Revised: February 15, 2013
Accepted: March 15, 2013
Published online: May 12, 2013
Processing time: 155 Days and 3.5 Hours
Viruses are extremely heterogeneous entities; the size and the nature of their genetic information, as well as the strategies employed to amplify and propagate their genomes, are highly variable. However, as obligatory intracellular parasites, replication of all viruses relies on the host cell. Having co-evolved with their host for several million years, viruses have developed very sophisticated strategies to hijack cellular factors that promote virus uptake, replication, and spread. Identification of host cell factors (HCFs) required for these processes is a major challenge for researchers, but it enables the identification of new, highly selective targets for anti viral therapeutics. To this end, the establishment of platforms enabling genome-wide high-throughput RNA interference (HT-RNAi) screens has led to the identification of several key factors involved in the viral life cycle. A number of genome-wide HT-RNAi screens have been performed for major human pathogens. These studies enable first inter-viral comparisons related to HCF requirements. Although several cellular functions appear to be uniformly required for the life cycle of most viruses tested (such as the proteasome and the Golgi-mediated secretory pathways), some factors, like the lipid kinase Phosphatidylinositol 4-kinase IIIα in the case of hepatitis C virus, are selectively required for individual viruses. However, despite the amount of data available, we are still far away from a comprehensive understanding of the interplay between viruses and host factors. Major limitations towards this goal are the low sensitivity and specificity of such screens, resulting in limited overlap between different screens performed with the same virus. This review focuses on how statistical and bioinformatic analysis methods applied to HT-RNAi screens can help overcoming these issues thus increasing the reliability and impact of such studies.
- Citation: Amberkar S, Kiani NA, Bartenschlager R, Alvisi G, Kaderali L. High-throughput RNA interference screens integrative analysis: Towards a comprehensive understanding of the virus-host interplay. World J Virol 2013; 2(2): 18-31
- URL: https://www.wjgnet.com/2220-3249/full/v2/i2/18.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i2.18
Viruses are obligate intracellular parasites causing more than 3 million deaths per year worldwide (http://www.cdc.gov/). Development of highly efficient vaccines to prevent infection, or antiviral compounds to promote viral clearance from infected patients is hindered by their high variability and mutation rate[1]. Given the small size of their genome, which can be as small as just a few kilobases[2], viruses necessarily rely on host cell factors (HCFs) in order to propagate their genetic information. Therefore key HCFs required for the viral life cycle might represent potential target for the development of new anti-viral compounds[3]. Indeed these factors can be ablated either pharmacologically or genetically, resulting in a drop of viral replication[4,5]. While pharmacological ablation necessarily relies on the availability of highly specific inhibitors, the discovery of RNA interference (RNAi) allows to genetically hinder the expression of virtually any human gene, by reducing its mRNA levels and therefore protein expression[6,7]. The availability of libraries of small interfering RNAs (siRNAs) directed towards almost every human gene (genome-wide libraries) enables to perform large scale, high-throughput RNAi (HT-RNAi) screens to identify key HCF involved in virtually any measurable cellular process. To this end, HT-RNAi technology has been extensively used to identify cellular factors involved in cell division[8], Wnt signaling[9], Janus kinase/signal transducers and activators of transcription signaling[10], extracellular signal-regulated kinases signaling[11], caspase activation[12], mitochondrial function[13] and many others. A similar approach could also be undertaken to search for HCFs required for a certain step of the life cycle of any given virus, which is able to replicate in cell culture. Because viral infection is a multi-step process that starts with the interaction between the parasite and the target cell and ends with the release of newly generated infectious particles, any of these steps is a potential target of therapeutic intervention through silencing of the involved HCFs. Therefore, several genome-wide HT RNAi screens have been performed to identify key factors involved in the life cycle of a number of viruses, including human major pathogens such as influenza virus (INF)[14-16], human immunodeficiency virus-1 (HIV-1)[17-19], and human hepatitis C virus (HCV)[20,21], the only constrain being the availability of a robust cell culture system to assay the outcome of infection. The very first genome-wide HT-RNAi screen performed on viruses was performed on Drosophila C virus (DCV)[22]. Indeed most of the first genome wide HT-RNAi screens were performed in Drosophila cells because of several reasons, including the fact that Drosophila Melanogaster’s genome was completely sequenced in 2000[23], allowing for synthesis of comprehensive Drosophila dsRNA libraries[24,25] and that long dsRNAs added to the medium of Drosophila tissue culture cells are rapidly taken up by the cells in the absence of any transfection reagents, mediating efficient and specific mRNAs knockdown[26]. The first genome-wide screen for viral HCFs relied on a very simple experimental set-up: cells were incubated with a single RNAi specific for each gene in 384 well plates for 3 d, infected with DCV, and 1 d later, processed for immunofluorescence against the capsid antigen before automated microscopic imaging. By visual inspection, the authors identified 210 dsRNA species that reduced the relative number of infected cells by > 40%. dsRNAs targeting these genes were re-synthesized and tested again for their ability to decrease DCV infection. This “validation” screening allowed identifying 112 host dependency factors (HDFs). Among them, 66 proteins were ribosomal proteins, specifically required for translation of DCV polyprotein but not for vesicular stomatitis virus, a pathogen whose genome, in contrast to that of DCV, does not contain a ribosomal entry site (IRES) mediating RNA translation in the absence of a 5’ cap. The authors therefore concluded that the ribosomal genes identified in their study are essential for DVC IRES mediated genome translation.
Since this very first example, it became rapidly clear that many sources of errors such as RNAi reagent design, an inhomogeneous staining, differences in cell growing properties as well as in transfection and infection efficiencies could negatively affect the outcome of such HT-RNAi screens. Therefore, HT-RNAi screens became more and more sophisticated (Table 1). Authors started to worry to strengthen the statistical reliability of their studies by including several replicas, and increasing the number of oligos tested per gene. The most popular approach so far has been to test four different oligos per gene, pooled in a single well in a primary screen, to reduce the so called “off targets effects”[27]. Subsequently hit genes from the primary screen are further tested in a secondary validation screen, where the four different oligos used in the primary screen are tested individually for their ability to reproduce the original phenotype[17,20,21,28]. Several studies started to include the possibility to distinguish genes important early in the viral life cycle (entry/replication phases) from those involved later on (assembly/release of new viral particles), by implementing a two-step procedure, according to which cells are incubated with the siRNA library before infection with the virus of interest. Measurement of viral replication at a given time point enables to identify gene products important for early phases of the viral life cycle such as virus entry and genome replication. Simultaneously, supernatants are collected from infected cells and used to re-infect naive cells, therefore enabling to identify genes important for late stages of viral life cycle such as viral assembly and release[14,17,20]. As far as the readout is concerned, some authors preferred to utilize reporter viruses carrying either the GFP[29]-to avoid issues related to antigen staining and detection - or the Luciferase (Luc) genes in their genome[15,16,18,21], the latter solution enabling an easier and more quantitative analysis of the levels of viral replication. Interpretation of HT-RNAi screening results is also complicated by the fact that different screens performed with the same virus yielded little overlap between HCFs, raising questions concerning the reliability and reproducibility of this approach[30]. Hence, several authors have implemented interesting bioinformatics and statistical approaches (see below) to strengthen the significance and reliability of their results by integrating RNAi data with protein-protein interaction (PPI) databases[18,20].
| Family | Virus | Readout | Viral life cycle step(s) studied | Partition (primary/validation) |
| Flaviviridae (ssRNA+) | DENV1[100] | IF | Single step | Primary: single oligo; validation, single oligo (the same) |
| Picornavirus (ssRNA+) | DCV1[22] | IF | Single step | Primary: single, validation, single (the same) |
| Flaviviridae (ssRNA+) | HCV[21] | Luc (reporter virus) | Single step (replicon) | Primary: pools of 4 oligos; validation: the 4 oligos forming the pool tested individually |
| Flaviviridae (ssRNA+) | HCV[20] | IF | Two step (full virus): entry/ replication; assembly/release | Primary: pools of 4 oligos; validation: the 4 oligos forming the pool tested individually |
| Retroviridae (dsRNA) | HIV-1[17] | Part I: IF; part II: Luc (reporter cell line) | Two steps: entry/replication; assembly/release | Primary: pools of 4 oligos; validation: the 4 oligos forming the pool tested individually |
| Retroviridae (dsRNA) | HIV-1[18] | Luc (reporter virus) | Single step | Primary only: 3 pools of 2 |
| Retroviridae (dsRNA) | HIV-1[19] | Beta-Gal activity | Two steps, but without distinction between them | Primary: pools of 3; validation; independent poools of 3 |
| Orthomixoviridae; (ssRNA- segmented) | INF[14] | Part I: IF; part II: Luc (reporter cell line) | Two steps: entry/replication; assembly/release | Primary: 4 oligoes |
| Orthomixoviridae; (ssRNA- segmented) | INF[15] | Luc (reporter virus) | Single step | Primary only: some genes 2 oligoes, other genes only one |
| Orthomixoviridae; (ssRNA- segmented) | INF1[16] | Luc (reporter virus) | Single step | Primary only: a single sirna x gene |
| Rhabdoviridae (ssRNA-) | VSV[29] | IF | Single step | Primary: 2 pools of 2 oligos; validation: pool of 4 oligos from different vendor |
| Flaviviridae (ssRNA+) | WNV[28] | IF | Single step | Primary: pools of 4 oligos; validation: the 4 oligos forming the pool tested individually |
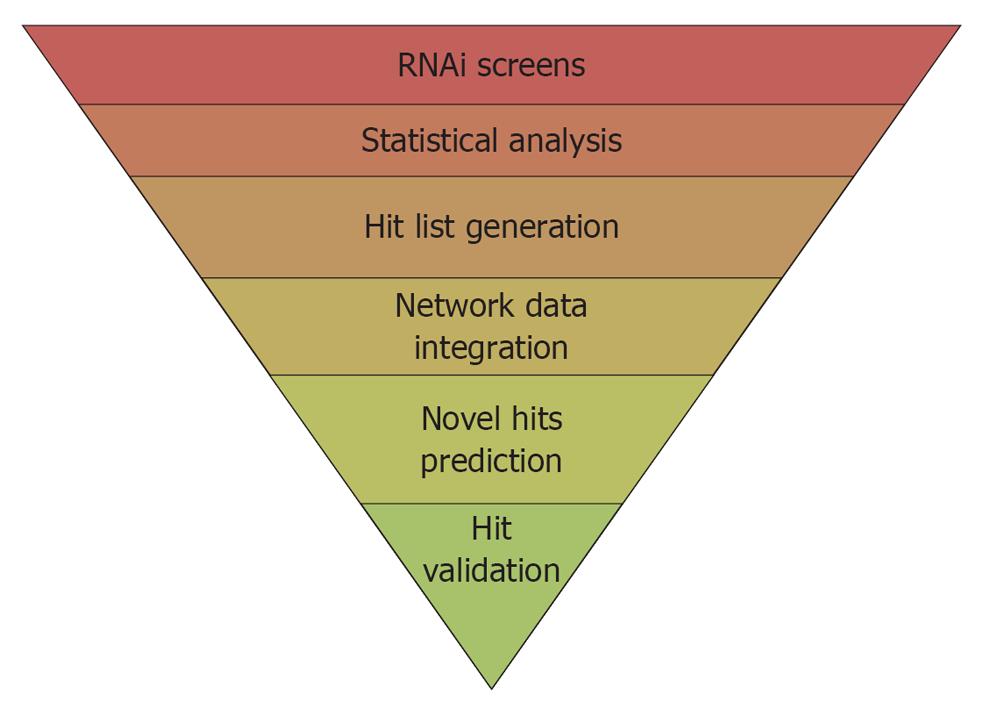
Overall the picture emerging from the above-mentioned studies is that different viruses rely on some common structures, such as the proteasome proteolytic pathway, the spliceosome complex, and the Golgi secretory system. Because of the “housekeeping” nature of the latter processes, these findings, although representing a crucial stating point to understand the molecular biology behind the virus-host cell interaction, might be of limited importance to the identification of anti-viral targets and to the understanding of how specific viruses differentially exploit the cell for their own purposes. However, a few virus specific HCFs have also been identified. Among them, a lipid kinase, the phosphatidylinositol-4-kinase IIIα (PI4K-IIIα) has been identified by several HT-RNAi screens as a crucial factor for HCV replication, in spite of differences in the HCV genotypes used and the experimental setup[20,21,31-33]. A recently proposed model hypothesized that during HCV infection, a viral protein recruits PI4K-IIIα to the sites of viral replication to increase local levels of phosphatidylinositol-4-phosphate, necessary for their integrity of the membraneous replication compartment and hence viral replication[34]. Importantly, a recent study reported that AL-9, a 4-anilino quinazoline specifically inhibiting HCV replication[35], acts a selective inhibitor of PI4K-IIIα[36]. This inhibitor could therefore represent the basis for the development of new-highly needed antiviral compounds to combat HCV infection. The next sections offer a brief overview on how bioinformatics and statistical approaches can overcome most limitations connected with HT-RNAi screens applied to the study the virus-host interaction, resulting in a simple workflow for the analysis of HT-RNAi screens aimed at identifying key host regulators of viral life cycles (Figure 1).
Readout systems for HT RNAi screens are extremely heterogeneous, ranging from bulk readouts of fluorescence reporters to high-content microscopy based assays. Basically any phenotype, either directly or indirectly measured through a reporter, can be used as readout in HT-RNAi screens. However, the main measurement types of cell-based screens in use are: Uniform well readouts: these assays usually use high throughput plate readers to produce their measurements. Absorbance, Luminescence, Fluorescence Intensity, Fluorescent Polarization and Resonance Energy Transfer are the most usual uniform well detection methods[37]; Reporter gene systems: these are mostly high throughput assays using Fluorescence-assisted cell sorting. They employ high throughput FACS to produce readouts of GFP, Luc, etc.[38]; High-Content Imaging Screens: they are designed to identify those genes that alter the cellular phenotype in a desired manner (i.e., decreases in the production of cellular products, nuclear and cellular morphology, proteins subcellular localization, etc.)[37,39].
The selection of the type of assay depends on the goal of screen and practical constrains. The analysis of arrayed screens can involve application of image analysis software or custom programs, as well as various methods of statistical analysis and Bioinformatics (see below). The aim of statistical analysis is to identify “hit” genes that are robust up- and down-regulators of viral replication. Overall, most of the methods currently used for statistical analysis of RNAi screens are reminiscent of those developed in the past for the statistical analysis of cell-based small-molecule screens, with considerable improvement having been implemented in several aspects - including data normalization, replicate tests, and selection of cut-off threshold to determine hits[40]. Cell death and cell clumping are among the most serious problems, which can be directly or indirectly linked to the silencing effect of individual siRNAs[39]. These phenomena can create background or saturation effects in the corresponding wells. In addition, viral infection might induce even more data variation since it can lead to a different cellular behaviour[41]. Errors of unknown origin may also occur over the entire process. These adverse effects can often be minimized by quality control procedures and statistical corrections. Data variation caused by stochastic reasons can be minimized performing additional experimental replicas.
Successful data analysis heavily depends on careful experimental design and assay development prior to the primary screen[42]. Therefore, for example, the optimization of transfection conditions is crucial to the success of experiments. Several factors play important roles in the development of a good assay. The nature of RNAi duplexes to be used (different companies offer RNAi with specific chemical modifications reducing off-target effects), the number of unique individual RNAi duplexes targeting each gene, the number of replicate tests, the number of “no treatment” controls (negative controls), the individual plates layout design (including the placement of negative controls) are the most important factors which should be considered during the experimental design[38,43,44]. Testing of two or more non-overlapping RNAi reagents per gene is nowadays a general standard for primary screens[38]. Once RNAi concentration and transfection conditions have been established, checking the sensitivity to accurately differentiate between positive “hits” and negative controls can be very useful[45]. During this hit selection process, two kinds of decision errors can occur, leading to “false positives” (FPs), experimental findings that cannot be subsequently confirmed, and to “false negatives” (FNs), factors which should have been identified were not. If the assay is not sensitive enough, a high frequency of FNs will be obtained. Conversely, if the readout is too sensitive, a significant number of FPs will be identified. The best way to ascertain the rates of FNs and FPs is to perform a pilot screen. For this purpose, two or more plates fully loaded with positive and negative controls should be used to test the outputs “robustness”[46]. Three measurements are commonly used to this end: signal-to-background ratio, coefficient of variation (CV) and the Z’ factor[45]. As assay variability increases, the signal-to-background ratio must increase for a screen to be successful.
Some candidates identified through a screen might generate the phenotype of interest; however, this might be due to the type of assay used for the readout or to an off-target effect. To overcome such problems, one can use an alternative, or orthogonal, screening procedure. The selected candidate forms the basis for further investigations, for example a secondary screen (also called validation screen). Secondary screens test a much smaller number of compounds (e.g., the 1% strongest hits from the primary screen) and typically use at least duplicate measurements. The magnitude of the statistical artefact can be minimized, e.g., by obtaining replicate measurements, and thus improving precision of the overall estimate. The assumptions that RNAi duplexes targeting specific genes randomly plated and the most of them do not have an effect on viral replication for secondary screens are not valid. Below, we present a sample workflow for analyzing RNAi screens.
The goal of HT-RNAi screens is normally to identify “hits”. To this aim, it is of crucial importance to separate FPs from bona fide “hits”. This is largely related to the quality of the assay used. It is therefore necessary to monitor each step, checking the quality of raw and normalized data. To increase the probability of success, quality assessment should be performed while the screen is in progress, and also after each step of the analysis pipeline, thus allowing the detection of potential issues as they occur. This will also help with the choice of analysis methods. In case of a failed quality control for individual wells or plates, these should be either removed from further analysis or repeated.
In biological experiments the use of controls - positive and negative - helps to assess the quality of obtained data. Negative controls can be used to assess plate-to-plate variability, and provide a means to measure background noise levels of an assay. Positive controls provide an estimate of expected effect strengths, and are used to establish if effects are observed at all, and if they are of the expected strength. Controls allow the calculation of several different quality metric such as signal to noise ratios, the dynamic range[47], CV or the Z’ factor. In contrast to simple signal/noise ratio, the dynamic range and the Z’ factor calculate the separability of positive and negative controls and use this criterion to evaluate assay quality. CV measures the data quality based on the reproducibility of results. In contrast to dynamic range and the Z’ factor, CV does not use controls and can be used in case that controls are not available or they did not work properly in some screens or at least on individual plates. Calculating the correlation among replicates by correlation measures such as Pearson’s correlation or Spearman’s rank also can be used to check the reproducibility and reliability of the data.
Apart from quality metric, plate visualization is one of the most effective techniques to find systematic sources of error or identify data with poor reproducibility due to suboptimal assay design or implementation[37,39,42,43,48,49]. Heat maps and plate-well scatter plots, which allow to display the overall screen performance, as well as replicate correlation plots to visualize overall reproducibility, are the most widely used methods used for plate visualization[43,48]. Box Plots of readouts of all plates can be used to detect systematic errors among the plates.
When RNAi duplexes are randomized between plates and experiments are performed under identical conditions, the box plots of raw data should show approximately the same location and scale. However, it is possible due to systematic variability that some of the plates have lower (higher) median intensities than the others, resulting in considerably higher (lower) hit rates on these plates. This can be the consequence of pipetting issues resulting in altered transfection or infection efficiencies: such deviations can be adjusted by normalization. For more details about the individual plots and their interpretation, please refer to[47,48,50,51]. Finally, wells with lowest and highest 1%-5% of cell counts in the entire screen are sometimes excluded from further analysis in particular in the case of image based screens, because of potential interference with viral replication readout and errors in image segmentation when cells are very dens[39].
Readout of each spot in a plate is a function of at least two factors: the siRNA’s real activity and random error. There are many sources of systematic errors (variations) that affect readouts of HT-RNAi screens. The ability of combination and comparison of all of the plates in a production run to each other is very important. Systematic errors can cause a high degree of intra-plate and plate-to-plate variability, which does not allow comparison and combination of data from different plates. Data normalization is a process intended to remove such variation from the data to allow comparison and combination of data from different plates of the screen. Intra-plate spatial effects and correlation between cell numbers and signal intensity are the most important sources of systematic errors[40]. A number of normalization methods have been developed to address these issues[42,47]. Normalization is generally performed at two levels: per-plate and per-experiment (intra- and between-plates normalization).
The per-plate (intra-plate) normalization aims at reducing systematic errors on individual plates, such as differing cell numbers over the plate, or edge-effects affecting the signal intensities. This can be the consequence of pipetting issues resulting in altered transfection or infection efficiencies, as well as of evaporation of media from the outer wells. The per-experiment (between-plate) normalization removes systematic bias that occurs between different plates. This bias might be due to measurements performed using diverse microscope settings or under different environmental situations such as different levels of humidity. Since data is varying across specific experimental setups, a standard normalization strategy that is appropriate for all of them does not exist. For example, normalization methods for primary and validation screens are different, due to the method of selection and distribution of the RNAi duplexes. Normalization methods can be categorized into two main groups: control-based and sample-based. Control-based normalization methods compare individual experimental sample values to aggregated values of negative controls, while sample-based method use the samples themselves as de facto negative controls[42]. The latter choice can provide more accurate measurements, because on each plate the number of experimental samples exceeds that of the negative controls by several folds. This approach is based on the assumption that most experimental samples will not display a biological effect in the assay being analyzed. Obviously, this assumption is not valid in the case of validation screens and therefore sample-based normalization methods should not be used in the case of validation screens. In this case, plates are made comparable by control-based normalization method. Additionally, the use of sample-based normalization methods is particularly problematic when dealing with statistical measures (such as mean and standard deviation) that are strongly sensitive to outliers in the data.
Including controls on every individual plate can help identifying plate-to-plate variability and establishing background levels of an assay. One common approach to for plate-to-plate normalization is to scale the intensity values based on the controls. Whether for the normalization the negative or the positive controls shall be chosen, it depends on the type of experiment. For RNAi data, negative controls are used in most cases. It should be noted that in this approach, any inaccuracies and random measurement errors in controls would lower the accuracy and precision of the normalized values through error propagation. Therefore, it is important to obtain as accurate and precise measurements as possible. Using a relatively large number of control measurements and omitting outliers among the controls before normalizing can improve the quality of normalized values.
In this approach, the mean or median of the controls of a plate is subtracted from the readout value of each spot in the same plate and the result is divided by the controls standard deviation or median absolute deviation (MAD).
As mentioned before, under the assumption that most siRNAs in plate would not cause an effect, it is possible to use experimental samples as controls. Z-score normalization is a well-known data scaling strategy, which uses this assumption. For each spot, the Z-score is defined as the number of standard deviations from the mean of the samples on the plate.
The readout of each spot rescaled relative to intra-plate variation by subtracting the average of the plate values and dividing the difference by the standard deviation estimated from all measurements of the plate. In this approach, the mean of all the samples on the plate is used instead of that of the negative controls, thus limiting the need for large numbers of controls. Z-score gives explicit information on the strength of each siRNA relative to the rest of the sample distribution. An advantage of Z-score is that it integrates information about the variability of replicate measurements in the score. The main disadvantage of this method is its non-robustness to outliers, that can strongly affect estimates of the mean and standard deviation used in the Z-score.
A modified version of it called the robust Z-score, is generally considered preferable for the analysis of HT-RNAi screens. It uses the median and MAD for mean and standard deviation in the Z-score calculation.
The B-score is known as a robust analogue of the Z-score. It is more robust to the presence of outliers, and also differences in the measurement error distributions of the different spots on a plate. If the quality control has identified the presence of within-plate systematic errors, the B-score normalization[52] may be applied to remove row and column effects within a single plate. The systematic measurement offsets for each row and column, row and column effects, is estimated using the Tukey median polish method. The resulting residuals within each plate are then divided by their MAD to standardize for plate-to-plate variability. This thus allows the comparison of different plates, since it scales the data according to the overall plate median. The B-score has three advantages: it is nonparametric (that is, it makes minimal distributional assumptions), it minimizes measurement bias due to positional effects, and it is resistant to statistical outliers[47].
Lowess (locally weighted least squares regression) normalization performs intra-plate corrections. If RNAi data is multi-parametric, different read-outs may depend on each other and these can cause a systematic bias. Lowess regression is a technique for fitting a smoothing curve to a data. Data points that are nearer to the estimated fit are weighted higher than more distant points. The degree of smoothing is determined by the window width parameter. A larger window width results in a smoother curve, a smaller window results in more local variation. The normalized signal intensities are the difference of the signal intensity values and the corresponding point on this curve[51]. For example, Lowess normalization can be applied to remove the correlation between signal intensities and cell count by adjusting the signal intensities for the effect of unequal cell numbers in wells. This should be done for each plate individually, since effects may be different from plate to plate[39].
Very recently it was shown that different cells in a population display heterogeneity in their cellular behaviours[53,54]. This heterogeneity implies that cellular responses to a particular stimulus or perturbation, such as virus uptake, may also be variable[41]. For example certain viruses prefer to infect cells that are in a less dense region, others preferentially infect densely packed cells[54]. Therefore analyzing certain phenotypes at the single-cell level instead of using population averages to measure an effect might completely change the results. Snijder et al[54-56] showed that the population context of a cell strongly affects its behaviour: factors such local cells density, their position within an islet, size, distance from cell-colony edges and population size are the main determinants of cell to cell variability in HT-RNAi screening. To address this issue they suggested normalizing data by considering the population context. They corrected population context effects using quantile multidimensional bin models[55]. Knapp et al[57] used a similar approach in normalizing data but they developed a statistical testing procedure that takes into account individual cell measurements in hit-scoring. They used gene set enrichment analysis (EA) on sets defined not by genes but by cells coming from one spot, one siRNA or one gene. These approaches suggest that normalization for population context can lead to a substantial decrease in experimental variability, and may to some extent underlie the low gene overlap and lack of reproducibility of RNAi screens targeting even the same virus.
Once data have been pre-processed with quality control checks and normalization procedures, the next critical step is the hit identification procedure to decide which siRNAs should be further tested in a secondary screen. The identification of “hits” or “screening positives” is the goal of any primary RNAi screen. Hit identification is, essentially, the selection process of those samples whose measured values for a given phenotype differ significantly from that of the negative controls[52]. A wide range of hit identification techniques is available. Hits can be identified as a percentage of the genes that generate the highest measured activity (e.g., top 1%), or as those whose activity exceeds a fixed “percent of control” threshold. Alternatively, the hit threshold may be defined as a number of standard deviations (typically 2) beyond the mean of the raw or processed data. This approach selects a standard deviation threshold relative to the mean or median normalized data and defines “hits” the samples that go beyond this threshold. However, hits (outliers) may cause the distribution of the siRNAs measurements to be skewed. The use of the median rather than of the mean is more robust to outliers, and has been shown to more effectively enable the identification of weak hits from RNAi data[58].
The threshold methods assume a common magnitude of random error for all measurements, but do not capture data variability effectively. To address this issue, researchers then turned to the Z-score method or strictly standardized mean difference (SSMD)[59], which can capture data variability in negative controls. According to the Z-score method, any compound whose score after Z-score normalization deviates from the bulk by a given threshold will be considered as hit. The Z-score method is based on the assumption that the measured values (usually fluorescent intensity in log scale) of all investigated siRNAs in a plate have a normal distribution. SSMD also works the best under the normality assumption. The drawback common to all of these metrics is that they rely on non-robust statistics, which may lead to inferential errors in hit detection. Because of the potential existence of true hits and strong assay artefacts, outliers are not uncommon in HT screens. The regular versions of Z-score and SSMD are sensitive to outliers[59]. In general, there are two major types of approaches for hit selection: analytic metrics and hypothesis testing. The methods belonging to the first approach (such as fold change, mean difference, SSMD, percent activity, percent viability and percent inhibition) assess and rank the size of RNAi effects, while the methods belonging to the second group (for instance t-test) test the null hypothesis that no difference exists among the means of particular well and negative controls or mean of plate[48,52,59,60]. If enough replicates are available, a statistical approach can be applied to assign a P value to each condition. If the P value is smaller than a given significance level, the null hypothesis can be rejected. A common practice is to use the t-test. It is a parametric testing method (assuming normally distributed data), which assesses the difference of the means between replicates for each condition.
If siRNA duplexes are randomly distributed on a plate and if it can be assumed that most of them have no effect, replicates in the test can be compared with the overall population. If this assumption is not valid, e.g., in a validation screen, the test is carried out against negative controls. This approach needs at least three replicates of each condition and that data follows a normal distribution[61]. In case of non-normal distribution, the Mann-Whitney test can be used as non-parametric test[39].
The methods for hit selection differ according to the experimental setup of the HT-RNAi screen, depending on the fact that replicates have been performed or not. For example, the Z-score method is suitable for screens where replicates have not been performed, whereas the t-test is suitable for screens where three or more replicates have been performed. It is not possible to directly estimate the data variability for each siRNA in screens without replication. Instead, it is indirectly possible to estimate data variability by making the assumption that every siRNA has the same variability as a negative reference in a plate in the screen. The Z-score, the B-score and the SSMD rely on this strong assumption for cases without replicates[62].
A typical outcome of any statistical analysis of a genome- wide HT-RNAi knockdown screen is a list of gene products that significantly differ from other genes in the same study, relatively to a given readout. Classically, these lists are then subjected to over-representation analysis (ORA) or EA over different known pathway datasets such as KEGG, Reactome, Wikipathways and gene ontologies (GO), in order to facilitate interpretation of the hits functional importance. A major caveat in such analyses is the fact that the datasets used for such analyses are far from being complete. Inconsistencies and lack of concurrency between these pathway databases reduces their reliability, thus hampering the coverage of ORA/EA. This problem is particularly evident in the case of HT-RNAi screens concerning the same virus[17-19], where the overlaps over these ontologies are minimal[30]. In order to overcome this problem, network approaches have been implemented to analyze HT-RNAi screens. This section describes studies that exemplify the usage of PPI network data for analyzing RNAi screens.
With the wealth of public repositories housing PPI data, and exponentially growing computational power to analyze such data, the need to integrate the outcome of HT-RNAi screens with PPI data is pressing. Protein interactions between viral and host proteins are a subset of this data type that can be created by combining previously published and experimentally newly identified interactions. VirHostNet[63], VirusMINT[64] and the HIV-1 Human Protein Interaction Database (HHPID) at National Institute of Allergy and Infectious Diseases (NIAID)[65] are examples of such resources.
A host of analysis pipelines has been developed to integrate PPI data with the HT-RNAi screens hits, which can be applied to add depth and significance to latter results. An example is the SinkSource algorithm described in a recent study[66]. In the latter, the authors used a semi-supervised machine learning approach to predict novel HIV-1 HDFs using known HDFs. In other words, by combining HDFs identified from recent studies[17-19] and PPI data, the authors developed a classification algorithm that would learn from the known HDFs in a network context to then predict novel ones. The host PPI network is modeled as a liquid flow network. Each node (protein) is a reservoir of fluid while an edge (connection between 2 nodes) is a pipe. The weight of an edge indicates the amount of fluid that can flow through the pipes per unit time. When the fluid network attains equilibrium (amount of liquid flowing into each node equals amount flowing out), the reservoir height at each node denotes the confidence that the node is a HDF. HDFs identified in three previously published HIV screens[17-19] were assigned a reservoir level at of 1 unit while non-HDFs nodes had a reservoir level of 0. This algorithm is similar to the one formulated in a previous study[67] for functional prediction of genes, except that SinkSource also accepts negative values in the form of non-HDFs which are non-lethal. These non-lethal, non-HDFs formed the negative set while HDFs identified in the three studies and their intersection formed the 4 positive datasets used for prediction of novel HDFs through a two-fold cross validation. The latter involved splitting of both the positive and negative datasets in halves and each half was used for prediction of the genes in the other half. SinkSource had higher specificity and precision-recall values when compared to six similar algorithms (used for functional gene prediction). SinkSource predicted 1394 HDFs in addition to the 908 from the above three screens, with an accuracy > 80% based on two-fold cross validation described earlier. After combining the known HDFs with those predicted by their algorithm, the authors then searched for dense subgraphs in an integrated protein network through MCODE[68]. Using this approach, they identified cellular processes and components essential for HIV replication. These included, as far as the GO cellular component are concerned: spliceosome, kinetochore and mitochondrion, whereas GTPase mediated signal transduction, DNA replication initiation and MHC protein complex were identified as enriched cellular processes.
Another example of a network based analysis between PPI and HT-RNAi data is from MacPherson et al[69]. In this study, the authors utilized the HIV-1 HHPID at NIAID[65] and applied a bi-clustering algorithm to identify clusters of genes enriched for HIV-1-Human PPIs. In order to establish a hierarchical overview of functions from the clusters, they were further linked to form a cladogram. The distance between 2 clusters was based on the number of overlaps between them; clusters with more overlaps were closer to each other than the ones with fewer overlaps. GO enrichment of these clusters then defined the cluster function and in turn allowed identification of 37 host subsystems potentially important for HIV-1 infection. Interestingly, hits previously identified in three published HIV HT-RNAi screens[17-19] were found in 10 of the 27 subsystems identified. These included proteasome core complex, regulation of apoptosis, mRNA transport, endosome, RNA polymerase activity, peptidase activity, regulation of transcription, ubiquitin camp-dependent protein kinase complex, and v-akt. Classically, the virus-host interaction dataset used in this study would only provide information about how a viral protein interacts with a host protein and its mechanism, as extracted from literature. In this study, the authors showed how a viral protein interacts with host cellular systems in contrast to a single protein. A theoretical validation was provided by highlighting systems enriched with hits from the 3 HIV-1 RNAi screens[17-19].
A very important consequence of viral-based RNAi screens could be the discovery of new potential targets for the development of anti-viral agents. Over the years, repositories holding detailed information on various drugs, including their cellular targets, have been publicly made available. de Chassey et al[70] used the DrugBank database, one of such resources (http://www.drugbank.ca) to identify potential drug targets for INF. By combining results from 6 different IFV HT-RNAi screens, the authors identified 925 essential host factors (EHFs), required for IFV replication[14-16,71-73]. Network analysis performed integrating these data with the PPI dataset from VirHostNet[63] revealed that 17 EHFs are directly targeted by a viral protein while the neighborhood of EHFs (proteins physically interacting with EHFs) included 204 proteins that were targeted by at least one viral protein.
In parallel, known drug molecules interacting with EHFs were further retrieved from DrugBank. This analysis revealed that 100 EHFs could be targeted by 298 different molecules comprised of 204 FDA-approved drugs and 94 experimental drugs. These 100 EHFs were further filtered down to 33 proteins, based on their ability to fulfill at least one of the following three criteria: the EHF was directly targeted by a viral protein, the EHF was connected to at least another EHF, and the EHF was connected to a non-EHF targeted by the virus. Of these, 32 EHFs could be targeted by 49 FDA approved molecules with an exception of one target, HSP90AA1, fulfilling the first 2 criteria mentioned above, is also directly interacting with a viral protein. Interestingly, the authors found that this EHF is the target of 1 FDA-approved molecule (Ribafutin) and 5 experimental molecules. Among the latter, Geldanamycin has already been proved to reduce IFV viral replication by 2 logs in cell culture[74]. Therefore the authors concluded that combination of Geldanamycin with Ribafutin (which is also used as a first line of treatment in tuberculosis) could represent a novel strategy to identify antiviral drugs to combat IFV infection.
As with any high-throughput study, the issue of false-positives and false-negatives also exist for HT-RNAi screens. FPs due to off-target effect of RNAi are common in genome-wide screens and confer ambiguity to the final hit-list selected from a screen. Hence, it is recommended to perform a multi-level validation and a functional analysis for hit genes. Even more critical and tricky is the issue of FNsof a screen. These are typically genes that have an effect but are missed due to the statistical selection criteria. Wang et al[75] addressed these problems by developing an algorithm based on machine learning principles, utilizing protein interaction data and network topology. Considering network centralities such as direct neighbour, shortest path, diffusion kernel and association analysis-based transformation[76] along with gene similarities, they developed a set of scoring functions called Network RNAi Phenotype (NePhe). Utilizing the guilt-by-association principle, Wang et al[75], reasoned that FNs would be scored higher by a scoring function over false-positives FPs, as they would be linked by a greater number of true hits. Thus, a near-ideal gene classifier would always rank FNs with a higher rank compared to a non-hit. When this methodology was applied over the Wnt and the Hedgehog signaling pathways, the NePhe scoring system was able to identify all regulators, which were missed even by the follow-up validation screens. This algorithm was tested on 24 screens to study different molecular aspects of the fruit fly, Drosophila. Its efficacy in recovering FN in screens devoted to identifying viral HDFs in human systems is yet to be determined.
In general, these studies highlighted how using virus-host PPI databases to integrate the outcome of HT-RNAi data can maximize the relevance of the latter results, reducing FPs and FNs, increasing number of HDFs identified and eventually lead to identification of new drugs to combat viral infection.
A minor shortcoming of these studies has been that they have been largely biased towards a particular virus. For instance, all studies from[66,77-79] have been based on HIV screens while there has been only one meta-analysis on IFV screens[70]. It would be worthwhile and interesting for the community to see these approaches applied to other viruses. With the availability of several HT-RNAi screens for different viruses, a multi species meta-analysis can highlight similarities and differences between host-virus interactions, based on RNAi screen hits. Therefore, much effort still needs to be done to perform reliable HT-RNAi hits for a large number of viruses, including those such as hepatitis B virus, for which a reliable cell culture system is still not available.
A complementary approach to integrate HT-RNAi with PPI datasets is to perform Network analyses. Particularly it is possible to characterize viral HDFs by computing several topological measures (network centralities) in addition to their biological function. These properties form the basis for interpreting the role of such hits from a network perspective. Furthermore, these scores also allow for a different level of hit prioritization for subsequent analysis. As mentioned earlier, specialized repositories host-pathogen interaction databases such as VirusMint, HIV-1-human protein interaction database, Host-Pathogen Interaction Database, VirHostNet, PHIDIAS, etc.[63-65,70,80,81] have fuelled these studies and shed new light on host-pathogen interactions. This section summarizes results from such studies and provides an overview of topological properties of viral HDFs.
In this context, the most comprehensive study has been recently published by Dyer et al[82]. The authors highlighted properties of host factors involved in the life cycle of 190 different pathogens from a network perspective. To this end, they collated experimentally identified human PPIs for 190 pathogen strains partitioned into 54 groups (35 viral, 17 bacterial, and two protozoan) pooled from 7 public databases[62,68,83-87], to determine properties of proteins targeted by most pathogens, including viruses. The main conclusion from this study was that pathogens preferentially target bottlenecks and hubs, implying that targeting central proteins is a common strategy shared by different pathogens. This study revealed that viral targeted host proteins also play a major role in different cancers of which some are induced by a viral infection itself (e.g., Herpesvirus and Papillomavirus). Gulbahce et al[88] showed that the neighborhood of HDFs is as important as the HDFs themselves. They formulated what they term as “local impact hypothesis” wherein they propose that genes associated with virally implicated diseases are located in the neighborhood of viral targets. They tested their hypothesis by calculating the mean shortest path between genes that are viral targets and the ones implicated in a viral disease. This mean value was significantly shorter than between random samples. Scanning for genes within these path lengths, and subsequent experimental validation in human keratinocyte populations for HPV16 expressing E6, E7 proteins revealed 104 genes regulated by the 15 targets of E6 and E7 (these genes were 2 connections or paths away from these 15 targets). Of these 104 genes, 22 were also differentially expressed in IMR90 cells expressing HPV16 E6 or E7 proteins. A novel link was predicted between HPV and Fanconi anemia, through the E6->TP53->FANCC pathway through the FANCC gene which was one of the 22 genes described above.
Similar studies have been conducted recently with a specific focus towards HIV-1 host protein interactions. van Dijk et al[78] analyzed the HIV-1-Human protein interaction data and also highlighted the fact that viral products preferentially interact with host proteins that represent hubs or bottlenecks. Furthermore, they also determined enriched network motifs, statistically significant patterns of interacting proteins, from this network that allowed dynamic interpretation of interactions. For example, one of the enriched motifs included the 2 nodes feedback loop found in the HIV-host activation/inhibition network. This suggests the inhibitory nature of HIV proteins on human proteins that in turn inhibit the HIV protein. This motif occurred mostly for HIV Tat and Gp120 proteins with the human interferon γ. GO enrichment of the observed network motif indicated that it is involved in immune response.
Another independent study, performed using the same HIV-1-Human protein interaction dataset, reiterated that HIV-1 proteins attack hubs and bottlenecks over others[77]. By implementing an ascertainment bias, that normalizes weightage given to the genes based on publication count in order to avoid false interpretations[89], the authors came to two striking conclusions. First, HIV-1 interacting proteins and gene essentiality didn’t have a strong correlation. Secondly, HIV-1 interacting proteins didn’t tend to be disease-associated. Still, GO enrichment analyses of HIV-1 interacting proteins suggested that proviral and antiviral interactions are highly complex.
These studies thus have further enhanced our knowledge of the intricacies involved in HIV-1 infection, and opened new doors for the development of novel hypotheses.
Similar studies have been performed to experimentally determine the virus-host interactome of HCV, DENV and HTLV-1/2[90-92]. More recently a comprehensive study focused on determining the interactome of 70 viral modulators of the innate immune response from 30 different viruses[93].
The common outcome of these studies is that viral proteins have the remarkable tendency to have significantly more targets, to be more central to the networks, to participate in more cellular pathways and are more likely to hold key positions in these pathways, as compared to an average human protein. On the same lines, both experimental and computational approaches helped identifying some common features of HDFs. These proteins have higher values of degree and of betweenness, which imply that viral proteins preferentially target proteins that are “central” to a given network. Smaller mean path length values of the HDFs, relative to the whole network also indicate that viral proteins target subnetworks that are “closely bound”. Future studies in this direction might delve a bit deeper to uncover more topological features beyond what is already known.
Indeed, given the limited size of their genomes, viral products are required to interact with a high number of host proteins, which usually represent key factors regulating several biological processes. Morever these approaches can also help us to identify new HDFs: using these topological features, computational algorithms can be formulated to predict potential “generic” HDFs. For e.g., PageRank centrality is one such feature. It is utilized by Google in order to decide the rank of the search hits. In the simplest sense, a node’s importance is determined by the importance of its neighbors. Thus, the more “important” a node is topologically, the more it might also be biologically important, and therefore likely to be the target of viruses to overtake cellular functions. Jaeger et al[79], used this centrality measure to identify 21 surface membrane proteins critical for HIV-1 infection of which 11 are novel predictions, 3 are confirmed hits (chemokine receptor CCR1, chemokine binding protein 2 and duffy antigen chemokine receptor) and 7 have been confirmed in other studies. These receptors are potentially involved in different phases of HIV infection and influence progression of AIDS.
Degree, betweenness, pagerank and shortest-paths are just few of the many network centralities that have been defined to date for HDFs. It would be interesting to compute some additional measures to characterize HDFs. Quantifying structural properties of viral HDFs can help researchers in developing efficient machine learning algorithms to predict novel HDFs with greater accuracy. In addition to such predictions, a further, crucial layer of analysis would be to check for mouse orthologs of such predicted HDFs and verify if they are lethal for mouse. This step allows filtering of candidate HDFs, to be used for secondary validation, which can produce a lethal phenotype. Specificity and tissue localization of these HDFs can then be determined by utilizing tissue specific expression data from Protein Atlas (http://www.proteinatlas.org/)[94]. These steps would give a comprehensive overview of all HDFs beyond function and thus would aid in hypothesizing regulatory mechanisms and interactions between viral proteins and HDFs. Moreover, this would also reduce time, effort and cost of experimentalists and would serve as a guide to a more directed approach for hit validation.
All the above mentioned studies, both those considering RNAi hits and those which do not, strongly underlined the importance of inclusion of PPI network information to propose better hypotheses as well as therapeutic targets. They also highlighted the fact that for increasing reliability and confidence in HT-RNAi screens, validation by computational approaches via multiple data-types and sources is as important as verification with biological assays. Indeed, combination of data generated by different screens performed using the same virus, has evidently shown to strengthen the statistical significance of hits and reduce FP. The upcoming virus-host interaction databases, together with the availability of expression data and powerful, public tools for integrating and analyzing HT-RNAi screens will undoubtedly provide a comprehensive understanding of virus-host interactions at a cellular level.
Despite the remarkable efforts done so far to apply the use of HT-RNAi screening approaches to the study of the host cell-virus relationship, a great body of work is still required before we reach a comprehensive overview of how different viruses selectively exploit the host cell. This will finally lead to the design of specific anti-viral compounds targeting host cell functions, which are therefore less prone to the selection of drug resistant viral strains. This process is strongly limited by the high number of different human pathogenic viruses, and that identification of HCFs required for viral replication necessarily relies on the availability of robust in vitro systems to propagate such viruses. Unfortunately, despite the tremendous advances made in the field, for example with the development of systems to propagate HCV[95-97], we are still lacking a system to efficiently propagate in vitro other important human pathogens (the most striking example being exemplified by Hepatitis B Virus, responsible for approximately 600 thousand causalities each year[98].
Beside this crucial shortcoming, it seems that the initial concerns related to the specificity and sensitivity of the HT-RNAi technology can be solved by combining data from different independent screens performed for the same virus, and by implementing sophisticated statistical algorithms that take into account differences within a cell population - an approach that have been proposed to strongly limit variance[54-56], as well as integrating HT-RNAi data with PPI datasets. In particular, the latter approach has been successfully used to reduce the number of FNs and FPs[75], to identify new HDFs[66], and also to identify new potential drug targets for treatment of viral infection[70]. Another major benefit of such integrative approaches relies in the possibility to perform network analysis of host factors and PPI datasets[77,82], thus enabling study the connections of viral products and the cellular effectors that are directly targeted by their action.
A third crucial point which needs to be considered is the growing need for in depth biochemical and biological characterization of the newly described hits. Indeed it is important not only to know the name and the molecular function of HDFs, but also the reason why exactly these factors are required for the life cycle of a given virus, for example, by enabling the formation of its replication compartments, or by being incorporated into the mature virion to mediate later on the recognition of a cellular receptor, to cite just a couple of examples of two well characterized viral HDFs for HCV, namely PIK4α and ApoE[34,60,99]. This knowledge enables at the same time to understand more in detail the mechanisms behind the usurpation of the host cell by viruses and to devise strategies to prevent this process.
In summary, progress still needs to be done in three directions before a complete understanding of the virus-host interplay: Development of appropriate cell culture systems to enable in vitro culture of human pathogenic viruses and their use to perform HT-RNAi screens, which should be rigorously analyzed by statistical analysis methods. Integration of data generated in different studies using the same virus, with other datasets, such as those deposited in PPI databases, to maximize sensitivity and specificity of the results. In depth characterization of identified hits of major relevance, including potential targets for the development of anti-viral drugs.
P- Reviewer Melero JA S- Editor Gou SX L- Editor A E- Editor Zheng XM
| 1. | Pond SL, Murrell B, Poon AF. Evolution of viral genomes: interplay between selection, recombination, and other forces. Methods Mol Biol. 2012;856:239-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 2. | Wang KS, Choo OL, Weiner AJ, Ou JH, Denniston KJ, Gerin JL, Houghton M. The viroid-like structure of the hepatitis delta (delta) genome: synthesis of a viral antigen in recombinant bacteria. Prog Clin Biol Res. 1987;234:71-82. [PubMed] |
| 3. | Lange CM, Zeuzem S. Diacylglycerol acyltransferase-1: a critical host factor for hepatitis C virus assembly and potential new drug target. Gastroenterology. 2011;140:1345-1347. [PubMed] |
| 4. | Chiu YL, Cao H, Jacque JM, Stevenson M, Rana TM. Inhibition of human immunodeficiency virus type 1 replication by RNA interference directed against human transcription elongation factor P-TEFb (CDK9/CyclinT1). J Virol. 2004;78:2517-2529. [PubMed] |
| 5. | Johnson RA, Wang X, Ma XL, Huong SM, Huang ES. Human cytomegalovirus up-regulates the phosphatidylinositol 3-kinase (PI3-K) pathway: inhibition of PI3-K activity inhibits viral replication and virus-induced signaling. J Virol. 2001;75:6022-6032. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 185] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 6. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10136] [Article Influence: 375.4] [Reference Citation Analysis (1)] |
| 7. | Timmons L, Fire A. Specific interference by ingested dsRNA. Nature. 1998;395:854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1427] [Cited by in RCA: 1379] [Article Influence: 51.1] [Reference Citation Analysis (0)] |
| 8. | Kittler R, Putz G, Pelletier L, Poser I, Heninger AK, Drechsel D, Fischer S, Konstantinova I, Habermann B, Grabner H. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432:1036-1040. [PubMed] |
| 9. | Furlong EE. A functional genomics approach to identify new regulators of Wnt signaling. Dev Cell. 2005;8:624-626. [PubMed] |
| 10. | Müller P, Kuttenkeuler D, Gesellchen V, Zeidler MP, Boutros M. Identification of JAK/STAT signalling components by genome-wide RNA interference. Nature. 2005;436:871-875. [PubMed] |
| 11. | Friedman A, Perrimon N. A functional RNAi screen for regulators of receptor tyrosine kinase and ERK signalling. Nature. 2006;444:230-234. [PubMed] |
| 12. | Yi CH, Sogah DK, Boyce M, Degterev A, Christofferson DE, Yuan J. A genome-wide RNAi screen reveals multiple regulators of caspase activation. J Cell Biol. 2007;179:619-626. [PubMed] |
| 13. | Chen J, Shi X, Padmanabhan R, Wang Q, Wu Z, Stevenson SC, Hild M, Garza D, Li H. Identification of novel modulators of mitochondrial function by a genome-wide RNAi screen in Drosophila melanogaster. Genome Res. 2008;18:123-136. [PubMed] |
| 14. | Karlas A, Machuy N, Shin Y, Pleissner KP, Artarini A, Heuer D, Becker D, Khalil H, Ogilvie LA, Hess S. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature. 2010;463:818-822. [PubMed] |
| 15. | König R, Stertz S, Zhou Y, Inoue A, Hoffmann HH, Bhattacharyya S, Alamares JG, Tscherne DM, Ortigoza MB, Liang Y. Human host factors required for influenza virus replication. Nature. 2010;463:813-817. [PubMed] |
| 16. | Hao L, Sakurai A, Watanabe T, Sorensen E, Nidom CA, Newton MA, Ahlquist P, Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature. 2008;454:890-893. [PubMed] |
| 17. | Brass AL, Dykxhoorn DM, Benita Y, Yan N, Engelman A, Xavier RJ, Lieberman J, Elledge SJ. Identification of host proteins required for HIV infection through a functional genomic screen. Science. 2008;319:921-926. [PubMed] |
| 18. | König R, Zhou Y, Elleder D, Diamond TL, Bonamy GM, Irelan JT, Chiang CY, Tu BP, De Jesus PD, Lilley CE. Global analysis of host-pathogen interactions that regulate early-stage HIV-1 replication. Cell. 2008;135:49-60. [PubMed] |
| 19. | Zhou H, Xu M, Huang Q, Gates AT, Zhang XD, Castle JC, Stec E, Ferrer M, Strulovici B, Hazuda DJ. Genome-scale RNAi screen for host factors required for HIV replication. Cell Host Microbe. 2008;4:495-504. [PubMed] |
| 20. | Li Q, Brass AL, Ng A, Hu Z, Xavier RJ, Liang TJ, Elledge SJ. A genome-wide genetic screen for host factors required for hepatitis C virus propagation. Proc Natl Acad Sci USA. 2009;106:16410-16415. [PubMed] |
| 21. | Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5:298-307. [PubMed] |
| 22. | Cherry S, Doukas T, Armknecht S, Whelan S, Wang H, Sarnow P, Perrimon N. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 2005;19:445-452. [PubMed] |
| 23. | Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF, George RA, Lewis SE, Richards S, Ashburner M, Henderson SN, Sutton GG, Wortman JR, Yandell MD, Zhang Q, Chen LX, Brandon RC, Rogers YH, Blazej RG, Champe M, Pfeiffer BD, Wan KH, Doyle C, Baxter EG, Helt G, Nelson CR, Gabor GL, Abril JF, Agbayani A, An HJ, Andrews-Pfannkoch C, Baldwin D, Ballew RM, Basu A, Baxendale J, Bayraktaroglu L, Beasley EM, Beeson KY, Benos PV, Berman BP, Bhandari D, Bolshakov S, Borkova D, Botchan MR, Bouck J, Brokstein P, Brottier P, Burtis KC, Busam DA, Butler H, Cadieu E, Center A, Chandra I, Cherry JM, Cawley S, Dahlke C, Davenport LB, Davies P, de Pablos B, Delcher A, Deng Z, Mays AD, Dew I, Dietz SM, Dodson K, Doup LE, Downes M, Dugan-Rocha S, Dunkov BC, Dunn P, Durbin KJ, Evangelista CC, Ferraz C, Ferriera S, Fleischmann W, Fosler C, Gabrielian AE, Garg NS, Gelbart WM, Glasser K, Glodek A, Gong F, Gorrell JH, Gu Z, Guan P, Harris M, Harris NL, Harvey D, Heiman TJ, Hernandez JR, Houck J, Hostin D, Houston KA, Howland TJ, Wei MH, Ibegwam C, Jalali M, Kalush F, Karpen GH, Ke Z, Kennison JA, Ketchum KA, Kimmel BE, Kodira CD, Kraft C, Kravitz S, Kulp D, Lai Z, Lasko P, Lei Y, Levitsky AA, Li J, Li Z, Liang Y, Lin X, Liu X, Mattei B, McIntosh TC, McLeod MP, McPherson D, Merkulov G, Milshina NV, Mobarry C, Morris J, Moshrefi A, Mount SM, Moy M, Murphy B, Murphy L, Muzny DM, Nelson DL, Nelson DR, Nelson KA, Nixon K, Nusskern DR, Pacleb JM, Palazzolo M, Pittman GS, Pan S, Pollard J, Puri V, Reese MG, Reinert K, Remington K, Saunders RD, Scheeler F, Shen H, Shue BC, Sidén-Kiamos I, Simpson M, Skupski MP, Smith T, Spier E, Spradling AC, Stapleton M, Strong R, Sun E, Svirskas R, Tector C, Turner R, Venter E, Wang AH, Wang X, Wang ZY, Wassarman DA, Weinstock GM, Weissenbach J, Williams SM, WoodageT KC, Wu D, Yang S, Yao QA, Ye J, Yeh RF, Zaveri JS, Zhan M, Zhang G, Zhao Q, Zheng L, Zheng XH, Zhong FN, Zhong W, Zhou X, Zhu S, Zhu X, Smith HO, Gibbs RA, Myers EW, Rubin GM, Venter JC. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185-2195. [PubMed] |
| 24. | Kiger AA, Baum B, Jones S, Jones MR, Coulson A, Echeverri C, Perrimon N. A functional genomic analysis of cell morphology using RNA interference. J Biol. 2003;2:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 319] [Cited by in RCA: 308] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 25. | Lum L, Yao S, Mozer B, Rovescalli A, Von Kessler D, Nirenberg M, Beachy PA. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science. 2003;299:2039-2045. [PubMed] |
| 26. | Caplen NJ, Fleenor J, Fire A, Morgan RA. dsRNA-mediated gene silencing in cultured Drosophila cells: a tissue culture model for the analysis of RNA interference. Gene. 2000;252:95-105. [PubMed] |
| 27. | Myers JW, Chi JT, Gong D, Schaner ME, Brown PO, Ferrell JE. Minimizing off-target effects by using diced siRNAs for RNA interference. J RNAi Gene Silencing. 2006;2:181-194. [PubMed] |
| 28. | Krishnan MN, Ng A, Sukumaran B, Gilfoy FD, Uchil PD, Sultana H, Brass AL, Adametz R, Tsui M, Qian F. RNA interference screen for human genes associated with West Nile virus infection. Nature. 2008;455:242-245. [PubMed] |
| 29. | Panda D, Das A, Dinh PX, Subramaniam S, Nayak D, Barrows NJ, Pearson JL, Thompson J, Kelly DL, Ladunga I. RNAi screening reveals requirement for host cell secretory pathway in infection by diverse families of negative-strand RNA viruses. Proc Natl Acad Sci USA. 2011;108:19036-19041. [PubMed] |
| 30. | Bushman FD, Malani N, Fernandes J, D’Orso I, Cagney G, Diamond TL, Zhou H, Hazuda DJ, Espeseth AS, König R. Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 2009;5:e1000437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 365] [Cited by in RCA: 344] [Article Influence: 21.5] [Reference Citation Analysis (0)] |
| 31. | Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci USA. 2009;106:7577-7582. [PubMed] |
| 32. | Trotard M, Lepère-Douard C, Régeard M, Piquet-Pellorce C, Lavillette D, Cosset FL, Gripon P, Le Seyec J. Kinases required in hepatitis C virus entry and replication highlighted by small interference RNA screening. FASEB J. 2009;23:3780-3789. [PubMed] |
| 33. | Borawski J, Troke P, Puyang X, Gibaja V, Zhao S, Mickanin C, Leighton-Davies J, Wilson CJ, Myer V, Cornellataracido I. Class III phosphatidylinositol 4-kinase alpha and beta are novel host factor regulators of hepatitis C virus replication. J Virol. 2009;83:10058-10074. [PubMed] |
| 34. | Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32-45. [PubMed] |
| 35. | Schmitz U, Tan SL. NS5A--from obscurity to new target for HCV therapy. Recent Pat Antiinfect Drug Discov. 2008;3:77-92. [PubMed] |
| 36. | Bianco A, Reghellin V, Donnici L, Fenu S, Alvarez R, Baruffa C, Peri F, Pagani M, Abrignani S, Neddermann P. Metabolism of phosphatidylinositol 4-kinase IIIα-dependent PI4P Is subverted by HCV and is targeted by a 4-anilino quinazoline with antiviral activity. PLoS Pathog. 2012;8:e1002576. [PubMed] |
| 37. | Campeau E, Gobeil S. RNA interference in mammals: behind the screen. Brief Funct Genomics. 2011;10:215-226. [PubMed] |
| 38. | Mohr SE, Perrimon N. RNAi screening: new approaches, understandings, and organisms. Wiley Interdiscip Rev RNA. 2012;3:145-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 107] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 39. | Börner K, Hermle J, Sommer C, Brown NP, Knapp B, Glass B, Kunkel J, Torralba G, Reymann J, Beil N. From experimental setup to bioinformatics: an RNAi screening platform to identify host factors involved in HIV-1 replication. Biotechnol J. 2010;5:39-49. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Mohr S, Bakal C, Perrimon N. Genomic screening with RNAi: results and challenges. Annu Rev Biochem. 2010;79:37-64. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 220] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 41. | Barr AR, Bakal C. A direct look at RNAi screens. Mol Syst Biol. 2012;8:580. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 42. | Birmingham A, Selfors LM, Forster T, Wrobel D, Kennedy CJ, Shanks E, Santoyo-Lopez J, Dunican DJ, Long A, Kelleher D. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6:569-575. [PubMed] |
| 43. | Boutros M, Ahringer J. The art and design of genetic screens: RNA interference. Nat Rev Genet. 2008;9:554-566. [PubMed] |
| 44. | Sharma S, Rao A. RNAi screening: tips and techniques. Nat Immunol. 2009;10:799-804. [PubMed] |
| 45. | Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user’s guide. Nat Rev Genet. 2006;7:373-384. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 276] [Cited by in RCA: 270] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 46. | Zhang XD. Novel analytic criteria and effective plate designs for quality control in genome-scale RNAi screens. J Biomol Screen. 2008;13:363-377. [PubMed] |
| 47. | Malo N, Hanley JA, Cerquozzi S, Pelletier J, Nadon R. Statistical practice in high-throughput screening data analysis. Nat Biotechnol. 2006;24:167-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 530] [Cited by in RCA: 513] [Article Influence: 27.0] [Reference Citation Analysis (0)] |
| 48. | Boutros M, Brás LP, Huber W. Analysis of cell-based RNAi screens. Genome Biol. 2006;7:R66. [PubMed] |
| 49. | Macarrón R, Hertzberg RP. Design and Implementation of High Throughput Screening Assays. High Throughput Screening. Totowa: Humana Press 2002; 1-29. |
| 50. | Gunter B, Brideau C, Pikounis B, Liaw A. Statistical and graphical methods for quality control determination of high-throughput screening data. J Biomol Screen. 2003;8:624-633. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 47] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 51. | Rieber N, Knapp B, Eils R, Kaderali L. RNAither, an automated pipeline for the statistical analysis of high-throughput RNAi screens. Bioinformatics. 2009;25:678-679. [PubMed] |
| 52. | Brideau C, Gunter B, Pikounis B, Liaw A. Improved statistical methods for hit selection in high-throughput screening. J Biomol Screen. 2003;8:634-647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 250] [Cited by in RCA: 253] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 53. | Altschuler SJ, Wu LF. Cellular heterogeneity: do differences make a difference. Cell. 2010;141:559-563. [PubMed] |
| 54. | Snijder B, Sacher R, Rämö P, Damm EM, Liberali P, Pelkmans L. Population context determines cell-to-cell variability in endocytosis and virus infection. Nature. 2009;461:520-523. [PubMed] |
| 55. | Snijder B, Sacher R, Rämö P, Liberali P, Mench K, Wolfrum N, Burleigh L, Scott CC, Verheije MH, Mercer J. Single-cell analysis of population context advances RNAi screening at multiple levels. Mol Syst Biol. 2012;8:579. [PubMed] |
| 56. | Snijder B, Pelkmans L. Origins of regulated cell-to-cell variability. Nat Rev Mol Cell Biol. 2011;12:119-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 258] [Article Influence: 18.4] [Reference Citation Analysis (0)] |
| 57. | Knapp B, Rebhan I, Kumar A, Matula P, Kiani NA, Binder M, Erfle H, Rohr K, Eils R, Bartenschlager R. Normalizing for individual cell population context in the analysis of high-content cellular screens. BMC Bioinformatics. 2011;12:485. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 58. | Chung N, Zhang XD, Kreamer A, Locco L, Kuan PF, Bartz S, Linsley PS, Ferrer M, Strulovici B. Median absolute deviation to improve hit selection for genome-scale RNAi screens. J Biomol Screen. 2008;13:149-158. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 143] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 59. | Zhang XD, Yang XC, Chung N, Gates A, Stec E, Kunapuli P, Holder DJ, Ferrer M, Espeseth AS. Robust statistical methods for hit selection in RNA interference high-throughput screening experiments. Pharmacogenomics. 2006;7:299-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 60. | Zhang XD, Lacson R, Yang R, Marine SD, McCampbell A, Toolan DM, Hare TR, Kajdas J, Berger JP, Holder DJ. The use of SSMD-based false discovery and false nondiscovery rates in genome-scale RNAi screens. J Biomol Screen. 2010;15:1123-1131. [PubMed] |
| 61. | Manly KF, Nettleton D, Hwang JT. Genomics, prior probability, and statistical tests of multiple hypotheses. Genome Res. 2004;14:997-1001. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 56] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 62. | Kerrien S, Alam-Faruque Y, Aranda B, Bancarz I, Bridge A, Derow C, Dimmer E, Feuermann M, Friedrichsen A, Huntley R. IntAct--open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561-D565. [PubMed] |
| 63. | Navratil V, de Chassey B, Meyniel L, Delmotte S, Gautier C, André P, Lotteau V, Rabourdin-Combe C. VirHostNet: a knowledge base for the management and the analysis of proteome-wide virus-host interaction networks. Nucleic Acids Res. 2009;37:D661-D668. [PubMed] |
| 64. | Chatr-aryamontri A, Ceol A, Peluso D, Nardozza A, Panni S, Sacco F, Tinti M, Smolyar A, Castagnoli L, Vidal M. VirusMINT: a viral protein interaction database. Nucleic Acids Res. 2009;37:D669-D673. [PubMed] |
| 65. | Fu W, Sanders-Beer BE, Katz KS, Maglott DR, Pruitt KD, Ptak RG. Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 2009;37:D417-D422. [PubMed] |
| 66. | Murali TM, Dyer MD, Badger D, Tyler BM, Katze MG. Network-based prediction and analysis of HIV dependency factors. PLoS Comput Biol. 2011;7:e1002164. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 67. | Nabieva E, Jim K, Agarwal A, Chazelle B, Singh M. Whole-proteome prediction of protein function via graph-theoretic analysis of interaction maps. Bioinformatics. 2005;21 Suppl 1:i302-i310. [PubMed] |
| 68. | Bader GD, Hogue CW. An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinformatics. 2003;4:2. [PubMed] |
| 69. | MacPherson JI, Dickerson JE, Pinney JW, Robertson DL. Patterns of HIV-1 protein interaction identify perturbed host-cellular subsystems. PLoS Comput Biol. 2010;6:e1000863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 49] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 70. | de Chassey B, Meyniel-Schicklin L, Aublin-Gex A, André P, Lotteau V. Genetic screens for the control of influenza virus replication: from meta-analysis to drug discovery. Mol Biosyst. 2012;8:1297-1303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 30] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 71. | Brass AL, Huang IC, Benita Y, John SP, Krishnan MN, Feeley EM, Ryan BJ, Weyer JL, van der Weyden L, Fikrig E. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell. 2009;139:1243-1254. [PubMed] |
| 72. | Shapira SD, Gat-Viks I, Shum BO, Dricot A, de Grace MM, Wu L, Gupta PB, Hao T, Silver SJ, Root DE. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell. 2009;139:1255-1267. [PubMed] |
| 73. | Ward SE, Kim HS, Komurov K, Mendiratta S, Tsai PL, Schmolke M, Satterly N, Manicassamy B, Forst CV, Roth MG. Host modulators of H1N1 cytopathogenicity. PLoS One. 2012;7:e39284. [PubMed] |
| 74. | Chase G, Deng T, Fodor E, Leung BW, Mayer D, Schwemmle M, Brownlee G. Hsp90 inhibitors reduce influenza virus replication in cell culture. Virology. 2008;377:431-439. [PubMed] |
| 75. | Wang L, Tu Z, Sun F. A network-based integrative approach to prioritize reliable hits from multiple genome-wide RNAi screens in Drosophila. BMC Genomics. 2009;10:220. [PubMed] |
| 76. | Pandey G, Steinbach M, Gupta R, Garg T, Kumar V. Association analysis-based transformations for protein interaction networks: a function prediction case study. Proceedings of the 13th ACM SIGKDD international conference on Knowledge discovery and data mining. 2007;540-549. |
| 77. | Dickerson JE, Pinney JW, Robertson DL. The biological context of HIV-1 host interactions reveals subtle insights into a system hijack. BMC Syst Biol. 2010;4:80. [PubMed] |
| 78. | van Dijk D, Ertaylan G, Boucher CA, Sloot PM. Identifying potential survival strategies of HIV-1 through virus-host protein interaction networks. BMC Syst Biol. 2010;4:96. [PubMed] |
| 79. | Jaeger S, Ertaylan G, van Dijk D, Leser U, Sloot P. Inference of surface membrane factors of HIV-1 infection through functional interaction networks. PLoS One. 2010;5:e13139. [PubMed] |
| 80. | Kumar R, Nanduri B. HPIDB--a unified resource for host-pathogen interactions. BMC Bioinformatics. 2010;11 Suppl 6:S16. [PubMed] |
| 81. | Xiang Z, Tian Y, He Y. PHIDIAS: a pathogen-host interaction data integration and analysis system. Genome Biol. 2007;8:R150. [PubMed] |
| 82. | Dyer MD, Murali TM, Sobral BW. The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 2008;4:e32. [PubMed] |
| 83. | Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572-D574. [PubMed] |
| 84. | Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes HW, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436-D441. [PubMed] |
| 85. | Joshi-Tope G, Gillespie M, Vastrik I, D’Eustachio P, Schmidt E, de Bono B, Jassal B, Gopinath GR, Wu GR, Matthews L. Reactome: a knowledgebase of biological pathways. Nucleic Acids Res. 2005;33:D428-D432. [PubMed] |
| 86. | Mishra GR, Suresh M, Kumaran K, Kannabiran N, Suresh S, Bala P, Shivakumar K, Anuradha N, Reddy R, Raghavan TM. Human protein reference database--2006 update. Nucleic Acids Res. 2006;34:D411-D414. [PubMed] |
| 87. | Xenarios I, Rice DW, Salwinski L, Baron MK, Marcotte EM, Eisenberg D. DIP: the database of interacting proteins. Nucleic Acids Res. 2000;28:289-291. [PubMed] |
| 88. | Gulbahce N, Yan H, Dricot A, Padi M, Byrdsong D, Franchi R, Lee DS, Rozenblatt-Rosen O, Mar JC, Calderwood MA. Viral perturbations of host networks reflect disease etiology. PLoS Comput Biol. 2012;8:e1002531. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 87] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 89. | Pfeiffer T, Hoffmann R. Temporal patterns of genes in scientific publications. Proc Natl Acad Sci USA. 2007;104:12052-12056. [PubMed] |
| 90. | de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. [PubMed] |
| 91. | Khadka S, Vangeloff AD, Zhang C, Siddavatam P, Heaton NS, Wang L, Sengupta R, Sahasrabudhe S, Randall G, Gribskov M. A physical interaction network of dengue virus and human proteins. Mol Cell Proteomics. 2011;10:M111.012187. [PubMed] |
| 92. | Simonis N, Rual JF, Lemmens I, Boxus M, Hirozane-Kishikawa T, Gatot JS, Dricot A, Hao T, Vertommen D, Legros S. Host-pathogen interactome mapping for HTLV-1 and -2 retroviruses. Retrovirology. 2012;9:26. [PubMed] |
| 93. | Pichlmair A, Kandasamy K, Alvisi G, Mulhern O, Sacco R, Habjan M, Binder M, Stefanovic A, Eberle CA, Goncalves A. Viral immune modulators perturb the human molecular network by common and unique strategies. Nature. 2012;487:486-490. [PubMed] |
| 94. | Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248-1250. [PubMed] |
| 95. | Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Kräusslich HG, Mizokami M. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791-796. [PubMed] |
| 96. | Lindenbach BD, Evans MJ, Syder AJ, Wölk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623-626. [PubMed] |
| 97. | Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci USA. 2005;102:9294-9299. [PubMed] |
| 98. | Guha C, Mohan S, Roy-Chowdhury N, Roy-Chowdhury J. Cell culture and animal models of viral hepatitis. Part I: hepatitis B. Lab Anim (NY). 2004;33:37-46. [PubMed] |