Published online Jun 25, 2024. doi: 10.5501/wjv.v13.i2.91580
Revised: February 8, 2024
Accepted: April 15, 2024
Published online: June 25, 2024
Processing time: 176 Days and 1.6 Hours
The hepatitis E virus (HEV), a member of the Hepeviridae family, is a small, non-enveloped icosahedral virus divided into eight distinct genotypes (HEV-1 to HEV-8). Only genotypes 1 to 4 are known to cause diseases in humans. Genotypes 1 and 2 commonly spread via fecal-oral transmission, often through the consum
Core Tip: Hepatitis E virus (HEV) typically causes a self-limiting infection, but it can establish a persistent infection progressing into chronic hepatitis, cirrhosis or acute liver failure, particularly in individuals with compromised immune systems and preexisting liver diseases. In recent decades, the prevalence of hepatitis E has increased dramatically, and this trend continues unabated. Thus, a better understanding of the pathogenesis of hepatitis E and the development of more effective prevention and treatment strategies have become an urgent medical problem. We herein discuss the major features of HEV, the conditions predisposing to severe hepatitis E, and the underlying pathological immune processes.
- Citation: Orosz L, Sárvári KP, Dernovics Á, Rosztóczy A, Megyeri K. Pathogenesis and clinical features of severe hepatitis E virus infection. World J Virol 2024; 13(2): 91580
- URL: https://www.wjgnet.com/2220-3249/full/v13/i2/91580.htm
- DOI: https://dx.doi.org/10.5501/wjv.v13.i2.91580
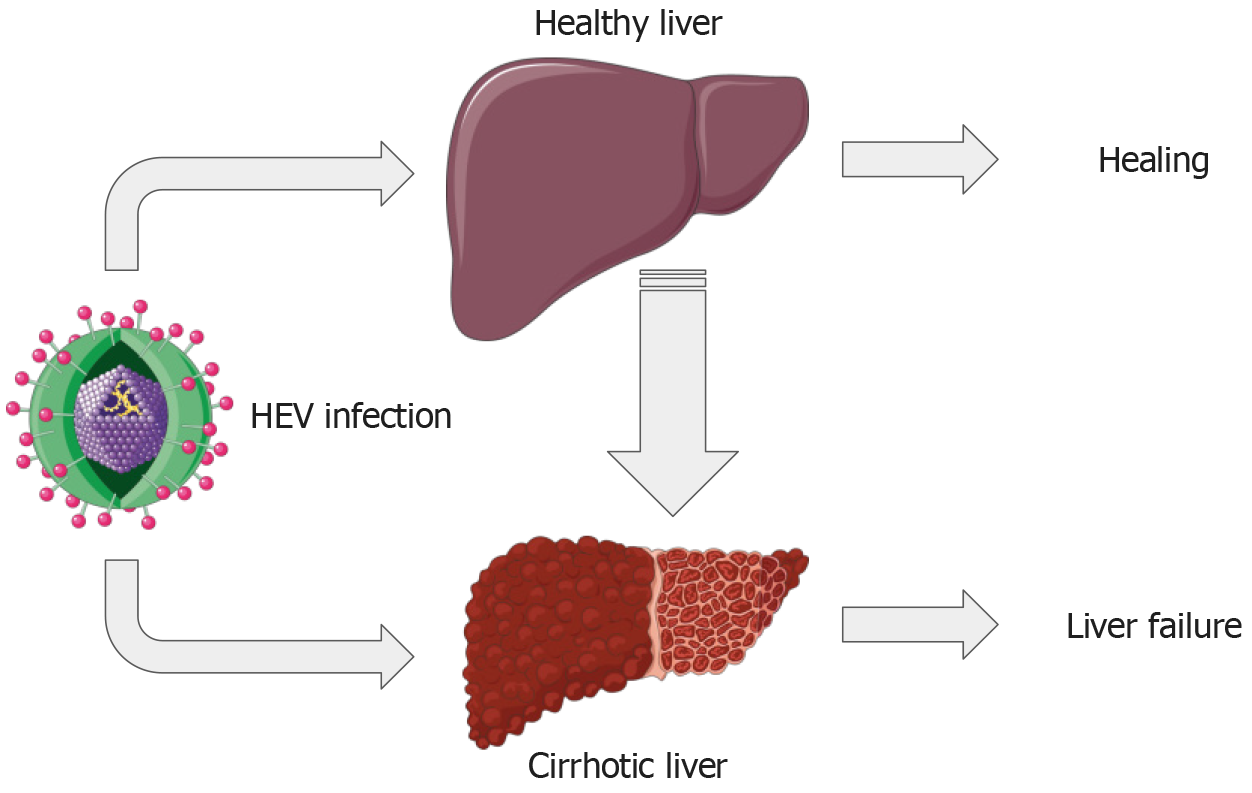
The hepatitis E virus (HEV), a non-enveloped virus characterized by its icosahedral capsid symmetry and approximately 27-34 nm size, belongs to the Hepevirus genus within the Hepeviridae family. Its genome is a positive-sense, single-stranded RNA strand of roughly 7.2 kb[1,2]. There are eight known genotypes of HEV, but genotypes 1-4 account for most human diseases[3]. HEV primarily spreads through the fecal-oral route, with contaminated water and undercooked meat being common infection sources[3,4]. Typically, HEV infections are self-limiting and resolve naturally within 6–8 wk[4]. However, in immunocompromised individuals, HEV infection can become chronic[3,4]. Persistent infections could lead to serious conditions such as liver cirrhosis, liver failure, and hepatocellular carcinoma (Figure 1)[5-11].
The interplay between HEV infection and conditions like liver cirrhosis, pregnancy, and immunosuppression warrants significant attention in hepatology. Liver disease progression is profoundly impacted when individuals with cirrhosis contract HEV, resulting in a greater risk of acute-on-chronic liver failure (ACLF). This necessitates careful monitoring and proactive strategies for HEV detection, prevention, and treatment among these at-risk individuals.
Pregnant women, particularly in their third trimester, also face a significant risk of severe complications and death from HEV infection[12]. The underlying factors for these complications during pregnancy remain unknown, highlighting an urgent need for more research in this area.
Immunosuppressed individuals, including organ transplant recipients and those with human immunodeficiency virus (HIV), elevate the possibility of chronic HEV infection and subsequently developing liver cirrhosis.
In summary, a strong connection exists between HEV infection and liver cirrhosis, especially in pregnant and immunosuppressed individuals. As such, acknowledging HEV infection as a potential risk factor and implementing prevention, detection, and treatment strategies are fundamental, particularly for those with liver disease or those undergoing immunosuppressive treatment. This review offers a broad perspective on the relationship between HEV infection and liver cirrhosis by consolidating available data on this matter.
We carried out a literature search through the PubMed database using the following search terms: “hepatitis E” limited to “open-reading frame”, “replication”, “innate immune response”, “cytokine”, “humoral immune response”, “cellular immune response”, “acute-on-chronic liver failure”, “pregnancy”, “immunosuppression”, and “cirrhosis”. We excluded articles pertaining to diagnostics, therapy, animal studies, and case reports already covered in other included studies. From the 5744 articles initially found, we selected 149 for our study. All chosen materials were in English.
The RNA genome of HEV is capped at the 5′ end and polyadenylated at the 3′ end. It contains three slightly overlapping open reading frames (ORF) 1–3 along with a recently discovered ORF4[1,13,14]. Additionally, infected cells produce a 2.2 sub-genomic RNA from which ORF2 and ORF3 are translated[1].
The 5079 base pairs (bp) ORF1 sequence encodes a polyprotein comprised of 1693 amino acids (aa) that weighs 190-kDa[15]. This ORF1 sequence encodes nonstructural proteins that are enzymatically active, including methyltransferase, papain-like cysteine protease (PCP), RNA helicase, RNA-dependent RNA polymerase (RdRp), macro domains or X/Y domains, and a hypervariable region. These proteins play crucial roles in RNA synthesis, RNA capping, RNA unwinding, tRNA metabolism, transcription, and replication[16].
The methyltransferase, located on ORF1 polyprotein positions 60 to 240, and a 110-kDa protein (on ORF1 polyprotein positions 1 to 979) with guanyltransferase and guanine-7-methyltransferase activity are involved in viral RNA capping (m7G cap: 27–35 nucleotides at the 5′ end). This is because the 7-methylguanine cap is critical for HEV infectivity[17,18].
The PCP (potentially located on ORF1 polyprotein positions 433 to 592) may also inhibit cellular antiviral immune function[19]. Meanwhile, RNA helicase (on ORF1 polyprotein positions 960 to 1204) is a member of the 5′→ 3′ class of the superfamily 1 of helicases and plays a vital role in HEV RNA replication[20,21].
Furthermore, RdRp is found on ORF1 polyprotein positions 1207 to 1693 and is an essential enzyme for RNA replication[22]. Macro domain proteins (on ORF1 polyprotein positions 775 to 960) hydrolyze adenosine diphosphate (ADP)-ribose 1”-phosphate and might contribute to RNA replication, posttranslational modification, and cellular apoptosis[23].
Lastly, the Polyproline region or Hypervariable region has several potential functions, such as providing peptide cleavage sites, areas modified by enzymes, and sites that can bind to proteins, nucleotides, and metal ions. This suggests a potential role in viral replication[24].
ORF2, featuring 1980 bp, encodes the capsid protein, which comprises 660 aa with an approximate molecular weight of 72 kDa[25]. ORF2 of the HEV genotype 3 consists of three sections: The N-terminal domain (aa 1–111), virus-like particle (VLP) (aa 112–608), and C-domain (aa 609–660).
The VLP can be further subdivided into the shell domain (S, aa 129–319), the middle domain (M, aa 320–455), and the protruding domain (P, aa 456–606)[26,27]. Each VLP subdivision contains neutralization epitopes recognized by anti-HEV immunoglobulins (IgM and IgG) responsible for attaching the virion to susceptible cells[28].
A separate product can also be identified-a glycoprotein weighing approximately 88 kDa with three potential glycosylation sites. This can be transferred through the endoplasmic reticulum and expressed on the surface[29].
Three variations of ORF2 from the HEV genotype 3 have been noted: ORF2i (the infectious form), ORF2g (the glycosylated and secreted version), and ORF2c (the cleaved and secreted type). ORF2c is theorized to be a cleavage product of the ORF2g protein.
All ORF2 proteins possess three potential N-glycosylation sites and several O-linked glycosylation sites. ORF2g and ORF2c are secreted and glycosylated (glycosylation sites: N1 and N3 in ORF2g/c) but do not associate with infecting virions. In contrast, ORF2i is not glycosylated but is the only version packaged into infectious particles[30].
Lastly, ORF2s is the secreted form of ORF2, which is glycosylated and does not contain the binding site of the cellular receptor[31].
The smaller third ORF is located at the end of ORF1, and it overlaps ORF2 at the 5′ end[2]. It is translated from bicistronic sub-genomic RNA. This ORF, termed ORF3, encodes a protein consisting of 123 aa known as Vp13, a non-glycosylated protein with an approximate size of 13.5-kDa[1,2].
Vp13 is notable for containing two proline-rich domains (P1 and P2) and two strong hydrophobic regions (D1 and D2) in the N-terminal half. It can be phosphorylated at a serine residue (ser-80), which is crucial for its interaction with the capsid protein[1,32]. This phosphorylation may be necessary for in vivo replication. Additionally, the palmitoylation of Vp13 plays a significant role in the release of virions[33].
Vp13 is known to interact with the cell’s cytoskeleton and aids in binding to microtubules[32,34]. The P2 domain specifically has two PXXP motifs, where P and X designate proline and an unspecified amino acid, respectively. PXXP motifs are instrumental in the interaction of the P2 domain with the SH3 domains of cellular proteins, such as the α-1-microglobulin and bikunin precursor protein. This interaction can activate the mitogen-activated protein kinase, which could disrupt cellular signal transduction[35,36].
ORF3’s functionality extends to its binding with hemopexin, a protein that protects against oxidative damage. An HEV infection is linked to a decline in the serum levels of hemopexin, which indicates the pivotal role of ORF3 in infection[37]. Vp13 also functions as a class I viroporin–its two PXXP motifs form an ion channel instrumental in viral discharge[38].
In a recent discovery, another ORF called ORF4 was found in HEV genotype 1[13]. This ORF4 transcription is only noticeable under the stress of the endoplasmic reticulum and plays a key role in viral replication by maintaining the proper function of RdRp[14].
The hepatocyte is the main cell type wherein HEV replicates, though HEV can also replicate in other cells and tissues like monocytes, the spleen, lymph nodes, and the small intestine[39]. The virus usually spreads through fecal-oral transmission and reaches the hepatocytes via the bloodstream. A cellular receptor for viral binding has not been identified yet. Still, non-enveloped virions need heparan sulfate proteoglycan for attachment, whereas quasi-enveloped virions attach to the cell surface independently[40].
Research by Holla et al[41] suggests that the virions may enter host cells through a process known as dynamin-2-dependent clathrin-mediated endocytosis[41]. The uncoating of HEV requires a low pH. The virus’s positive-sense RNA genome is subsequently released into the cytosol and serves as the template for the translation of ORF1[42].
Transcription begins when RdRp binds to the 3′ untranslated region of the viral RNA to produce the negative-sense intermediate RNA. This serves as a template for synthesizing positive-sense RNA and is translated into ORF2 and ORF3 proteins[43]. Structural proteins might be expressed from two sub-genomic RNAs (3.7 kb and 2.0 kb). The ORF2 protein packages the genomic positive-sense RNA into progeny virions[30].
The HEV virions assembled in this manner bud into the lumen of multivesicular endosomes, thus acquiring a quasi-envelope. These loaded endosomes then move to the cytoplasmic membrane, fusing with it and releasing the quasi-enveloped virions into the space outside the cells.
In the bile canaliculi, these quasi-enveloped HEV (eHEV) virions lose their envelopes to the bile and convert into non-enveloped particles, though a small number of eHAV are released into the blood via the hepatocytes’ basolateral side. The removal of the lipid layer makes the eHEV particles more infectious[44].
Innate immune cells interact with HEV by using their pattern recognition receptors (PRRs), such as Toll-like receptor (TLR) 2 and TLR4, which recognize the capsid. Other receptors, like TLR7/8, Retinoic Inducible Gene-I (RIG-I)-like receptors, and Melanoma Differentiation-Associated Protein 5 (MDA5), attach to the viral RNA[45]. It has been shown that TLR2, 3, 4, 7, and 8 expression levels heighten in the peripheral blood mononuclear cells (PBMC) of HEV patients[46,47]. Studies also suggest that A549 cells show increased PRR levels following HEV infection[48].
The interaction of HEV with TLRs, RIG-I, and MDA5 activates the interferon regulatory factor (IRF) 3 and nuclear factor-κB (NF-κB) transcription factors through signaling pathways that involve Toll/interleukin (IL)-1 receptor domain-containing adaptor inducing interferon (IFN) β, tumor necrosis factor (TNF) receptor-associated factor (TRAF) 3, TRAF6, myeloid differentiation primary response 88, and mitochondrial antiviral-signaling protein (MAVS) adaptors[47,48]. These activated transcription factors, in turn, stimulate the transcription of numerous pro- and anti-inflammatory cytokine genes.
During self-limited acute HEV infection, among the pro-inflammatory cytokines and chemokines, the levels of TNF-α, IFN-α, IFN-ω, IFN-β, IFN-γ, IFN-λ, IL-1β, IL-4, IL-6, IL-18, C-X-C motif chemokine ligand (CXCL) 8 (IL-8), C-C motif chemokine ligand (CCL) 5 protein [Regulated upon activation, normal T cell expressed and secreted (RANTES)], interferon-stimulated gene (ISG) 15, and ISG20 were increased in sera or PBMC obtained from HEV-infected patients[47,49,50]. In hepatocytes and enterocytes, HEV virions or RNA triggered the production of IFN-β, IFN-λ1, IFN-λ2 and IFN-λ3 in a cell type-dependent manner. Human and swine hepatocytes mainly secreted type I IFNs, whereas enterocytes predominantly produced type III IFNs[51,52]. Interestingly, IRF3 and IRF7 were essential for the HEV genomic RNA-mediated induction of IFNs, while the cellular RNA sensing pathways were not required[52].
IFNs bind to their corresponding receptors and activate the Janus kinase (JAK)-signal transducer and activator of transcription (STAT) signaling pathway, stimulating the expression of several hundred genes, whose products exert antiviral, immunomodulatory, and antiproliferative actions. IFNs induced by HEV infection increase the expression of ISG15, IFN-α inducible protein 6, MX1, 2′, 5′-oligoadenylate synthetase 1, and interferon-induced proteins with tetratricopeptide repeats 2, all of which belong to the IFN-stimulated genes[48,52]. All types of IFNs inhibit HEV replication directly, but HEV can impair the function of the IFN system.
HEV encodes a microRNA (miRNA), HEV-miRNA-A6, which can inhibit the phosphorylation of IRF3, thereby reducing the production of type I IFNs[53]. Moreover, HEV ORF-1 interferes with the recognition of viral RNA by RIG-I and impairs the activation of the JAK-STAT signaling pathway by inhibiting STAT-1 activation and nuclear translocation[54,55]. Hence, ORF-1 inhibits the transcriptional activation of ISGs and the antiviral effect of type I IFNs (IFN-α, IFN-β and IFN-ω)[54,55]. Consequently, HEV induces the synthesis of numerous pro- and anti-inflammatory cytokines while inhibiting the production and effects of type I interferons, resulting in a cytokine environment with a weak antiviral effect. This environment is unable to effectively control virus replication in the early stages of infection.
During an acute, self-limited infection, anti-HEV IgM is found concurrently with a rise in transaminase levels in the bloodstream and starts to decrease after recovery. On average, the persistence of anti-HEV IgM lasts for around 20 wk, although in some cases, it can be detected even after 3 years[56-58]. The reasons for this prolonged presence are still unclear. In individuals having HIV, those who have undergone transplantation, patients with cancer and those who are immunosuppressed, the emergence of HEV-specific IgM may take months[57-59].
During the early stage of infection, anti-HEV IgA can also be detected in the bloodstream for at least 4 months[60]. Following the emergence of IgM, anti-HEV IgG becomes noticeable in the bloodstream. Its titer and avidity increase and can persist for at least 5 years before gradually decreasing[58,61-63]. HEV-specific antibodies might have a potentially protective role by neutralizing effects and initiating some antiviral immune mechanisms.
Immunodominant conformational and non-immunodominant linear epitopes have been identified on the ORF2 protein of HEV[64-67]. These neutralizing antibodies work by inhibiting the virus from binding to cell surface receptors and entering the cells. Interestingly, while immune sera can neutralize virion infectivity in feces[68], there’s been no such neutralizing effect on HEV particles in the bloodstream. The blood-circulating HEV particles are quasi-enveloped, surrounded by a host cell-derived membrane lacking viral proteins[69]. Thus, neutralizing antibodies cannot bind with these eHEV particles. As a result, less than 10% of HEV virions bind to circulating antibodies, partially restricting antibody-mediated neutralization[68].
HEV-specific non-neutralizing antibodies may also contribute to immune defense through other mechanisms like epitope unmasking, intracellular neutralization, antibody-dependent cellular cytotoxicity, activation of the classical complement pathway, and phagocytosis[70]. The role of these non-neutralizing antibodies in immune protection against HEV, however, remains unclear. Although humoral immunity helps slow down the rate of viral replication and reduce the severity of symptoms, it is not enough for full protection. Therefore, breakthrough infections can still occur in both immunocompetent and immunosuppressed individuals with measurable humoral immunity levels.
HEV-specific cellular immune responses have been extensively studied by analyzing the number, immune phenotype, and function of PBMC or hepatic immune cells. Stimulation of PBMC with a HEV genotype 3a peptide library, encompassing 616 peptides spanning ORF1-3, or a recombinant capsid protein, resulted in robust T-cell responses in seropositive healthy individuals, as evidenced by IFN-γ production[71-73]. While peptides derived from all three ORFs triggered strong cellular immune responses, ORF2 was identified as carrying the immune-dominant epitopes of HEV[71]. However, both CD4+ and CD8+ T-cell responses against peptides from ORF2 and ORF3 were absent, and IFN-γ production was diminished in organ transplant recipients with chronic hepatitis E[73]. The activation, proliferation, and cytokine secretion of lymphocytes is regulated by inhibitory receptors like programmed cell death protein 1 (PD1) and cytotoxic T-lymphocyte-associated protein 4 (CTLA4). Blockade of the PD1 and CTLA4 pathways using anti-PDL1 and anti-CTLA4 antibodies restored the proliferative responses of CD4+ and CD8+ lymphocytes stimulated with an ORF2 and ORF3 peptide pool[73].
Kemming et al[74] identified several HEV-specific CD8+ T-cell epitopes restricted by nine different HLA class I alleles, with 17 and eight out of 25 epitopes located on the ORF1 and ORF2 proteins, respectively[74]. The immune-dominant cytotoxic T lymphocyte (CTL) epitopes of HEV were found within the ORF2 protein[74]. During self-limited acute infection, elevated expressions of activation and proliferation markers (CD38 and Ki67), Tbet transcription factor, and granzyme B were observed, indicating the development of an intense effector CD8+ T-cell response[74]. In the recovery phase, these markers were downregulated, and a stable memory cell pool was established[74]. In contrast, patients with chronic hepatitis E exhibited significantly diminished CD8+ T-cell responses. These CD8+ T-cells co-expressed activation and proliferation markers along with the PD1 inhibitory receptor, reflecting their terminally exhausted phenotype and profound functional deterioration[74]. Thus, HEV-specific CD8+ T cells are key cellular elements of protective immunity, playing an essential role in virus elimination.
Another study revealed higher numbers of CD4+CD25+Foxp3+ and CD4+CD25ˉFoxp3+ regulatory T-cells in the blood of patients with self-limited acute HEV infection and recovered individuals compared to seronegative controls[75]. Increased IL-10 production was also detected in PBMC stimulated with recombinant ORF2 in these patient groups[75]. Transient differences were observed in the number and activation state of natural killer (NK) and NKT cells, as well as in the cytotoxic activity of NK cells in the peripheral blood of acute hepatitis E patients[76].
El Costa et al[77] compared the CD8+ T-cell responses between asymptomatic and symptomatic hepatitis E patients[77]. Symptomatic elderly patients exhibited a comprehensive global expansion of an activated effector memory CD8+ T-cell compartment, consisting of both HEV genotype 3-specific and non-specific cell populations[77]. The cells isolated from symptomatic elderly patients were HLA-DR/CD38/PD1 triple-positive, showed increased C-X-C motif chemokine receptor (CXCR) 3, IL-4, and granzyme B expression, and decreased IFN-γ production[77]. The levels of CXCL9 and CXCL10, known ligands of CXCR3, were also markedly elevated in symptomatic patients[77]. Further investigations of immune cell phenotypes in biopsy specimens obtained from patients with HEV-induced acute liver failure revealed the accumulation of activated CD8+ T-cells containing granzyme in the liver[78]. These studies, which highlight the emergence of a CD8+ T-cell population with high cytotoxic potential and the ability to infiltrate the liver during symptomatic infections, support the idea that liver damage caused by HEV infection is mediated by pathological immune processes[77,78].
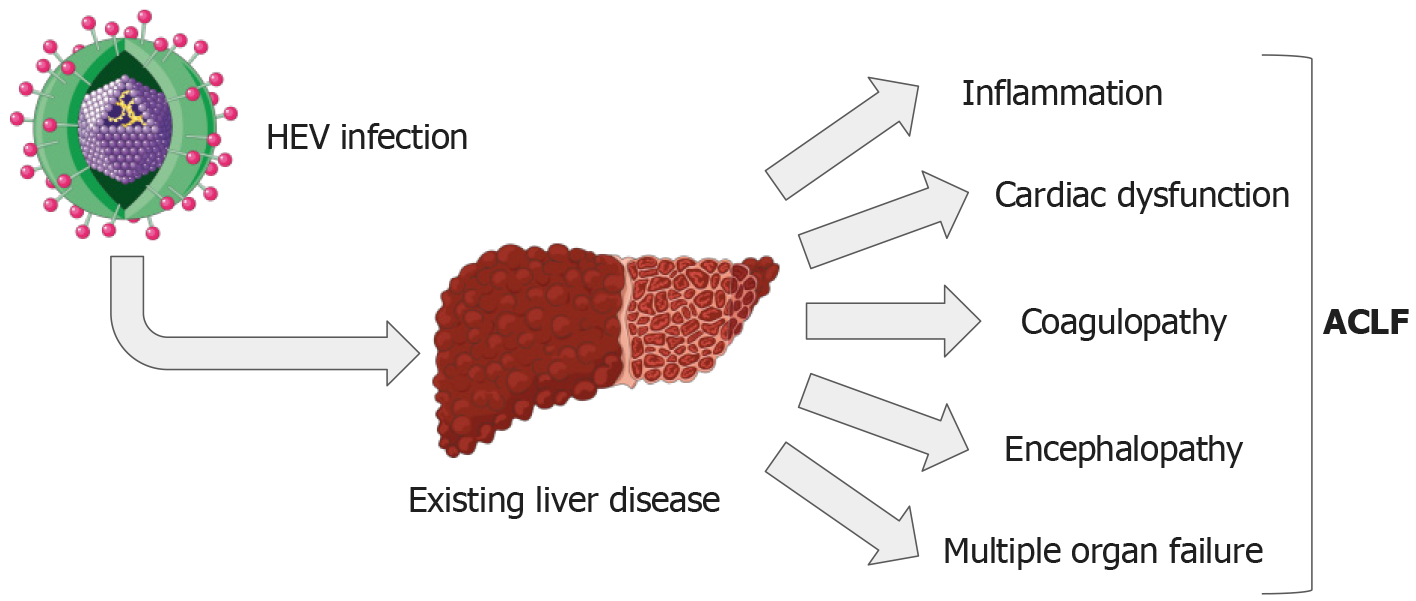
ACLF is a disorder marked by sudden liver damage, evident through jaundice and coagulopathy. ACLF differs from chronic hepatic decompensation in several ways. Notably, liver failure and the resulting organ dysfunction progress faster in ACLF. Furthermore, the 3-month death rate during ACLF is much higher than predicted in chronic hepatic decompensation cases[79,80]. Usually, ACLF develops after cirrhosis of the liver, with additional factors possibly exacerbating the liver damage. These can include co-infection with other forms of viral hepatitis (like HEV), ischemic hepatitis, alcoholic hepatitis, drug-induced hepatitis, and metabolic dysfunction-associated steatotic liver disease and steatohepatitis (MASLD/MASH). Recent studies suggest that a combination of viral hepatitis and surgery are the primary factors causing ACLF[81].
The transition from stable cirrhosis to ACLF is triggered by the production of pro-inflammatory cytokines due to a systemic inflammatory response. Inflammatory mediators have been shown to enhance hyperammonemia’s induction of encephalopathy[82]. Previous studies have noted a decrease in peripheral CD4+ T-cell levels and an increase in CD8+ T-cell levels in individuals with HEV infection, causing an alteration in the CD4/CD8 ratio[78]. A systemic inflammatory response also considered a negative prognostic factor, is associated with the development of encephalopathy, thereby reducing the potential for successful organ transplantation[83]. Moreover, hepatic synthesis often weakens, causing hypoalbuminemia, which subsequently results in edema and ascites. Hyperbilirubinemia and clinical jaundice are typically unavoidable, while thrombocytopenia can cause hemorrhagic diathesis (Figure 2). Altogether, given the well-known link between HEV infection and ACLF, it is essential to take proactive steps to manage this public health issue and shield vulnerable populations from the severe implications of liver complications due to HEV.
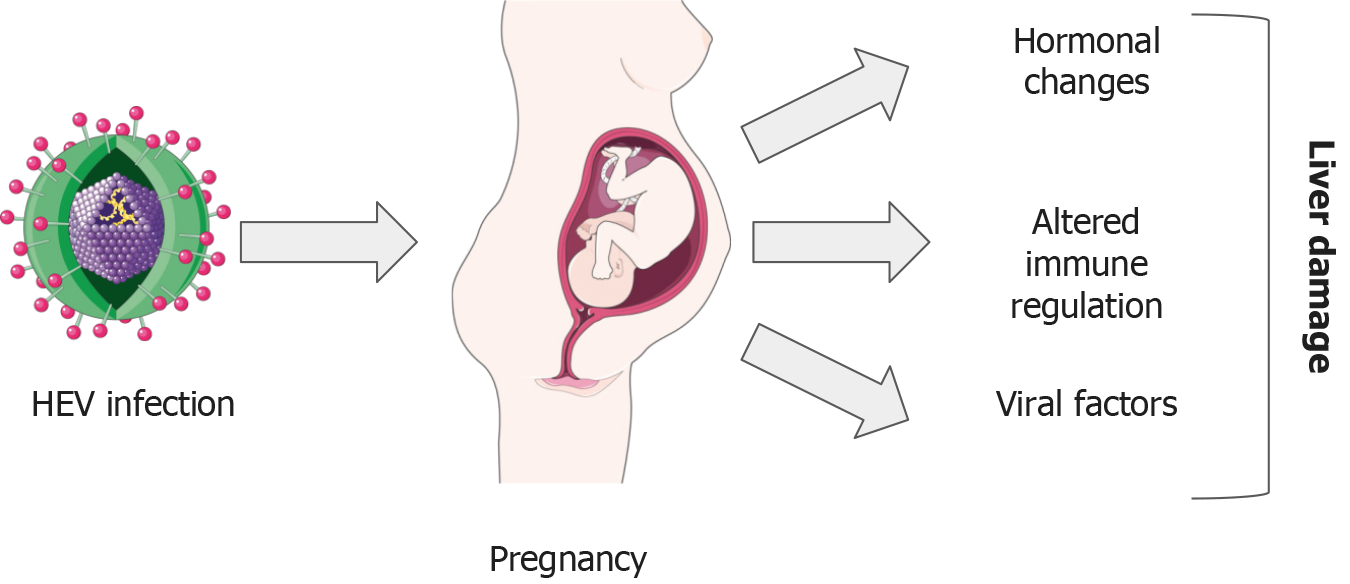
Pregnancy can complicate the course of an infection, making it more severe. Changes in hormone concentrations and immune responses induced by pregnancy can affect the immune system’s ability to fight infections like hepatitis E. Other than these factors, specific viral elements have been identified that could exacerbate the infection’s progression (Figure 3).
The severe liver damage caused by a HEV infection during pregnancy can be attributed to unclear pathophysiological mechanisms. Past studies have suggested that the unique conditions in pregnant women, like altered immune responses, hormone levels, and viral aspects, such as the diversity and changes in the HEV genome, could potentially affect the disease’s severity[84]. Throughout pregnancy, the mother’s immune system is challenged to maintain a robust response to ward off harmful pathogens for both her and the developing fetus. Immune system changes in pregnancy are complex, including reductions in the quantity and function of NK cells and T-cells and increases in monocytes, granulocytes, and dendritic cells in the peripheral blood. Th2 cells enhance B lymphocyte activation and antibody production while concurrently suppressing the response of CTLs, resulting in a weakened cell-mediated immunity.
The development of liver damage resulting from HEV infection in the general population is primarily associated with the activation of immune cells, specifically CD8+ T-cells and NK cells, both within the liver and in the peripheral circulation. In pregnant patients experiencing fulminant hepatic failure due to HEV, the presence of a ‘Th2 bias’ has been documented, although its exact influence on the severity of the illness remains uncertain[84].
During pregnancy, specific hormones such as progesterone, estrogen, and human chorionic gonadotropin experience significant changes in their levels. A study found that these hormonal levels were higher in pregnant patients with acute liver failure who tested positive for HEV compared to those who tested negative[85]. Notably, the increased estradiol levels in the bloodstream of HEV-infected pregnant women promote the virus’s multiplication[86-88]. Moreover, high estrogen levels are linked to premature birth, underweight newborns, and fetal death due to impaired placental functioning in HEV-infected pregnant women[86-88]. Both progesterone and estrogen were found to disturb the balance between Th1 and Th2 responses[12]. Moreover, estrogen plays a direct role in reducing the cytotoxicity of CD8+ T-cells [89]. It can adjust the survival and activation of B cells and can hinder the production of B cells during pregnancy[89].
Differences in maternal illness and death rates vary across HEV genotypes. The most prevalent genotypes associated with these issues in pregnancy are HEV-1 and HEV-2. However, this correlation between increased incidence and severity of hepatitis E in pregnant women exists with HEV-1 or HEV-2 but not HEV-3 and HEV-4[90-95]. In a recent study using a model based on maternal and fetal organ culture, HEV-1 exhibited more effective replication, produced more infectious particles, and induced more severe tissue changes than HEV-3[96]. This suggests that genotype might significantly impact HEV infection severity, especially in pregnant women. However, due to a lack of appropriate in vivo and in vitro models and difficulties cultivating the virus in vitro, a comprehensive study investigating susceptibility, infectivity, replication capacity, and pathogenicity across different HEV genotypes has not been conducted. Additionally, genetic variations in HEV can affect viral shape, development, clinical outcomes, and antiviral resistance[3,97,98]. However, it remains uncertain if these variations are linked to maternal illness or death, warranting further research.
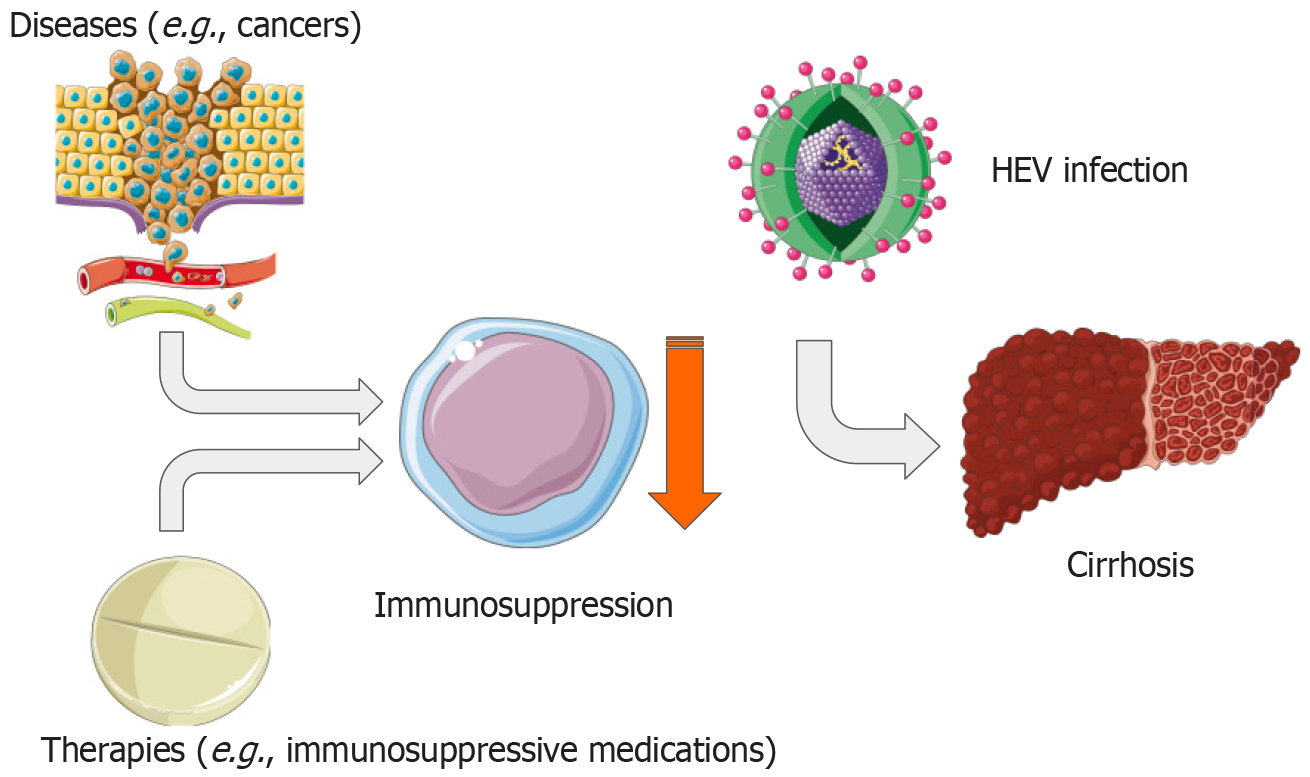
The damaging effects of HEV infection can be amplified due to the increased prevalence of disease-related or treatment-induced immunosuppression in contemporary healthcare (Figure 4).
Research by Pischke et al[99] revealed that transplant recipients and immunosuppressed patients, after exposure to HEV in blood products, showed symptoms of liver disease within the standard incubation period of 50–60 d[99]. These patients are at risk of chronic HEV infection, typically defined as the ongoing detection of HEV RNA in blood and/or feces for over 3–6 months[100].
The presence of long-term HEV infection was first observed in European transplant recipients in 2008[101-103]. Initially, 14 cases of acute HEV infection were reported among liver or kidney transplant recipients[104]. From these, eight developed persistent HEV infections marked by high alanine aminotransferase levels, inflammation in the portal vein, minor necrosis, and evidence of fibrosis within a year post-infection.
Nevertheless, the exact prevalence of lingering acute HEV infection among transplant recipients remains disputed. According to a retrospective, multicenter European study, it was found that 66% of transplant recipients suffering from symptomatic acute HEV infection later developed chronic hepatitis E[105]. However, the study’s retrospective nature suggests possible undetected asymptomatic HEV infections. Previous studies accounting for both symptomatic and asymptomatic HEV infections among transplant recipients suggest progression to chronicity ranges from 21%–50%[106-108].
The chance of transplant recipients developing a long-term HEV infection can vary based on the specific immunosuppressive treatment and the degree of immunosuppression[73]. The use of Tacrolimus has been associated with a higher risk of chronic hepatitis compared to Cyclosporine A[105]. In contrast, a lower risk of chronic hepatitis was observed with the use of Mycophenolic acid[107]. Interestingly, laboratory tests reveal that calcineurin inhibitors could potentially increase HEV replication, while Mycophenolic acid seems to inhibit it[109]. A large-scale study involving Japanese liver transplant recipients noted a very low prevalence of chronic HEV infection, suggesting possible variations in HEV subtypes, strains, or host genetic factors influencing the persistence of the virus in the body[110,111]. While no instances of fulminant liver failure linked to HEV infection in transplant patients have been reported, there is a risk of ongoing infection and chronic hepatitis. Chronic HEV infections are mostly seen with genotype 3, with a few recorded cases involving genotype 4[112,113]. Recently, persistent HEV infections from genotypes 7 and 8, originating from camels, were identified in transplant recipients[114]. To date, no chronic HEV infections have been reported resulting from genotypes 1 or 2.
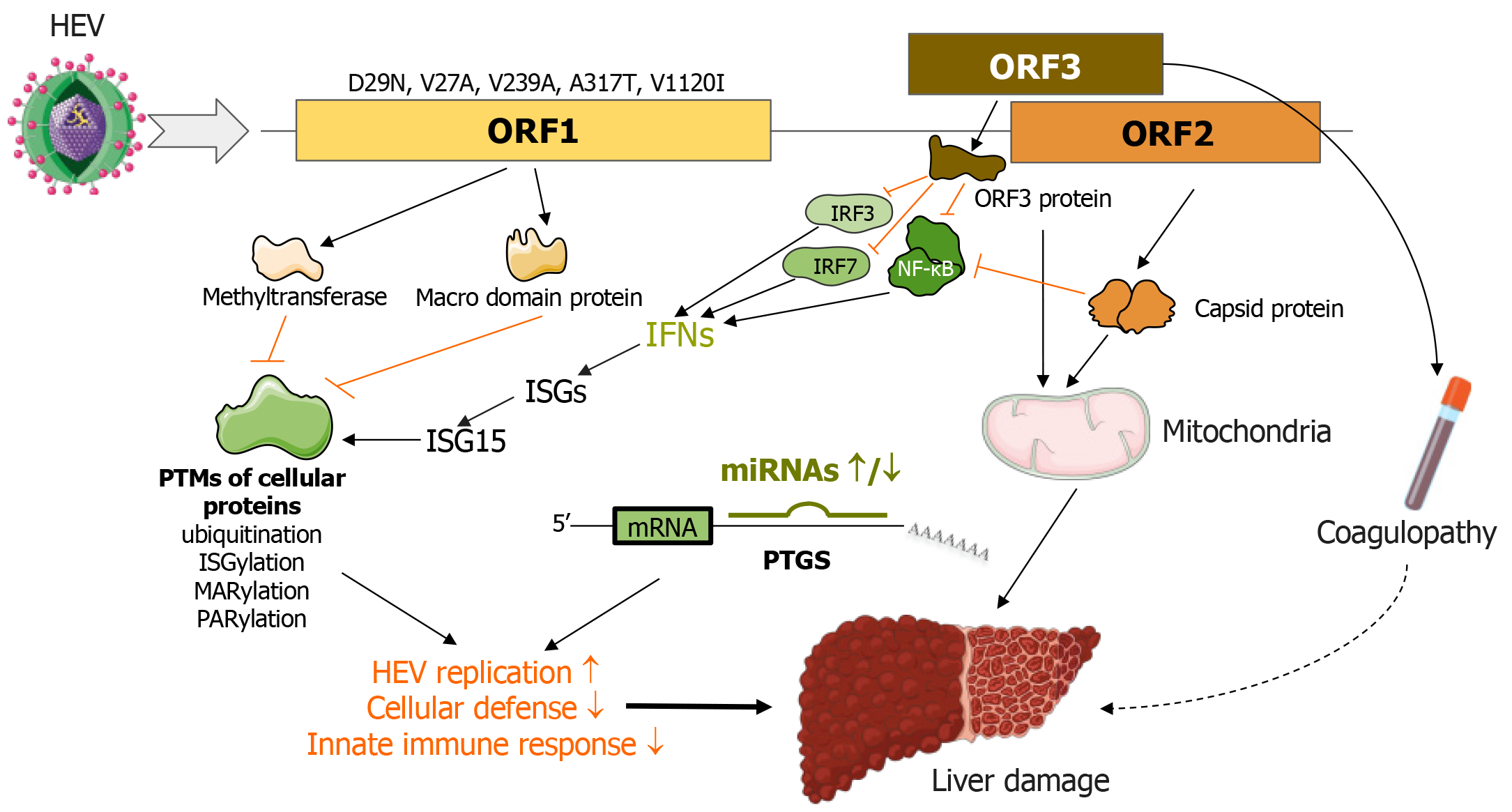
Despite the unclear roles of viral proteins in HEV infection and pathogenesis, intriguing evidence suggests that the genotype and specific mutations in the ORF1 region could affect factors such as the virus’s replicative potential, viremia levels, immune response characteristics, cell damage degree, and disease progression.
It has been reported that genotype 1 HEV shows more efficient replication, causing a greater cytopathic effect and higher levels of IL-6, CCL-3, CCL-4, and CXCL10 than genotype 3. This difference was observed in lab-cultured tissue explants from the decidua basalis and placenta[96]. Genotype 1 reduced the production of type III IFNs, while genotype 3 had no impact. The impact of more efficient replication by genotype 1 HEV might explain the severe progression of HEV infection during pregnancy[96].
In addition, the V239A mutation in the helicase produced by ORF1 has been associated with acute hepatitis cases, potentially suggesting greater virulence of genotype 3[115]. Similarly, 11 mutations in various ORF1 regions that seem to affect HEV infection severity have been identified. These include V27A, D29N, H105R, F179S, A317T, T735I, L1110F, V1120I, F1439, C1483, and N1530T[116-119].
Particularly, the D29N and V27A mutations in the ORF1-encoded methyltransferase are strongly linked with adverse outcomes in acute liver failure cases[117]. Genotype 1 HEV strains carrying the A317T and V1120I mutations–found in the Y and helicase regions of ORF1, respectively–exhibit enhanced replicative potential and are frequently seen in patients experiencing fulminant hepatic failure (Figure 5)[119].
ORF1 encodes multiple proteins responsible for managing the post-translational modification (PTM) of cellular proteins, which impacts protein stability and function. Notably, these proteins can influence deconjugation of a variety of substrates, such as a specific fluorescent probe (7-amino-4-methylcoumarin)-labeled ubiquitin, ISG15, Nedd8, and Small Ubiquitin-like Modifier (SUMO)[19]. Although the HEV methyltransferase-PCP displayed marginally impactful deneddylation and deSUMOylation effects, its deubiquitination and deISGylation activities were potent, potentially severely hampering the antiviral effects of IFNs[19].
Separate studies have revealed that the Macro domain protein, encoded by ORF1, interferes with cellular ADP-ribosylation[23]. There are two known forms of ADP-ribosylation: Poly (ADP-ribosyl) ation and mono (ADP-ribosyl) ation, commonly referred to as PARylation and MARylation, respectively[120,121]. These modifications regulate a myriad of cellular processes, including DNA damage response, unfolded protein response, cell cycle progression, cell death, metabolism, and immune responses[122-125]. Enzymes in both mammals and microbes can attach, erase, or recognize ADP-ribosyl moieties of target proteins[126].
The Macro domain protein in HEV operates as an ADP-ribose-protein hydrolase that removes mono-ADP-ribose and/or poly (ADP-ribose) modifications from target proteins, processes known as deMARylation and dePARylation, respectively[23]. Given the extensive effects of ADP-ribosylation, it is hypothesized that the Macro domain protein is a key player in the way HEV neutralizes the antiviral effect of IFNs and disrupts immune responses (Figure 5)[23].
These findings present a potential new therapeutic target, and inhibitors of the HEV Macro domain protein are deemed promising treatments for severe hepatitis E4[127-129]. Furthermore, PTMs have been associated with the disease mechanisms of several conditions, including alcohol-induced liver injury, MASLD/MASH, and cirrhosis[128-133]. Additional research is needed to explore how HEV infection influences the abnormal PTM patterns linked with preexisting liver diseases and its potential effects on disease progression.
The capsid protein produced by ORF2 can interfere with IFN activation by preventing IRF-3 phosphorylation. This obstruction stems from its interaction with certain proteins, including MAVS, TANK-binding kinase 1, and IRF3 (Figure 5)[134]. It has recently been discovered that ORF2 encodes a unique protein that undergoes glycosylation and can be released as a dimer into either the patient’s bloodstream or the supernatants of HEV-infected cell cultures[135]. The proteins that ORF2 expresses may interfere with or evade the host’s innate immunity. Furthermore, research suggests the capsid protein aids in HEV’s survival or replication within infected hepatocytes. For example, it prevents cellular NF-κB activity in human liver cancer cells by obstructing the ubiquitin-mediated proteasomal degradation of IκBα, potentially enhancing the survival of HEV-infected hepatocytes (Figure 5)[136].
The protein produced by ORF3 is believed to cause coagulopathy linked with HEV, disturbing the balance of the coagulation and fibrinolysis processes[137]. In addition, the ORF3 protein has a role in controlling carbohydrate metabolism and mitochondrial function[138]. Importantly, it also contributes to the disruption of immune response. This HEV protein downregulates the expression of certain cytokine genes, such as IFNs, by reducing the levels of TLR3 and TLR7 and the activation of NF-κB, IRF3, and IRF7[139]. It also inhibits STAT-1 phosphorylation, which dampens IFN signaling and antiviral ISG induction (Figure 5)[54,55].
Cellular miRNAs are non-coding RNA molecules that regulate gene expression by inducing post-transcriptional gene silencing. With a broad impact range, miRNAs are crucial regulators in many physiological processes and pathological conditions. They are involved in liver regeneration, metabolism, fibrosis, inflammation, and carcinogenesis, with contributions to the pathogenesis of diseases such as cirrhosis, ACLF, MASLD/MASH, and liver cancer[140-143].
Several viruses, including HEV, are known to encode miRNAs[53]. To illustrate, HEV-miR-A6 aids viral replication and deters the host’s innate immune response by inhibiting the expression of type I IFNs[53]. Additionally, the cellular miR-122 could boost HEV replication[144].
Studies on pregnant women’s PBMCs reveal that HEV infection alters cellular miRNAs’ expression patterns. They identified miR-450b as a significant indicator of mortality in acute liver failure amongst HEV-infected pregnant women[145]. Consistent with this, miRNA profiling shows that miRNAs circulating in HEV-infected patients’ plasma could indicate self-resolving acute and chronic hepatitis E, as well as acute liver failure[146,147].
Furthermore, miRNA signatures identified in patients’ sera could link with hepatitis E-specific immune dysfunction[146,148]. It is noted that miRNAs can be packed in exosomes, which HEV virion particles use for egress[44]. Comparing the miRNA profiles of exosomes from healthy and HEV-infected blood donors, it was evident that HEV infection significantly alters the miRNA content of exosomes[149].
These fascinating studies have stirred interest in the potential use of miRNAs-targeting compounds as therapeutic options for severe hepatitis E.
These viral proteins play complex roles in HEV infection and disease development, highlighting the need for more research to fully understand their functions and possible treatment targets. More studies in this field could help develop effective methods for managing HEV-related liver failure and other complications. A deeper understanding of the virus also encourages investigations into new treatment targets, like miRNAs and enzymes involved in PTMs, thus expanding the range of potential drug targets.
Cirrhosis is associated with profound liver, intestinal and immune dysfunction. In patients with cirrhosis, the composition of the intestinal microbiota changes significantly, the ratio of commensal bacteria decreases, whereas the proportion of pathogenic species increases. The intestinal barrier function is damaged, leaky gut syndrome develops, and the continuous flow of intestinal bacteria and their various products into the bloodstream leads to persistent stimulation of the immune system. As a result, a profound functional impairment develops in the innate and adaptive immunity. HEV is a common cause of acute hepatitis, in healthy individuals, the infection is mostly asymptomatic or associated with minor symptoms. However, HEV can establish a chronic infection in immunosuppressed individuals. The most surprising and frightening feature of hepatitis E is that it can lead to acute decompensation of cirrhosis, and fulminant hepatic failure with a high mortality rate in pregnant women. Interesting observations suggest that the joint effects of immunopathological processes are responsible for the severe course of hepatitis E among at-risk individuals. The discovery of unknown elements of the HEV-specific immune response may facilitate the development of more efficient monitoring and therapeutic strategies.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Virology
Country/Territory of origin: Hungary
Peer-review report’s classification
Scientific Quality: Grade B, Grade C
Novelty: Grade B, Grade C
Creativity or Innovation: Grade B, Grade C
Scientific Significance: Grade B, Grade C
P-Reviewer: Dietrich CF, Switzerland; Gaman MA, Romania S-Editor: Liu H L-Editor: A P-Editor: Wang WB
| 1. | Graff J, Torian U, Nguyen H, Emerson SU. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol. 2006;80:5919-5926. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 218] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 2. | Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology. 1991;185:120-131. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 837] [Cited by in RCA: 807] [Article Influence: 23.7] [Reference Citation Analysis (0)] |
| 3. | Horvatits T, Schulze Zur Wiesch J, Lütgehetmann M, Lohse AW, Pischke S. The Clinical Perspective on Hepatitis E. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 78] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 4. | Aslan AT, Balaban HY. Hepatitis E virus: Epidemiology, diagnosis, clinical manifestations, and treatment. World J Gastroenterol. 2020;26:5543-5560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 61] [Cited by in RCA: 112] [Article Influence: 22.4] [Reference Citation Analysis (4)] |
| 5. | Péron JM. Hepatitis E Virus Infection and Cirrhosis of the Liver. Gastroenterol Hepatol (N Y). 2016;12:565-567. [PubMed] |
| 6. | Akyüz F, Çavuş B, Pınarbaşı B, Bozacı M, Baran B, Akyuz U, Uyanıkoglu A, Demir K, Beşışık F, Özdil S, Boztaş G, Mungan Z, Badur S, Yenen S, Kaymakoglu S. Cryptogenic liver cirrhosis and hepatitis E virus (HEV): Are they related? Ann Hepatol. 2019;18:585-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 7. | Fantilli A, López Villa SD, Zerega A, Di Cola G, López L, Wassaf Martínez M, Pisano MB, Ré VE. Hepatitis E virus infection in a patient with alcohol related chronic liver disease: a case report of acute-on-chronic liver failure. Virol J. 2021;18:245. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 8. | Wang Y, Liu H, Jiang Y, Pan Q, Zhao J. Poor Outcomes of Acute Hepatitis E in Patients With Cirrhotic Liver Diseases Regardless of Etiology. Open Forum Infect Dis. 2020;7:ofaa107. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 9. | Kumar Acharya S, Kumar Sharma P, Singh R, Kumar Mohanty S, Madan K, Kumar Jha J, Kumar Panda S. Hepatitis E virus (HEV) infection in patients with cirrhosis is associated with rapid decompensation and death. J Hepatol. 2007;46:387-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 206] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 10. | Kamar N, Mansuy JM, Cointault O, Selves J, Abravanel F, Danjoux M, Otal P, Esposito L, Durand D, Izopet J, Rostaing L. Hepatitis E virus-related cirrhosis in kidney- and kidney-pancreas-transplant recipients. Am J Transplant. 2008;8:1744-1748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 167] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 11. | Choi JW, Son HJ, Lee SS, Jeon H, Cho JK, Kim HJ, Cha RR, Lee JM, Jung WT, Lee OJ. Acute hepatitis E virus superinfection increases mortality in patients with cirrhosis. BMC Infect Dis. 2022;22:62. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 12. | Kourtis AP, Read JS, Jamieson DJ. Pregnancy and infection. N Engl J Med. 2014;370:2211-2218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 459] [Cited by in RCA: 495] [Article Influence: 45.0] [Reference Citation Analysis (0)] |
| 13. | Yadav KK, Boley PA, Fritts Z, Kenney SP. Ectopic Expression of Genotype 1 Hepatitis E Virus ORF4 Increases Genotype 3 HEV Viral Replication in Cell Culture. Viruses. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 14. | Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, Shalimar, Nayak B, Ranjith Kumar CT, Surjit M. Endoplasmic Reticulum Stress Induced Synthesis of a Novel Viral Factor Mediates Efficient Replication of Genotype-1 Hepatitis E Virus. PLoS Pathog. 2016;12:e1005521. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 132] [Cited by in RCA: 184] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 15. | Ropp SL, Tam AW, Beames B, Purdy M, Frey TK. Expression of the hepatitis E virus ORF1. Arch Virol. 2000;145:1321-1337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 16. | Holla RP, Ahmad I, Ahmad Z, Jameel S. Molecular virology of hepatitis E virus. Semin Liver Dis. 2013;33:3-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 85] [Cited by in RCA: 91] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 17. | Magden J, Takeda N, Li T, Auvinen P, Ahola T, Miyamura T, Merits A, Kääriäinen L. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol. 2001;75:6249-6255. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 128] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Hooda P, Chaudhary M, Parvez MK, Sinha N, Sehgal D. Inhibition of Hepatitis E Virus Replication by Novel Inhibitor Targeting Methyltransferase. Viruses. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 19. | Karpe YA, Lole KS. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol. 2011;92:2088-2092. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 51] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 20. | Karpe YA, Lole KS. NTPase and 5' to 3' RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol. 2010;84:3595-3602. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 21. | Fairman-Williams ME, Guenther UP, Jankowsky E. SF1 and SF2 helicases: family matters. Curr Opin Struct Biol. 2010;20:313-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 810] [Cited by in RCA: 713] [Article Influence: 47.5] [Reference Citation Analysis (0)] |
| 22. | Koonin EV, Gorbalenya AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A. 1992;89:8259-8263. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 381] [Cited by in RCA: 386] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 23. | Li C, Debing Y, Jankevicius G, Neyts J, Ahel I, Coutard B, Canard B. Viral Macro Domains Reverse Protein ADP-Ribosylation. J Virol. 2016;90:8478-8486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 105] [Cited by in RCA: 139] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 24. | Smith DB, Vanek J, Ramalingam S, Johannessen I, Templeton K, Simmonds P. Evolution of the hepatitis E virus hypervariable region. J Gen Virol. 2012;93:2408-2418. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 25. | Zhou Z, Xie Y, Wu C, Nan Y. The Hepatitis E Virus Open Reading Frame 2 Protein: Beyond Viral Capsid. Front Microbiol. 2021;12:739124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 26. | Li TC, Takeda N, Miyamura T, Matsuura Y, Wang JC, Engvall H, Hammar L, Xing L, Cheng RH. Essential elements of the capsid protein for self-assembly into empty virus-like particles of hepatitis E virus. J Virol. 2005;79:12999-13006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 102] [Cited by in RCA: 104] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 27. | Xing L, Wang JC, Li TC, Yasutomi Y, Lara J, Khudyakov Y, Schofield D, Emerson SU, Purcell RH, Takeda N, Miyamura T, Cheng RH. Spatial configuration of hepatitis E virus antigenic domain. J Virol. 2011;85:1117-1124. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 72] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 28. | Yamashita T, Mori Y, Miyazaki N, Cheng RH, Yoshimura M, Unno H, Shima R, Moriishi K, Tsukihara T, Li TC, Takeda N, Miyamura T, Matsuura Y. Biological and immunological characteristics of hepatitis E virus-like particles based on the crystal structure. Proc Natl Acad Sci U S A. 2009;106:12986-12991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 201] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 29. | Zafrullah M, Ozdener MH, Kumar R, Panda SK, Jameel S. Mutational analysis of glycosylation, membrane translocation, and cell surface expression of the hepatitis E virus ORF2 protein. J Virol. 1999;73:4074-4082. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 101] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 30. | Montpellier C, Wychowski C, Sayed IM, Meunier JC, Saliou JM, Ankavay M, Bull A, Pillez A, Abravanel F, Helle F, Brochot E, Drobecq H, Farhat R, Aliouat-Denis CM, Haddad JG, Izopet J, Meuleman P, Goffard A, Dubuisson J, Cocquerel L. Hepatitis E Virus Lifecycle and Identification of 3 Forms of the ORF2 Capsid Protein. Gastroenterology. 2018;154:211-223.e8. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 154] [Article Influence: 22.0] [Reference Citation Analysis (0)] |
| 31. | Chen Z, Guo S, Li G, Ying D, Wen G, Fang M, Wang Y, Tang Z, Zheng Z, Xia N. A Secreted Form of the Hepatitis E Virus ORF2 Protein: Design Strategy, Antigenicity and Immunogenicity. Viruses. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 32. | Kannan H, Fan S, Patel D, Bossis I, Zhang YJ. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol. 2009;83:6375-6382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 84] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 33. | Emerson SU, Nguyen H, Torian U, Purcell RH. ORF3 protein of hepatitis E virus is not required for replication, virion assembly, or infection of hepatoma cells in vitro. J Virol. 2006;80:10457-10464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 98] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 34. | Zafrullah M, Ozdener MH, Panda SK, Jameel S. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol. 1997;71:9045-9053. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 195] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 35. | Korkaya H, Jameel S, Gupta D, Tyagi S, Kumar R, Zafrullah M, Mazumdar M, Lal SK, Xiaofang L, Sehgal D, Das SR, Sahal D. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem. 2001;276:42389-42400. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 122] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 36. | Tyagi S, Surjit M, Roy AK, Jameel S, Lal SK. The ORF3 protein of hepatitis E virus interacts with liver-specific alpha1-microglobulin and its precursor alpha1-microglobulin/bikunin precursor (AMBP) and expedites their export from the hepatocyte. J Biol Chem. 2004;279:29308-29319. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 65] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 37. | Ratra R, Kar-Roy A, Lal SK. The ORF3 protein of hepatitis E virus interacts with hemopexin by means of its 26 amino acid N-terminal hydrophobic domain II. Biochemistry. 2008;47:1957-1969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 38. | Ding Q, Heller B, Capuccino JM, Song B, Nimgaonkar I, Hrebikova G, Contreras JE, Ploss A. Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc Natl Acad Sci U S A. 2017;114:1147-1152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 174] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 39. | Sayed IM, Seddik MI, Gaber MA, Saber SH, Mandour SA, El-Mokhtar MA. Replication of Hepatitis E Virus (HEV) in Primary Human-Derived Monocytes and Macrophages In Vitro. Vaccines (Basel). 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 40. | Kalia M, Chandra V, Rahman SA, Sehgal D, Jameel S. Heparan sulfate proteoglycans are required for cellular binding of the hepatitis E virus ORF2 capsid protein and for viral infection. J Virol. 2009;83:12714-12724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 167] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 41. | Holla P, Ahmad I, Ahmed Z, Jameel S. Hepatitis E virus enters liver cells through a dynamin-2, clathrin and membrane cholesterol-dependent pathway. Traffic. 2015;16:398-416. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 55] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 42. | Panda SK, Ansari IH, Durgapal H, Agrawal S, Jameel S. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol. 2000;74:2430-2437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 115] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 43. | Oechslin N, Da Silva N, Szkolnicka D, Cantrelle FX, Hanoulle X, Moradpour D, Gouttenoire J. Hepatitis E virus RNA-dependent RNA polymerase is involved in RNA replication and infectious particle production. Hepatology. 2022;75:170-181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 44. | Nagashima S, Takahashi M, Kobayashi T; Tanggis, Nishizawa T, Nishiyama T, Primadharsini PP, Okamoto H. Characterization of the Quasi-Enveloped Hepatitis E Virus Particles Released by the Cellular Exosomal Pathway. J Virol. 2017;91. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 167] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 45. | Devhare P, Madiyal M, Mukhopadhyay C, Shetty S, Shastry S. Interplay between Hepatitis E Virus and Host Cell Pattern Recognition Receptors. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 46. | Arya RP, Arankalle VA. Toll like receptors in self-recovering hepatitis E patients with or without pregnancy. Hum Immunol. 2014;75:1147-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 47. | Majumdar M, Ratho RK, Chawla Y, Singh MP. Role of TLR gene expression and cytokine profiling in the immunopathogenesis of viral hepatitis E. J Clin Virol. 2015;73:8-13. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 48. | Devhare PB, Chatterjee SN, Arankalle VA, Lole KS. Analysis of antiviral response in human epithelial cells infected with hepatitis E virus. PLoS One. 2013;8:e63793. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 59] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 49. | Murata K, Kang JH, Nagashima S, Matsui T, Karino Y, Yamamoto Y, Atarashi T, Oohara M, Uebayashi M, Sakata H, Matsubayashi K, Takahashi K, Arai M, Mishiro S, Sugiyama M, Mizokami M, Okamoto H. IFN-λ3 as a host immune response in acute hepatitis E virus infection. Cytokine. 2020;125:154816. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 50. | Tripathy AS, Das R, Rathod SB, Arankalle VA. Cytokine profiles, CTL response and T cell frequencies in the peripheral blood of acute patients and individuals recovered from hepatitis E infection. PLoS One. 2012;7:e31822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 51. | Sooryanarain H, Heffron CL, Meng XJ. The U-Rich Untranslated Region of the Hepatitis E Virus Induces Differential Type I and Type III Interferon Responses in a Host Cell-Dependent Manner. mBio. 2020;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 52. | Wang W, Wang Y, Qu C, Wang S, Zhou J, Cao W, Xu L, Ma B, Hakim MS, Yin Y, Li T, Peppelenbosch MP, Zhao J, Pan Q. The RNA genome of hepatitis E virus robustly triggers an antiviral interferon response. Hepatology. 2018;67:2096-2112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 41] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 53. | Qian Z, Yang C, Xu L, Mickael HK, Chen S, Zhang Y, Xia Y, Li T, Yu W, Huang F. Hepatitis E virus-encoded microRNA promotes viral replication by inhibiting type I interferon. FASEB J. 2022;36:e22104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 54. | Bagdassarian E, Doceul V, Pellerin M, Demange A, Meyer L, Jouvenet N, Pavio N. The Amino-Terminal Region of Hepatitis E Virus ORF1 Containing a Methyltransferase (Met) and a Papain-Like Cysteine Protease (PCP) Domain Counteracts Type I Interferon Response. Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 12] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 55. | Kang S, Choi C, Choi I, Han KN, Rho SW, Choi J, Kwon J, Park MK, Kim SJ, Myoung J. Hepatitis E Virus Methyltransferase Inhibits Type I Interferon Induction by Targeting RIG-I. J Microbiol Biotechnol. 2018;28:1554-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 17] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 56. | Hoofnagle JH, Nelson KE, Purcell RH. Hepatitis E. N Engl J Med. 2012;367:1237-1244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 376] [Cited by in RCA: 382] [Article Influence: 29.4] [Reference Citation Analysis (0)] |
| 57. | Riveiro-Barciela M, Rando-Segura A, Barreira-Díaz A, Bes M, P Ruzo S, Piron M, Quer J, Sauleda S, Rodríguez-Frías F, Esteban R, Buti M. Unexpected long-lasting anti-HEV IgM positivity: Is HEV antigen a better serological marker for hepatitis E infection diagnosis? J Viral Hepat. 2020;27:747-753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 58. | Walker CM. Adaptive Immune Responses in Hepatitis A Virus and Hepatitis E Virus Infections. Cold Spring Harb Perspect Med. 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 59. | Dalton HR, Bendall RP, Keane FE, Tedder RS, Ijaz S. Persistent carriage of hepatitis E virus in patients with HIV infection. N Engl J Med. 2009;361:1025-1027. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 408] [Cited by in RCA: 414] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 60. | Tian DY, Chen Y, Xia NS. Significance of serum IgA in patients with acute hepatitis E virus infection. World J Gastroenterol. 2006;12:3919-3923. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 27] [Cited by in RCA: 23] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 61. | Clayson ET, Myint KS, Snitbhan R, Vaughn DW, Innis BL, Chan L, Cheung P, Shrestha MP. Viremia, fecal shedding, and IgM and IgG responses in patients with hepatitis E. J Infect Dis. 1995;172:927-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 120] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 62. | Schemmerer M, Rauh C, Jilg W, Wenzel JJ. Time course of hepatitis E-specific antibodies in adults. J Viral Hepat. 2017;24:75-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 63. | Lu J, Huang Y, Wang P, Li Q, Li Z, Jiang J, Guo Q, Gui H, Xie Q. Dynamics of Hepatitis E Virus (HEV) Antibodies and Development of a Multifactorial Model To Improve the Diagnosis of HEV Infection in Resource-Limited Settings. J Clin Microbiol. 2021;59. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 64. | Tang ZM, Tang M, Zhao M, Wen GP, Yang F, Cai W, Wang SL, Zheng ZZ, Xia NS. A novel linear neutralizing epitope of hepatitis E virus. Vaccine. 2015;33:3504-3511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 65. | You M, Xin L, Yang Y, Zhang X, Chen Y, Yu H, Li S, Zhang J, An Z, Luo W, Xia N. Investigation of a special neutralizing epitope of HEV E2s. Protein Cell. 2014;5:950-953. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 66. | Zhao M, Li XJ, Tang ZM, Yang F, Wang SL, Cai W, Zhang K, Xia NS, Zheng ZZ. A Comprehensive Study of Neutralizing Antigenic Sites on the Hepatitis E Virus (HEV) Capsid by Constructing, Clustering, and Characterizing a Tool Box. J Biol Chem. 2015;290:19910-19922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 49] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 67. | Tang X, Yang C, Gu Y, Song C, Zhang X, Wang Y, Zhang J, Hew CL, Li S, Xia N, Sivaraman J. Structural basis for the neutralization and genotype specificity of hepatitis E virus. Proc Natl Acad Sci U S A. 2011;108:10266-10271. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 96] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 68. | Takahashi M, Tanaka T, Takahashi H, Hoshino Y, Nagashima S, Jirintai, Mizuo H, Yazaki Y, Takagi T, Azuma M, Kusano E, Isoda N, Sugano K, Okamoto H. Hepatitis E Virus (HEV) strains in serum samples can replicate efficiently in cultured cells despite the coexistence of HEV antibodies: characterization of HEV virions in blood circulation. J Clin Microbiol. 2010;48:1112-1125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 191] [Cited by in RCA: 221] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 69. | Das A, Rivera-Serrano EE, Yin X, Walker CM, Feng Z, Lemon SM. Cell entry and release of quasi-enveloped human hepatitis viruses. Nat Rev Microbiol. 2023;21:573-589. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 32] [Article Influence: 16.0] [Reference Citation Analysis (0)] |
| 70. | Chandler TL, Yang A, Otero CE, Permar SR, Caddy SL. Protective mechanisms of nonneutralizing antiviral antibodies. PLoS Pathog. 2023;19:e1011670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 29] [Article Influence: 14.5] [Reference Citation Analysis (0)] |
| 71. | Brown A, Halliday JS, Swadling L, Madden RG, Bendall R, Hunter JG, Maggs J, Simmonds P, Smith DB, Vine L, McLaughlin C, Collier J, Bonsall D, Jeffery K, Dunachie S, Klenerman P, Izopet J, Kamar N, Dalton HR, Barnes E. Characterization of the Specificity, Functionality, and Durability of Host T-Cell Responses Against the Full-Length Hepatitis E Virus. Hepatology. 2016;64:1934-1950. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 72. | Srivastava R, Aggarwal R, Jameel S, Puri P, Gupta VK, Ramesh VS, Bhatia S, Naik S. Cellular immune responses in acute hepatitis E virus infection to the viral open reading frame 2 protein. Viral Immunol. 2007;20:56-65. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 73. | Suneetha PV, Pischke S, Schlaphoff V, Grabowski J, Fytili P, Gronert A, Bremer B, Markova A, Jaroszewicz J, Bara C, Manns MP, Cornberg M, Wedemeyer H. Hepatitis E virus (HEV)-specific T-cell responses are associated with control of HEV infection. Hepatology. 2012;55:695-708. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 138] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 74. | Kemming J, Gundlach S, Panning M, Huzly D, Huang J, Lütgehetmann M, Pischke S, Schulze Zur Wiesch J, Emmerich F, Llewellyn-Lacey S, Price DA, Tanriver Y, Warnatz K, Boettler T, Thimme R, Hofmann M, Fischer N, Neumann-Haefelin C. Mechanisms of CD8+ T-cell failure in chronic hepatitis E virus infection. J Hepatol. 2022;77:978-990. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 75. | Tripathy AS, Das R, Rathod SB, Gurav YK, Arankalle VA. Peripheral T regulatory cells and cytokines in hepatitis E infection. Eur J Clin Microbiol Infect Dis. 2012;31:179-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 76. | Srivastava R, Aggarwal R, Bhagat MR, Chowdhury A, Naik S. Alterations in natural killer cells and natural killer T cells during acute viral hepatitis E. J Viral Hepat. 2008;15:910-916. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 77. | El Costa H, Gouilly J, Abravanel F, Bahraoui E, Peron JM, Kamar N, Jabrane-Ferrat N, Izopet J. Effector memory CD8 T cell response elicits Hepatitis E Virus genotype 3 pathogenesis in the elderly. PLoS Pathog. 2021;17:e1009367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 15] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 78. | Prabhu SB, Gupta P, Durgapal H, Rath S, Gupta SD, Acharya SK, Panda SK. Study of cellular immune response against Hepatitis E virus (HEV). J Viral Hepat. 2011;18:587-594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 79. | Olson JC, Kamath PS. Acute-on-chronic liver failure: what are the implications? Curr Gastroenterol Rep. 2012;14:63-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 80. | Verbeke L, Nevens F, Laleman W. Bench-to-beside review: acute-on-chronic liver failure - linking the gut, liver and systemic circulation. Crit Care. 2011;15:233. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 37] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 81. | Jalan R, Gines P, Olson JC, Mookerjee RP, Moreau R, Garcia-Tsao G, Arroyo V, Kamath PS. Acute-on chronic liver failure. J Hepatol. 2012;57:1336-1348. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 454] [Article Influence: 34.9] [Reference Citation Analysis (1)] |
| 82. | Wasmuth HE, Kunz D, Yagmur E, Timmer-Stranghöner A, Vidacek D, Siewert E, Bach J, Geier A, Purucker EA, Gressner AM, Matern S, Lammert F. Patients with acute on chronic liver failure display "sepsis-like" immune paralysis. J Hepatol. 2005;42:195-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 366] [Cited by in RCA: 393] [Article Influence: 19.7] [Reference Citation Analysis (0)] |
| 83. | Rolando N, Wade J, Davalos M, Wendon J, Philpott-Howard J, Williams R. The systemic inflammatory response syndrome in acute liver failure. Hepatology. 2000;32:734-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 506] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 84. | Wu C, Wu X, Xia J. Hepatitis E virus infection during pregnancy. Virol J. 2020;17:73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 85. | Jilani N, Das BC, Husain SA, Baweja UK, Chattopadhya D, Gupta RK, Sardana S, Kar P. Hepatitis E virus infection and fulminant hepatic failure during pregnancy. J Gastroenterol Hepatol. 2007;22:676-682. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 159] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 86. | Yang C, Yu W, Bi Y, Long F, Li Y, Wei D, Hao X, Situ J, Zhao Y, Huang F. Increased oestradiol in hepatitis E virus-infected pregnant women promotes viral replication. J Viral Hepat. 2018;25:742-751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 87. | Bi Y, Yang C, Yu W, Zhao X, Zhao C, He Z, Jing S, Wang H, Huang F. Pregnancy serum facilitates hepatitis E virus replication in vitro. J Gen Virol. 2015;96:1055-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 27] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 88. | Lhomme S, Marion O, Abravanel F, Chapuy-Regaud S, Kamar N, Izopet J. Hepatitis E Pathogenesis. Viruses. 2016;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 69] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 89. | Pazos M, Sperling RS, Moran TM, Kraus TA. The influence of pregnancy on systemic immunity. Immunol Res. 2012;54:254-261. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 146] [Cited by in RCA: 170] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 90. | Capai L, Charrel R, Falchi A. Hepatitis E in High-Income Countries: What Do We Know? Viruses. 2018;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 91. | Lachish T, Erez O, Daudi N, Shouval D, Schwartz E. Acute hepatitis E virus in pregnant women in Israel and in other industrialized countries. J Clin Virol. 2015;73:20-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 29] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 92. | Anty R, Ollier L, Péron JM, Nicand E, Cannavo I, Bongain A, Giordanengo V, Tran A. First case report of an acute genotype 3 hepatitis E infected pregnant woman living in South-Eastern France. J Clin Virol. 2012;54:76-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 70] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 93. | Tabatabai J, Wenzel JJ, Soboletzki M, Flux C, Navid MH, Schnitzler P. First case report of an acute hepatitis E subgenotype 3c infection during pregnancy in Germany. J Clin Virol. 2014;61:170-172. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 94. | Andersson MI, Hughes J, Gordon FH, Ijaz S, Donati M. Of pigs and pregnancy. Lancet. 2008;372:1192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 22] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 95. | Renou C, Gobert V, Locher C, Moumen A, Timbely O, Savary J, Roque-Afonso AM; Association Nationale des Hépato-Gastroentérologues des Hôpitaux Généraux (ANGH). Prospective study of Hepatitis E Virus infection among pregnant women in France. Virol J. 2014;11:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 96. | Gouilly J, Chen Q, Siewiera J, Cartron G, Levy C, Dubois M, Al-Daccak R, Izopet J, Jabrane-Ferrat N, El Costa H. Genotype specific pathogenicity of hepatitis E virus at the human maternal-fetal interface. Nat Commun. 2018;9:4748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 98] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 97. | van Tong H, Hoan NX, Wang B, Wedemeyer H, Bock CT, Velavan TP. Hepatitis E Virus Mutations: Functional and Clinical Relevance. EBioMedicine. 2016;11:31-42. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 98. | Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology. 2014;147:1008-11.e7; quiz e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 153] [Cited by in RCA: 162] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 99. | Pischke S, Hiller J, Lütgehetmann M, Polywka S, Rybczynski M, Ayuk F, Lohse AW. Blood-borne Hepatitis E Virus Transmission: A Relevant Risk for Immunosuppressed Patients. Clin Infect Dis. 2016;63:569-570. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 100. | Kamar N, Rostaing L, Legrand-Abravanel F, Izopet J. How should hepatitis E virus infection be defined in organ-transplant recipients? Am J Transplant. 2013;13:1935-1936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 103] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 101. | Gérolami R, Moal V, Colson P. Chronic hepatitis E with cirrhosis in a kidney-transplant recipient. N Engl J Med. 2008;358:859-860. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 259] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 102. | Haagsma EB, Niesters HG, van den Berg AP, Riezebos-Brilman A, Porte RJ, Vennema H, Reimerink JH, Koopmans MP. Prevalence of hepatitis E virus infection in liver transplant recipients. Liver Transpl. 2009;15:1225-1228. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 103. | Pischke S, Suneetha PV, Baechlein C, Barg-Hock H, Heim A, Kamar N, Schlue J, Strassburg CP, Lehner F, Raupach R, Bremer B, Magerstedt P, Cornberg M, Seehusen F, Baumgaertner W, Klempnauer J, Izopet J, Manns MP, Grummer B, Wedemeyer H. Hepatitis E virus infection as a cause of graft hepatitis in liver transplant recipients. Liver Transpl. 2010;16:74-82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 156] [Cited by in RCA: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 104. | Kamar N, Selves J, Mansuy JM, Ouezzani L, Péron JM, Guitard J, Cointault O, Esposito L, Abravanel F, Danjoux M, Durand D, Vinel JP, Izopet J, Rostaing L. Hepatitis E virus and chronic hepatitis in organ-transplant recipients. N Engl J Med. 2008;358:811-817. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1005] [Cited by in RCA: 995] [Article Influence: 58.5] [Reference Citation Analysis (0)] |
| 105. | Kamar N, Garrouste C, Haagsma EB, Garrigue V, Pischke S, Chauvet C, Dumortier J, Cannesson A, Cassuto-Viguier E, Thervet E, Conti F, Lebray P, Dalton HR, Santella R, Kanaan N, Essig M, Mousson C, Radenne S, Roque-Afonso AM, Izopet J, Rostaing L. Factors associated with chronic hepatitis in patients with hepatitis E virus infection who have received solid organ transplants. Gastroenterology. 2011;140:1481-1489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 488] [Article Influence: 34.9] [Reference Citation Analysis (0)] |
| 106. | Legrand-Abravanel F, Kamar N, Sandres-Saune K, Lhomme S, Mansuy JM, Muscari F, Sallusto F, Rostaing L, Izopet J. Hepatitis E virus infection without reactivation in solid-organ transplant recipients, France. Emerg Infect Dis. 2011;17:30-37. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 121] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 107. | Pischke S, Stiefel P, Franz B, Bremer B, Suneetha PV, Heim A, Ganzenmueller T, Schlue J, Horn-Wichmann R, Raupach R, Darnedde M, Scheibner Y, Taubert R, Haverich A, Manns MP, Wedemeyer H, Bara CL. Chronic hepatitis e in heart transplant recipients. Am J Transplant. 2012;12:3128-3133. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 132] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 108. | Pischke S, Hardtke S, Bode U, Birkner S, Chatzikyrkou C, Kauffmann W, Bara CL, Gottlieb J, Wenzel J, Manns MP, Wedemeyer H. Ribavirin treatment of acute and chronic hepatitis E: a single-centre experience. Liver Int. 2013;33:722-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 135] [Cited by in RCA: 139] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 109. | Wang Y, Zhou X, Debing Y, Chen K, Van Der Laan LJ, Neyts J, Janssen HL, Metselaar HJ, Peppelenbosch MP, Pan Q. Calcineurin inhibitors stimulate and mycophenolic acid inhibits replication of hepatitis E virus. Gastroenterology. 2014;146:1775-1783. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 145] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 110. | Inagaki Y, Oshiro Y, Tanaka T, Yoshizumi T, Okajima H, Ishiyama K, Nakanishi C, Hidaka M, Wada H, Hibi T, Takagi K, Honda M, Kuramitsu K, Tanaka H, Tohyama T, Ikegami T, Imura S, Shimamura T, Nakayama Y, Urahashi T, Yamagishi K, Ohnishi H, Nagashima S, Takahashi M, Shirabe K, Kokudo N, Okamoto H, Ohkohchi N. A Nationwide Survey of Hepatitis E Virus Infection and Chronic Hepatitis E in Liver Transplant Recipients in Japan. EBioMedicine. 2015;2:1607-1612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 41] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 111. | Pischke S, Wedemeyer H. Hepatitis E In Transplant Recipients: Why Is This Not A Problem In Japan? EBioMedicine. 2015;2:1564-1565. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 112. | Geng Y, Zhang H, Huang W, J Harrison T, Geng K, Li Z, Wang Y. Persistent hepatitis e virus genotype 4 infection in a child with acute lymphoblastic leukemia. Hepat Mon. 2014;14:e15618. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 56] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 113. | Geng Y, Zhao C, Huang W, Harrison TJ, Zhang H, Geng K, Wang Y. Detection and assessment of infectivity of hepatitis E virus in urine. J Hepatol. 2016;64:37-43. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 128] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 114. | Lee GH, Tan BH, Teo EC, Lim SG, Dan YY, Wee A, Aw PP, Zhu Y, Hibberd ML, Tan CK, Purdy MA, Teo CG. Chronic Infection With Camelid Hepatitis E Virus in a Liver Transplant Recipient Who Regularly Consumes Camel Meat and Milk. Gastroenterology. 2016;150:355-7.e3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 329] [Cited by in RCA: 406] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 115. | Takahashi K, Okamoto H, Abe N, Kawakami M, Matsuda H, Mochida S, Sakugawa H, Suginoshita Y, Watanabe S, Yamamoto K, Miyakawa Y, Mishiro S. Virulent strain of hepatitis E virus genotype 3, Japan. Emerg Infect Dis. 2009;15:704-709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 46] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 116. | Borkakoti J, Ahmed G, Kar P. Report of a novel C1483W mutation in the hepatitis E virus polymerase in patients with acute liver failure. Infect Genet Evol. 2016;44:51-54. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 117. | Borkakoti J, Ahmed G, Rai A, Kar P. Report of novel H105R, D29N, V27A mutations in the methyltransferase region of the HEV genome in patients with acute liver failure. J Clin Virol. 2017;91:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 118. | Mishra N, Walimbe AM, Arankalle VA. Hepatitis E virus from India exhibits significant amino acid mutations in fulminant hepatic failure patients. Virus Genes. 2013;46:47-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 119. | Wang B, Tian D, Sooryanarain H, Mahsoub HM, Heffron CL, Hassebroek AM, Meng XJ. Two mutations in the ORF1 of genotype 1 hepatitis E virus enhance virus replication and may associate with fulminant hepatic failure. Proc Natl Acad Sci U S A. 2022;119:e2207503119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Reference Citation Analysis (0)] |
| 120. | Daugherty MD, Young JM, Kerns JA, Malik HS. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet. 2014;10:e1004403. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 118] [Cited by in RCA: 140] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 121. | Challa S, Stokes MS, Kraus WL. MARTs and MARylation in the Cytosol: Biological Functions, Mechanisms of Action, and Therapeutic Potential. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 122. | Du Q, Miao Y, He W, Zheng H. ADP-Ribosylation in Antiviral Innate Immune Response. Pathogens. 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 123. | Lüscher B, Verheirstraeten M, Krieg S, Korn P. Intracellular mono-ADP-ribosyltransferases at the host-virus interphase. Cell Mol Life Sci. 2022;79:288. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 14] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 124. | Rosado MM, Pioli C. ADP-ribosylation in evasion, promotion and exacerbation of immune responses. Immunology. 2021;164:15-30. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 125. | Kang M, Park S, Park SH, Lee HG, Park JH. A Double-Edged Sword: The Two Faces of PARylation. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 25] [Reference Citation Analysis (0)] |
| 126. | Olzscha H. Posttranslational modifications and proteinopathies: how guardians of the proteome are defeated. Biol Chem. 2019;400:895-915. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 127. | Fu W, Yao H, Bütepage M, Zhao Q, Lüscher B, Li J. The search for inhibitors of macrodomains for targeting the readers and erasers of mono-ADP-ribosylation. Drug Discov Today. 2021;26:2547-2558. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 128. | Szántó M, Gupte R, Kraus WL, Pacher P, Bai P. PARPs in lipid metabolism and related diseases. Prog Lipid Res. 2021;84:101117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 85] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 129. | Wu X, Xu M, Geng M, Chen S, Little PJ, Xu S, Weng J. Targeting protein modifications in metabolic diseases: molecular mechanisms and targeted therapies. Signal Transduct Target Ther. 2023;8:220. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 91] [Article Influence: 45.5] [Reference Citation Analysis (0)] |
| 130. | Gligorijević N, Minić S, Nedić O. Structural changes of proteins in liver cirrhosis and consequential changes in their function. World J Gastroenterol. 2022;28:3780-3792. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 2] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (1)] |
| 131. | Osna NA, Carter WG, Ganesan M, Kirpich IA, McClain CJ, Petersen DR, Shearn CT, Tomasi ML, Kharbanda KK. Aberrant post-translational protein modifications in the pathogenesis of alcohol-induced liver injury. World J Gastroenterol. 2016;22:6192-6200. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 26] [Cited by in RCA: 18] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 132. | Rodríguez-Sanabria JS, Escutia-Gutiérrez R, Rosas-Campos R, Armendáriz-Borunda JS, Sandoval-Rodríguez A. An Update in Epigenetics in Metabolic-Associated Fatty Liver Disease. Front Med (Lausanne). 2021;8:770504. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 20] [Reference Citation Analysis (0)] |
| 133. | Zhong Q, Xiao X, Qiu Y, Xu Z, Chen C, Chong B, Zhao X, Hai S, Li S, An Z, Dai L. Protein posttranslational modifications in health and diseases: Functions, regulatory mechanisms, and therapeutic implications. MedComm (2020). 2023;4:e261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 58] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 134. | Lin S, Yang Y, Nan Y, Ma Z, Yang L, Zhang YJ. The Capsid Protein of Hepatitis E Virus Inhibits Interferon Induction via Its N-terminal Arginine-Rich Motif. Viruses. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 135. | Yin X, Ying D, Lhomme S, Tang Z, Walker CM, Xia N, Zheng Z, Feng Z. Origin, antigenicity, and function of a secreted form of ORF2 in hepatitis E virus infection. Proc Natl Acad Sci U S A. 2018;115:4773-4778. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 137] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 136. | Surjit M, Varshney B, Lal SK. The ORF2 glycoprotein of hepatitis E virus inhibits cellular NF-κB activity by blocking ubiquitination mediated proteasomal degradation of IκBα in human hepatoma cells. BMC Biochem. 2012;13:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 46] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 137. | Geng Y, Yang J, Huang W, Harrison TJ, Zhou Y, Wen Z, Wang Y. Virus host protein interaction network analysis reveals that the HEV ORF3 protein may interrupt the blood coagulation process. PLoS One. 2013;8:e56320. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 138. | Zhou Y, Zhao C, Tian Y, Xu N, Wang Y. Characteristics and Functions of HEV Proteins. Adv Exp Med Biol. 2016;948:17-38. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 139. | Lei Q, Li L, Zhang S, Li T, Zhang X, Ding X, Qin B. HEV ORF3 downregulates TLR7 to inhibit the generation of type I interferon via impairment of multiple signaling pathways. Sci Rep. 2018;8:8585. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 24] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 140. | Li QY, Gong T, Huang YK, Kang L, Warner CA, Xie H, Chen LM, Duan XQ. Role of noncoding RNAs in liver fibrosis. World J Gastroenterol. 2023;29:1446-1459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 141. | Ramezani M, Zobeiry M, Abdolahi S, Hatami B, Zali MR, Baghaei K. A crosstalk between epigenetic modulations and non-alcoholic fatty liver disease progression. Pathol Res Pract. 2023;251:154809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 12] [Reference Citation Analysis (0)] |
| 142. | Li H, Liu T, Yang Y, Cho WC, Flynn RJ, Harandi MF, Song H, Luo X, Zheng Y. Interplays of liver fibrosis-associated microRNAs: Molecular mechanisms and implications in diagnosis and therapy. Genes Dis. 2023;10:1457-1469. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 8] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 143. | Blake MJ, Steer CJ. Liver Regeneration in Acute on Chronic Liver Failure. Clin Liver Dis. 2023;27:595-616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 144. | Haldipur B, Bhukya PL, Arankalle V, Lole K. Positive Regulation of Hepatitis E Virus Replication by MicroRNA-122. J Virol. 2018;92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 145. | Trehanpati N, Sehgal R, Patra S, Vyas A, Vasudevan M, Khosla R, Khanam A, Kumar G, Maiwall R, Ramakrishna G, Kottilil S, Sarin SK. miRNA signatures can predict acute liver failure in hepatitis E infected pregnant females. Heliyon. 2017;3:e00287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 12] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 146. | Costafreda MI, Sauleda S, Riveiro-Barciela M, Rico A, Llorens-Revull M, Guix S, Pintó RM, Bosch A, Rodríguez-Frías F, Rando A, Piron M, Bes M. Specific Plasma MicroRNA Signatures Underlying the Clinical Outcomes of Hepatitis E Virus Infection. Microbiol Spectr. 2023;11:e0466422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 147. | Harms D, Choi M, Allers K, Wang B, Pietsch H, Papp CP, Hanisch L, Kurreck J, Hofmann J, Bock CT. Specific circulating microRNAs during hepatitis E infection can serve as indicator for chronic hepatitis E. Sci Rep. 2020;10:5337. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 148. | Gupta S, Singh P, Tasneem A, Almatroudi A, Rahmani AH, Dohare R, Parveen S. Integrative Multiomics and Regulatory Network Analyses Uncovers the Role of OAS3, TRAFD1, miR-222-3p, and miR-125b-5p in Hepatitis E Virus Infection. Genes (Basel). 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 12] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 149. | McGowan K, Simpson KJ, Petrik J. Expression Profiles of Exosomal MicroRNAs from HEV- and HCV-Infected Blood Donors and Patients: A Pilot Study. Viruses. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |