HuH-7 cells were washed with ice-cold PBS and incubated with 500 μL lysis buffer for 30 min, at room temperature. After centrifugation, the supernatants were collected, and 200 μL were added to individual wells of the ELISA plate. CAT detection was performed with a polyclonal anti-CAT antibody conjugated with digoxigenin followed by incubation with a monoclonal anti-digoxigenin antibody conjugated with peroxidase (Roche), as indicated by the manufacturer. The concentration of unknown samples was determined from a standard curve constructed from 1:2 serial dilutions of the standards.
To normalize for transfection efficiencies HuH-7 cells were cotransfected with plasmid pSV-β-galactosidase (Promega) and β-galactosidase (β-Gal) expression was monitored using a commercial ELISA kit (Roche). Briefly, 200 μL of the same cell lysis supernatants obtained as above described were loaded onto individual wells of the ELISA plate. The ELISA assay was performed according to the specifications of the manufacturer and β-Gal concentrations were determined from a standard curve obtained from 1:2 serial dilutions of the standards. All assays were performed in triplicate.
It was earlier reported that both HDV gRNA and agRNA are exported to the cytoplasm in HuH-7 cells[15,17]. This export is independent of the presence of HDAgs, and thus it is likely to rely on the direct interaction of the virus RNA with host factors[15]. Furthermore, Northern blot analysis of HuH-7 transfected cells seemed to indicate that the relative amounts of gRNA and agRNA in the nucleus and cytoplasm remained nearly equimolar up to 28 h after transfection[17].
In order to confirm that HDV agRNA is efficiently exported to the cytoplasm of HuH-7 cells we made use of plasmid pDL481[18], which codes exclusively for HDV agRNA, in transfection experiments. Plasmid pDL542[18], which codes exclusively for gRNA, was used in parallel experiments. Preliminary in situ hybridization analysis confirmed that both HDV gRNA and agRNA can be detected in the nuclear and cytoplasmic compartments of HuH-7 cells (Figure 1) 24 h post-transfection.
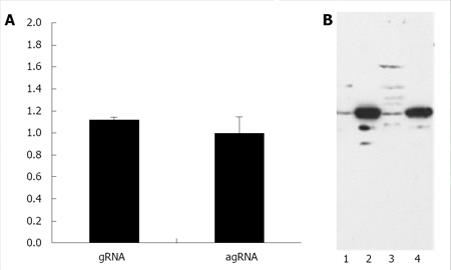
Since this approach did not allow us to determine if gRNA and agRNA are exported to the cytoplasm with similar efficiency with decided to quantify, by Real-time PCR, the amounts of both molecules in the nuclear and cytoplasmic compartments. To do this, HuH-7 cells were transfected with plasmids pDL481 and pDL542 respectively. After 24 h incubation, RNA samples were obtained from nuclear and cytoplasmic fractions for subsequent use in Real-time qPCR experiments. The possible cross contamination of nuclear and cytoplasmic fractions was monitored by Western blotting using an anti-GADPH antibody (Figure 2B). The reference gene in qPCR experiments was β-2-microglobulin (Genbank accession number P61769).The obtained results are displayed, as the cytoplasm to nuclear ratio of gRNA and agRNA, respectively, in Figure 2A. The HDV agRNA was found to be distributed in equimolar amounts between the nucleus and cytoplasm of HuH-7 cells 24 h after transfection. At the same time point the HDV gRNA was found in slightly higher amounts in the cytoplasm. These results are consistent with previously reported data and clearly indicate that the virus agRNA is efficiently exported to the cytoplasm.
After establishing that HDV gRNA and agRNA are exported to the cytoplasm, in the absence of HDAgs, with similar efficiency, until at least 24 h after transfection, we decided to investigate the eventual presence of a cis-acting nuclear export element in both RNA molecules. To do this, we made use of plasmid pDM138[25,26]. This plasmid codes for the second half of the HIV-1/SF2 genome under the control of the SV40 promoter. The DNA sequence coding for the CAT gene was inserted in the HIV-1 envelope gene intron, and the RRE was substituted by a linker containing a unique ClaI restriction site. Nuclear export of mRNAs derived from pDM138, and subsequent expression of the reporter CAT protein, is thus dependent on the insertion of a functional transport element in the ClaI restriction site.
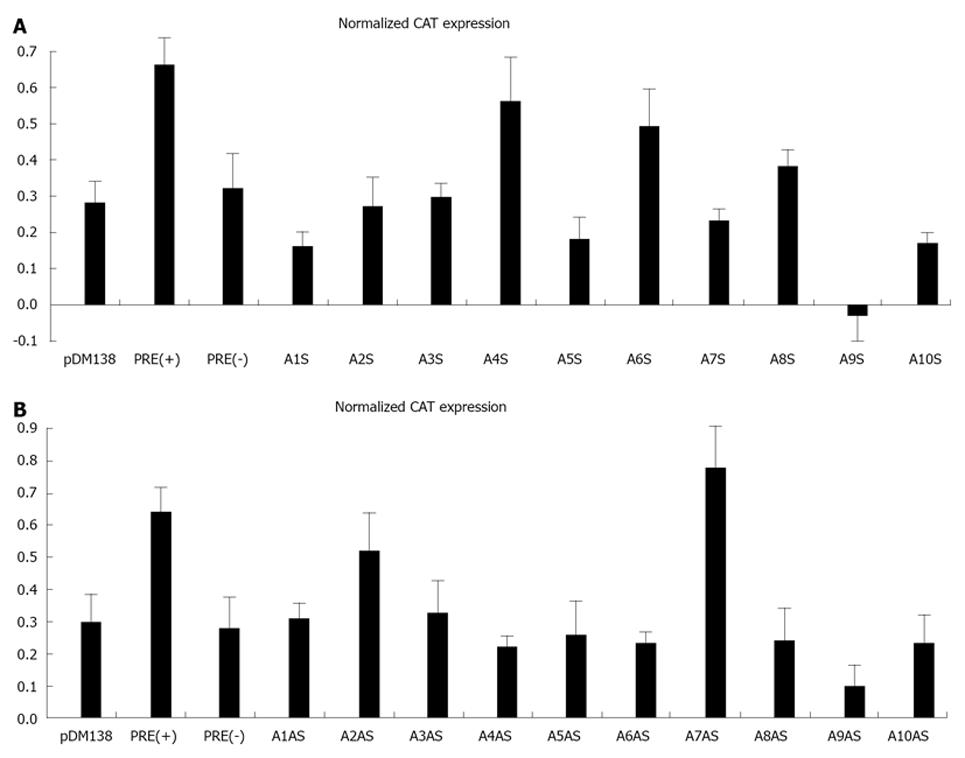
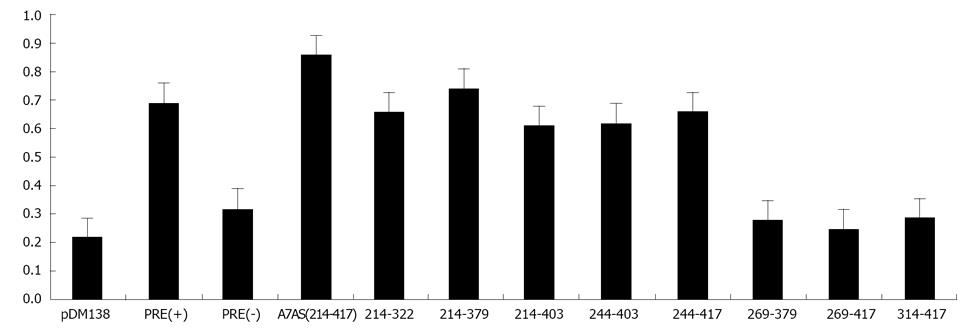
Initially, we amplified by PCR 20 cDNA fragments covering the entire HDV genome (10 fragments) and antigenome (also 10 fragments). The size of the obtained fragments ranged from 167 to 337 nt and the respective location in the genome is displayed in Table 1. Each fragment was subsequently cloned in the unique ClaI site of plasmid pDM138 in both sense and antisense orientations. We thus obtained 20 different constructs which, after being sequenced to confirm the correct insertion and orientation, were used to transfect HuH-7 cells. As positive and negative controls in these experiments we used plasmids pDM138-PRE(+) and pDM138-PRE(-), respectively. These plasmids contain the HBV post-transcriptional regulatory element, cloned in sense and antisense orientations, respectively[27]. In all experiments, plasmid pSV-β-Gal (Promega) was used to cotransfect HuH-7 cells in order to normalize for transfection efficiencies. Twenty four hours after transfection, total protein extracts were prepared and the production of CAT and β-Gal was determined by ELISA. Figure 3 displays the obtained results.
When analyzing gRNA all the tested sequences were found to be unable to promote the export of CAT mRNAs with efficiency as high as that determined for the positive pDM138-PRE(+) control. Nevertheless, two of the analyzed gRNA sequences, corresponding to nucleotides 266-489 and 584-920 (A4S, and A6S, respectively), showed an export-promoting ability slightly lower than the positive control albeit clearly higher than the negative control pDM138-PRE(-).
The analysis of agRNA coding sequences, however, showed that in HuH-7 cells transfected with plasmid pDM138-A7AS, which contains the HDV agRNA sequence corresponding to nucleotides 214-417, the expression levels of the reporter CAT protein are higher than those detected for the pDM138-PRE(+) positive control.
With the exception of A7AS and A2AS sequences, all the remaining tested constructs were found to be unable to promote the export of heterologous intron-containing mRNAs since the detected CAT expression levels were comparable or even lower than those observed in negative pDM138-PRE(-) transfected HuH-7 cell controls. The CAT expression values obtained for the A2AS sequence, however, were found to be intermediate between those obtained for the positive pDM138-PRE(+) and negative pDM138-PRE(-) controls.
According to what has been previously reported for the HBV PRE[27], the export promoting activity of the identified A7AS sequence in the HDV antigenome is dependent on its orientation relative to the ORF of the reporter gene. In fact, when cloned in opposite orientation (A7S), the A7AS sequence (nt 214-417) was not functional, since the observed CAT expression levels were similar to those found for the pDM138-PRE(-) negative control (data not shown).
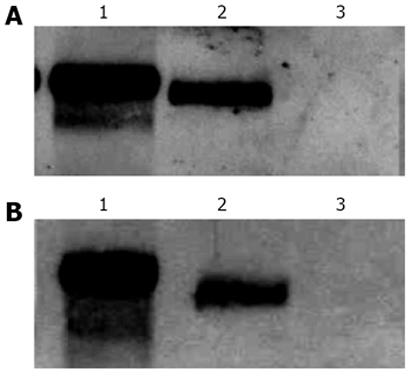
Since the quantification of CAT expression levels by ELISA represents an indirect approach for the determination of intron-containing mRNAs accumulation in the cytoplasm, we decided to investigate the presence of CAT mRNAs in cytoplasmic fractions, after transfection, by Northern blot. To do this, HuH-7 cells were transfected with plasmid pDM138-A7AS or plasmids pDM138-PRE(+) pDM138-A9AS as positive and negative controls, respectively, and after 24 h incubation total and cytoplasmic fractions were prepared and used for further mRNA extraction. After electrophoresis, the mRNA samples were transferred to nylon membranes, and a single-stranded dig-11-dUTP labeled DNA probe, which specifically hybridizes with CAT mRNA, was used in Northern blot assays. As expected, it was possible to detect the presence of CAT mRNA in cytoplasmic fractions of pDM138-PRE(+) and pDM138-A7AS transfected HuH-7 cells but not in pDM138-A9AS transfected cells (Figure 4).
The absence in Northern blot experiments of detected CAT mRNA in total and cytoplasmic fractions of pDM138-A9AS transfected cells is compatible with the data obtained by ELISA. The A9AS sequence includes the agRNA autocatalytic ribozyme domain. It is, thus, possible that CAT mRNAs that include the A9AS sequence are degraded in the nucleus before export to the cytoplasm. In conclusion, these results indicate that the HDV agRNA sequence located between nucleotides 214-417 can efficiently promote the export of heterologous intron-less RNAs. Moreover, the increase in CAT expression levels observed in pDM138-A7AS transfected cells is a consequence of the export and accumulation of the respective reporter mRNAs in the cytoplasm.
Having established that the HDV agRNA sequence corresponding to nucleotides 214-417 (A7AS) is able to promote the nuclear export of heterologous intron-containing RNAs, we next decided to analyze it in more detail. First, we generated by PCR eight truncated forms of the A7AS motif and cloned them in the unique ClaI site of plasmid pDM138. These truncated forms correspond to the A7AS sequence from which several nucleotides were removed from the 5’ and 3’ ends. The constructs were subsequently used to transfect HuH-7 cells, and after 24 h CAT expression was determined by ELISA. The obtained CAT expression values were normalized for transfection efficiency by cotransfection with plasmid pSV-β-Gal followed by determination of β-Gal expression. The obtained results allowed us to conclude that the agRNA sequences comprised between nucleotides 214-322, 214-379, 214-403, 244-403, and 244-417 induce an increase in CAT expression comparable to the observed for the HBV PRE(+) positive control (Figure 5) suggesting that these sequences are sufficient to promote export of heterologous RNAs.
Additionally, the first 30 nucleotides localized 3’ in the A7AS sequence seem not to be crucial to promote nuclear export since its deletion did not significantly affect the detected amounts of CAT expression. All the remaining analyzed sequences, 269-417, 314-417, and 269-379 were found to be considerably less efficient in promoting nuclear export.
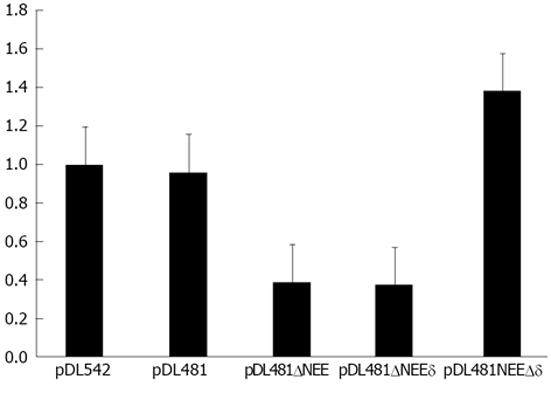
The identified NEE is localized in the central region of the rod-like full-length agRNA molecule. It could be possible that the complementary RNA sequence that pairs with the NEE in the antigenome is possibly also involved in the nuclear export of the agRNA. To test this hypothesis we constructed three deletion mutants of plasmid pDL481, which codes for the complete HDV agRNA molecule: pDL481ΔNEE in which the NEE was deleted (nt 214-417), pDL481Δδ in which the complementary to the NEE sequence was deleted (nt 1179-1385), and pDL481ΔNEEδ which does not contain the NEE and the corresponding complementary sequence (nt 214-417 and 1179-1385, respectively).
Plasmids pDL542, pDL481, and the obtained deletion constructs were used to transfect HuH-7 cells. Twenty four hours post-transfection, nuclear and cytoplasmic fractions were prepared and the RNAs derived from pDL542, pDL481, pDL481ΔNEE, pDL481Δδ, and pDL481ΔNEEδ, respectively, were quantified by qRT-PCR. Possible cross contaminations of nuclear and cytoplasmic fractions were monitored by western blot using an anti-GAPDH antibody as described before. The obtained results confirmed that both gRNA and agRNA are efficiently exported to the cytoplasm 24 h post-transfection. Additionally, the RNA derived from plasmid pDL481Δδ, in which the sequence coding for the region complementary to the NEE was deleted, showed to be efficiently exported (Figure 6). In contrast, the RNAs coded by plasmids pDL481ΔNEEδ and pDL481ΔNEE were mostly retained in the nucleus. Only about 40% of the total amount of these RNAs was found in the cytoplasm of transfected cells when compared to wt pDL481. These results indicate that the complementary to the NEE sequence in the HDV antigenome (nt 1179-1385) is not involved in nuclear export since its deletion did not reduce the amounts of agRNA detected in the cytoplasm.
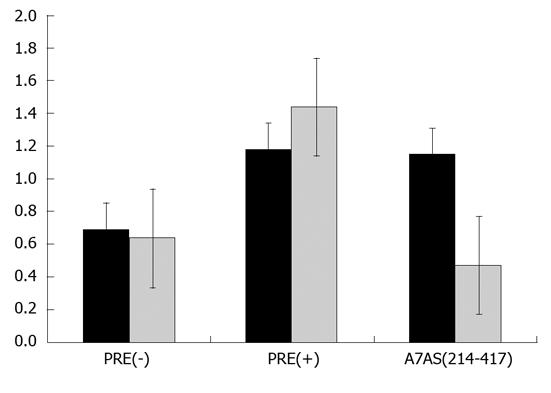
Export of cellular RNAs is accomplished using different pathways which involve the participation of distinct transport receptors. Typical examples include the export of mRNAs, which is mediated by members of NXF family of proteins, namely the TAP protein, and the export of UsnRNAs which is mediated by the exportin CRM1[20]. This specific export pathway can be inhibited in the presence of leptomycin B (LMB)[33]. In order to investigate whether HDV agRNA is exported to the cytoplasm using the pathway mediated by CRM1, we decided to analyze the effect of LMB on CAT protein expression in pDM138-A7AS transfected HuH-7 cells. To do this, 10 nmol/L LMB was added to the culture medium 18 h after transfection, and cells were further incubated for 6 h. Total protein extracts were then prepared and used to determine the concentration of CAT by ELISA. As negative control pDM138-PRE(+) transfected HuH-7 cells were used since it was previously reported that export of HBV PRE(+) is not sensitive to LMB[23,34]. Figure 7 displays the obtained results. As expected, in the absence of LMB both the HDV agRNA A7AS and HBV PRE(+) sequences promote the export of CAT mRNA thus confirming our previous data. In the presence of LMB the export capacity of the HBV PRE(+) sequence is not affected, and we could detect a slight increase in CAT expression. This observation is in accordance with the data obtained by others when measuring CAT enzyme activity in similar experiments[34]. In contrast, the export promoting activity of the HDV agRNA A7AS sequence was found to be affected in the presence of LMB. In fact, we observed a 60% reduction in CAT production when HuH-7 cells were transfected with pDM138-A7AS in the presence of 10 nmol/L LMB. These results seem to indicate that the nuclear export mediated by the A7As agRNA sequence is dependent, at least partially, on CRM1 activity and suggest the involvement of this cellular protein in HDV agRNA export.
Although HDV RNA replication occurs in the nucleus of liver cells, virus packaging takes place in the cytoplasm where HDV RNPs meet HBsAgs to assemble newly synthesized virions. Noteworthy, only gRNA molecules were found, until now, to be packaged into mature hepatitis delta virions. This restriction could be due to a possible impairment of export of agRNA molecules to the cytoplasm. However, agRNA was found in HDV virus-like particles secreted by cells expressing agRNA, L-HDAg, and HBsAgs[18].
A previous work showed that HDV RNPs are exported to the cytoplasm independent of the presence of HBsAgs[15]. Additionally, it was demonstrated that export of HDV RNPs is not mediated by a putative nuclear export signal present in delta antigens but is rather promoted by cis elements in virus RNA. In fact, both HDV gRNA and agRNA are exported to the cytoplasm in the absence of HDAgs, an observation also reported by Macnaughton et al[17].
Our first approach was designed to confirm that both HDV genomic and antigenomic RNA molecules are exported to the cytoplasm of HuH-7 cells. Using plasmids pDL542 and pDL481 which express exclusively HDV gRNA and agRNA, respectively, we were able to detect both molecules, by in situ hybridization, in the nuclear and cytoplasmic compartments of HuH-7 transfected cells. Furthermore, nuclear export of HDV gRNA and agRNA seems to occur with the same efficiency since similar amounts of genomic and antigenomic RNA molecules could be detected, by real time-PCR, in the nuclear and cytoplasmic compartments of HuH-7 cells transiently transfected with plasmids pDL542 and pDL481. The finding that both genomic and antigenomic HDV RNA molecules are exported with similar efficiency suggests a possible biological function associated with the presence of HDV agRNA in the cytoplasm. However, further research is mandatory to clarify the biological significance of these findings.
The above described observations are suggestive of a possible presence of cis-acting nuclear export elements in HDV RNA, both genomic and antigenomic. In an attempt to identify putative cis elements in the HDV gRNA and agRNA capable of mediating the export to the cytoplasm, we used a well characterized CAT reporter system previously used by others to investigate the role of the HBV PRE in export of intronless mRNAs[27]. This system is based on plasmid pDM138 which was generated in order to contain the second half of HIV-1 cDNA under the control of the SV40 promoter[25,26]. This vector was further engineered in order to remove the initiation codons for the Rev and Env proteins, to include the cDNA sequence coding for the CAT protein, and to substitute the HIV-1 RRE sequence by a linker containing a ClaI restriction site. After transcription, an mRNA containing the ORF for the CAT protein inserted in the intron of the HIV-1 Env proteins is produced. As positive and negative controls we used plasmids pDM138-PRE(+) and pDM138-PRE(-), respectively. These plasmids contain the HBV post-transcriptional regulatory element (PRE) inserted in the unique ClaI site of pDM138, in sense and antisense orientations, respectively[27]. We cloned several cDNA fragments, covering the entire HDV gRNA and agRNA, in plasmid pDM138. After transfection of HuH-7 cells, CAT production was determined by ELISA. This approach did not allow identifying unequivocally a nuclear export element in gRNA. Although two gRNA fragments corresponding to nucleotides 266-489 and 584-920, (A4S and A6S, respectively) were found to promote export of CAT mRNAs, the efficiency was in all experiments lower than the determined for the positive control pDM138-PRE(+). This could be possibly due to the lack of crucial nucleotides in the ends of the analyzed sequences. Clearly, additional experiments are mandatory to clarify this point.
However, we were able to identify a region (A7AS) in the HDV agRNA, located between positions 214-417, which promoted the export of CAT mRNA with slightly higher efficiency than that observed for the pDM138-PRE(+) positive control. This result was further confirmed by Northern blot analysis of total and cytoplasmic mRNAs prepared from pDM138-PRE(+) and pDM138A7AS transfected HuH-7 cells. Moreover, when inserted in the opposite orientation in pDM138, the fragment A7AS was not able to mediate export of CAT mRNAs (data not shown). All the other fragments tested were not capable of promoting export of CAT mRNA at levels comparable to those observed for the pDM138-PRE(+) positive control. In particular, the fragment A9AS which includes the HDV RNA ribozyme sequence displayed a CAT mRNA export capacity significantly lower than the calculated for the negative control. This may be a consequence of instability of the produced mRNA molecules due to the self-cleavage activity of the HDV ribozyme. Besides the A7AS sequence, the only exception to the low export promoting capacity of the analyzed fragments, concerns a cDNA fragment corresponding to positions 1301-1514 in the antigenome (A2AS). This agRNA fragment was able to promote CAT mRNA export with efficiency higher than the negative control but still lower than the determined for the positive control. This fragment, A2AS, includes part of the HDAg ORF. This observation may allow speculating about a possible presence of a cis element in the HDV mRNA involved in export to the cytoplasm. Huang and Carmichael have previously shown that export on intronless histone H2a mRNAs is mediated by a signal present in the coding region[35]. However, further experiments are needed to clarify the possible involvement of a similar signal in export of HDAg mRNA.
In an attempt to analyze in more detail the A7AS sequence in the HDV agRNA we generated, by PCR, eight truncated forms of this motif which were subsequently cloned in pDM138 and used in transfection experiments to determine CAT expression by ELISA. These truncated forms corresponded to nucleotides 314-417, 269-417, 244-417, 244-403, 214-403, 269-379, 214-379, and 214-322. The obtained results showed that deletion of the first 30 nucleotides in the 5’ end of the A7AS sequence did not significantly affect the export promoting capacity. However, deletion of the first 55 nucleotides in the 5’ end results in loss of the export capacity of the A7AS sequence. In contrast, deletions in the 3’ end of the A7AS motif, did not significantly affect the ability to promote nuclear export. In fact, removal of as much as the first 95 nucleotides in the 3’ end still resulted in the production of the CAT protein at intermediate levels between the negative and positive controls. Noteworthy, all the analyzed A7AS truncated forms were less efficient in promoting RNA export when compared to the wild-type sequence.
However, analysis of the secondary structure of the entire agRNA molecule did not allow predicting a similar branched structure in the region where the A7AS motif is located. In order to clarify a possible role of a, at least partially, complementary to the A7AS motif sequence in the antigenome, in nuclear export, we constructed deletion mutants of plasmid pDL481. These constructs were designed as follows: pDL481ΔNEE lacks the nuclear export element (A7AS sequence), pDL481Δδ lacks the complementary to the NEE sequence in the antigenome, and pDL481ΔNEEδ lacks both the NEE and the respective complementary sequence. After transfection of HuH-7 cells, the relative amounts of agRNA in nuclear and cytoplasmic fractions were determined by qRT-PCR. The obtained results showed that deletion of the NEE (pDL481ΔNEE) reduces export of agRNA by 60% when compared with wt pDL481. Additionally, deletion of both the NEE and the respective complementary sequence (pDL481ΔNEEδ) results in a similar reduction (62%) of detected cytoplasmic RNA. Finally, deletion of only the complementary to the NEE sequence did not impair the capacity of the agRNA to be exported to the cytoplasm. Taken together, these results indicate that the identified NEE is important to promote nuclear export of the HDV antigenome, and that export efficiency is not diminished by deletion of the respective putative complementary sequence.
Nuclear export of host and virus RNAs may be promoted by several cellular factors that participate in distinct pathways. One of these pathways is mediated by the exportin CRM1 which belongs to the kariopherin-β family of proteins. CRM1 mediates the export of the majority of proteins containing a nuclear export signal (NES) and of two classes of cellular non-coding RNAS, rRNAs and UsnRNAs[21]. The CRM1 export pathway may be specifically inhibited in the presence of LMB which binds to a cysteine residue in the central region of the protein[33]. In order to investigate a possible involvement of CRM 1 in export of HDV agRNA we used LMB to inhibit this pathway in pDM138-A7AS transfected HuH-7 cells. As positive and negative controls we used plasmids pDM138-PRE(+) and pDM138-PRE(-), respectively, since export of the HBV PRE was previously reported to be insensitive to LMB. The obtained results showed that RNA export mediated by the NEE of HDV agRNA is partially inhibited in the presence of LMB, displaying a 50% reduction of CAT expression when compared with the parental pDM138 vector. This suggests an involvement of the CRM1 protein in export of HDV agRNA. However, since CRM1 binds to RNA molecules indirectly through interaction with other NES-containing proteins this implies the participation of other not yet identified host factors in HDV agRNA export. Not surprisingly, using coimmunoprecipitation assays we couldn’t detect complexes between HDV RNPs and CRM1 (our unpublished data).
In this work we attempted to identify nuclear export elements present HDV gRNA and agRNA. Although two regions in gRNA were found to be able to promote export of heterologous RNAs, an unequivocal NEE could not be identified in gRNA. However, it was possible to identify a NEE in HDV agRNA located in positions 214-417. This cis element is not only capable of promoting the nuclear export of heterologous intronless RNAs but is also involved in export of HDV antigenomes. Analysis of the export capacity of several truncated forms of the NEE showed that the two mini-helices seem to play a crucial role in mediating RNA export. Cytoplasmic export of HDV agRNA was found to be sensitive to leptomycin B suggesting a possible involvement of a CRM1 mediated pathway.
We thank Dr. Tristram Parslow (Emory University School of Medicine, Atlanta, United States) for kindly providing plasmid pDM138, Dr. Benedict Yen (University of California, San Francisco, United States) for the generous gift of plasmids pDM138-PRE(-) and pDM138-PRE(+), and Dr. John Taylor (Fox Chase Cancer Center, Philadelphia, United States) for plasmid pDL481.
Hepatitis delta virus (HDV) is a satellite of hepatitis B virus (HBV). The two viruses share the same envelope which consists of HBV surface antigens (HBsAg). HBsAgs are coded exclusively by the HBV genome and are localized in the cytoplasm of infected cells. In contrast HDV replicates in the nucleus and assembly of mature virions takes place in the cytoplasm. Accordingly, HDV RNPs need to be exported to the cytoplasm and this process is thought to be mediated by cis-elements present in the virus RNA.
Export of RNA molecules to the cytoplasm depends on the interaction of cellular transport receptors with specific nucleotide sequences. These sequences were previously studied in simple and complex retroviruses as well as in HBV. The cellular proteins and pathways involved in virus RNA export were also identified. Here, the research hotspot is the identification of nucleotide sequences in HDV RNA capable of promoting export of heterologous RNAs and thus being able to function as nuclear export elements.
This is the first report describing a comprehensive analysis of HDV genomic and antigenomic RNA sequences aiming to identify cis-elements capable of promoting export to the cytoplasm. Two possible regions corresponding to nucleotides 266-489 and 584-920, respectively, were identified in HDV genomic RNA. In addition, analysis of HDV antigenomic RNA sequences allowed finding a nuclear export element in positions 214-417. Furthermore, export mediated by the nuclear export element of HDV antigenomic RNA was found to be sensitive to leptomycin B suggesting a possible role of the cellular protein CRM1 in this transport pathway.
The results herein described may contribute to a better understanding of an essential step in the HDV replication cycle-the export of virus RNPs to the cytoplasm where they interact with HBsAgs in order to assemble new virions.
Nuclear export element is an RNA sequence capable to interact with cellular proteins that promote export from the nucleus to the cytoplasm.
The paper is of good quality as research article, its english is perfect and its scientific content is original.
P- Reviewer Kamal SA S- Editor Zhai HH L- Editor A E- Editor Lu YJ
| 1. | Smedile A, Rizzetto M. HDV: thirty years later. Dig Liver Dis. 2011;43 Suppl 1:S15-S18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 2. | Govindarajan S, Chin KP, Redeker AG, Peters RL. Fulminant B viral hepatitis: role of delta agent. Gastroenterology. 1984;86:1417-1420. [PubMed] |
| 3. | Jacobson IM, Dienstag JL, Werner BG, Brettler DB, Levine PH, Mushahwar IK. Epidemiology and clinical impact of hepatitis D virus (delta) infection. Hepatology. 1985;5:188-191. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 4. | Smedile A, Rizzetto M, Gerin JL. Advances in hepatitis D virus biology and disease. Prog Liver Dis. 1994;12:157-175. [PubMed] |
| 5. | Taylor JM. Replication of human hepatitis delta virus: recent developments. Trends Microbiol. 2003;11:185-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 65] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 6. | Gerin JL, Casey JL, Purcell RH. Hepatitis delta virus. In: Knipe DM, Howley PM, editors. Fields Virology. 4th edition. Philadelphia, Lippincott Williams & Wilkins 2001; chapter 88. |
| 7. | Chang J, Nie X, Chang HE, Han Z, Taylor J. Transcription of hepatitis delta virus RNA by RNA polymerase II. J Virol. 2008;82:1118-1127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 8. | Sharmeen L, Kuo MY, Taylor J. Self-ligating RNA sequences on the antigenome of human hepatitis delta virus. J Virol. 1989;63:1428-1430. [PubMed] |
| 9. | Moraleda G, Taylor J. Host RNA polymerase requirements for transcription of the human hepatitis delta virus genome. J Virol. 2001;75:10161-10169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 58] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 10. | Casey JL, Gerin JL. Hepatitis D virus RNA editing: specific modification of adenosine in the antigenomic RNA. J Virol. 1995;69:7593-7600. [PubMed] |
| 11. | Chang J, Gudima SO, Tarn C, Nie X, Taylor JM. Development of a novel system to study hepatitis delta virus genome replication. J Virol. 2005;79:8182-8188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Jeng KS, Su PY, Lai MM. Hepatitis delta antigens enhance the ribozyme activities of hepatitis delta virus RNA in vivo. J Virol. 1996;70:4205-4209. [PubMed] |
| 13. | Chang FL, Chen PJ, Tu SJ, Wang CJ, Chen DS. The large form of hepatitis delta antigen is crucial for assembly of hepatitis delta virus. Proc Natl Acad Sci USA. 1991;88:8490-8494. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 252] [Cited by in RCA: 265] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 14. | Glenn JS, Watson JA, Havel CM, White JM. Identification of a prenylation site in delta virus large antigen. Science. 1992;256:1331-1333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 247] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 15. | Tavanez JP, Cunha C, Silva MC, David E, Monjardino J, Carmo-Fonseca M. Hepatitis delta virus ribonucleoproteins shuttle between the nucleus and the cytoplasm. RNA. 2002;8:637-646. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 47] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 16. | Chou HC, Hsieh TY, Sheu GT, Lai MM. Hepatitis delta antigen mediates the nuclear import of hepatitis delta virus RNA. J Virol. 1998;72:3684-3690. [PubMed] |
| 17. | Macnaughton TB, Lai MM. Genomic but not antigenomic hepatitis delta virus RNA is preferentially exported from the nucleus immediately after synthesis and processing. J Virol. 2002;76:3928-3935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 18. | Lazinski DW, Taylor JM. Expression of hepatitis delta virus RNA deletions: cis and trans requirements for self-cleavage, ligation, and RNA packaging. J Virol. 1994;68:2879-2888. [PubMed] |
| 19. | Grüter P, Tabernero C, von Kobbe C, Schmitt C, Saavedra C, Bachi A, Wilm M, Felber BK, Izaurralde E. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649-659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 436] [Cited by in RCA: 443] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 20. | Izaurralde E. A novel family of nuclear transport receptors mediates the export of messenger RNA to the cytoplasm. Eur J Cell Biol. 2002;81:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 39] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Cullen BR. Nuclear RNA export. J Cell Sci. 2003;116:587-597. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 160] [Cited by in RCA: 164] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 22. | Fornerod M, Ohno M, Yoshida M, Mattaj IW. CRM1 is an export receptor for leucine-rich nuclear export signals. Cell. 1997;90:1051-1060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1642] [Cited by in RCA: 1684] [Article Influence: 60.1] [Reference Citation Analysis (0)] |
| 23. | Zang WQ, Yen TS. Distinct export pathway utilized by the hepatitis B virus posttranscriptional regulatory element. Virology. 1999;259:299-304. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 24. | Paca RE, Ogert RA, Hibbert CS, Izaurralde E, Beemon KL. Rous sarcoma virus DR posttranscriptional elements use a novel RNA export pathway. J Virol. 2000;74:9507-9514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 57] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 25. | Hope TJ, Huang XJ, McDonald D, Parslow TG. Steroid-receptor fusion of the human immunodeficiency virus type 1 Rev transactivator: mapping cryptic functions of the arginine-rich motif. Proc Natl Acad Sci USA. 1990;87:7787-7791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 175] [Cited by in RCA: 196] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 26. | Huang XJ, Hope TJ, Bond BL, McDonald D, Grahl K, Parslow TG. Minimal Rev-response element for type 1 human immunodeficiency virus. J Virol. 1991;65:2131-2134. [PubMed] |
| 27. | Huang ZM, Yen TS. Role of the hepatitis B virus posttranscriptional regulatory element in export of intronless transcripts. Mol Cell Biol. 1995;15:3864-3869. [PubMed] |
| 28. | Sambrook J, Fritsch EF, Maniatis T. Molecular cloning, a laboratory manual. New York: Cold Spring Harbor Laboratory Press 1989; 9-14. |
| 29. | Wang Y, Zhu W, Levy DE. Nuclear and cytoplasmic mRNA quantification by SYBR green based real-time RT-PCR. Methods. 2006;39:356-362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 94] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 30. | Mota S, Mendes M, Penque D, Coelho AV, Cunha C. Changes in the proteome of Huh7 cells induced by transient expression of hepatitis D virus RNA and antigens. J Proteomics. 2008;71:71-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 19] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 31. | Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402-408. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149116] [Cited by in RCA: 133708] [Article Influence: 5571.2] [Reference Citation Analysis (1)] |
| 32. | Cunha C, Monjardino J, Cheng D, Krause S, Carmo-Fonseca M. Localization of hepatitis delta virus RNA in the nucleus of human cells. RNA. 1998;4:680-693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 29] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Kudo N, Matsumori N, Taoka H, Fujiwara D, Schreiner EP, Wolff B, Yoshida M, Horinouchi S. Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci USA. 1999;96:9112-9117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 853] [Cited by in RCA: 864] [Article Influence: 33.2] [Reference Citation Analysis (0)] |
| 34. | Otero GC, Harris ME, Donello JE, Hope TJ. Leptomycin B inhibits equine infectious anemia virus Rev and feline immunodeficiency virus rev function but not the function of the hepatitis B virus posttranscriptional regulatory element. J Virol. 1998;72:7593-7597. [PubMed] |
| 35. | Huang Y, Carmichael GG. The mouse histone H2a gene contains a small element that facilitates cytoplasmic accumulation of intronless gene transcripts and of unspliced HIV-1-related mRNAs. Proc Natl Acad Sci USA. 1997;94:10104-10109. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |