INTRODUCTION
Viruses are minute parasitic particles available in various shapes and sizes. These viral particles contain genetic material in the form of RNA or deoxyribonucleic acid (DNA), which can be single-stranded (ss) or double-stranded (ds), enclosed within a viral-encoded proteinaceous capsid coat. RNA viruses, characterized by exceptionally high genetic variability and phenotypic diversity, are intracellular obligatory parasites, capable of infecting a wide range of hosts[1,2]. They primarily target Eukarya and replicate using virally encoded RNA-dependent RNA polymerase (RdRp). In this type of viral replication, the synthesized RNA can serve as the genome, a copy of the genome, or messenger RNAs (mRNAs)[3]. Depending on the type of RNA acting as the genome, RNA viruses can be positive or plus strand or negative or minus strand.
RNA viruses are responsible for recurrent epidemics and occasional pandemics. Infections caused by respiratory and vector-borne RNA viruses, such as Influenza A virus, Zika virus, and West Nile virus, are of significant concern[4-6]. Other pathogenic RNA viruses affecting humans include Orthomyxoviruses, hepatitis C virus (HCV), Ebola virus, SARS, influenza, poliovirus, measles virus, and retroviruses such as adult human T-cell lymphotropic virus type 1 (HTLV-1) and human immunodeficiency virus (HIV). HIV, the causative agent of acquired immunodeficiency syndrome (AIDS), is a serious and prevalent viral disease. Currently, approximately 39 million people are living with HIV, and tens of millions have succumbed to AIDS since the beginning of the epidemic[7].
Recent outbreaks, such as the emergence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in 2019, have underscored the importance of understanding the molecular mechanisms governing the evolution of RNA genomes and the potential for viral exposure[8]. Studies in this area have shed light on novel findings related to viral replication, host-virus interactions, and the development of antiviral therapies. Researchers continue to explore these aspects to develop strategies for combating RNA viral infections and mitigating their impact on global health.
RNA viruses are responsible for recurrent epidemics and occasional pandemics, causing significant global human morbidities and mortalities through virally induced emerging infectious diseases. The emergence and re-emergence of these viral infections have profound implications for public health, the overall economy, and the quality of life of affected populations. To mitigate their impact, it is essential to prevent the replication of RNA viruses, including retroviruses. Therefore, a comprehensive understanding of the replication and transcription processes of these pathogens is crucial[9].
Research on RNA viruses and the infections they cause serves as a pivotal foundation for vaccine development and the formulation of strategies for prevention, control, and corresponding therapeutic interventions. A deep comprehension of virus-host interactions is paramount to understanding the mechanisms governing viral replication and the associated pathological consequences. Advances in sequencing methods have been instrumental in revealing the significance of RNA-protein and RNA-RNA interactions during infections, providing valuable insights into the development of targeted therapies[10].
This review article aims to explore the current understanding of various RNA virus infections, focusing on their pathogenesis and the latest therapeutic interventions. Recent research in this field has led to the identification of novel drug targets, the development of antiviral agents, and the exploration of innovative vaccination strategies. Additionally, studies have elucidated the role of host immune responses and the viral factors contributing to disease severity, paving the way for personalized treatment approaches. Understanding the genetic diversity of RNA viruses, their evolutionary dynamics, and the mechanisms of viral transmission is essential for devising effective public health measures and preparedness strategies against future outbreaks. Moreover, ongoing research efforts continue to unravel the intricate interactions between RNA viruses and host cells, providing valuable information for the development of next-generation antiviral therapies and vaccines.
RNA VIRUS REPLICATION AND TRANSMISSION
The viral DNA is protected and transported from cell to cell by very basic macromolecular structures known as envelopes or capsids in the extracellular environment. Only at the intracellular stage of their life viruses can produce distinctive compounds and engage in activities that are unique to living things. Creating a platform for genome replication and morphogenesis is one of these activities[11]. Due to the error-prone nature of RNA-dependent RNA polymerases of RNA viruses, they live as quasispecies with several variants within their populations[12]. A crucial phase in the life cycle of a virus is the replication of its genome. This procedure involves an intermediary complementary RNA strand for both plus- and minus-strand RNA viruses, but DNA for retroviruses[13].
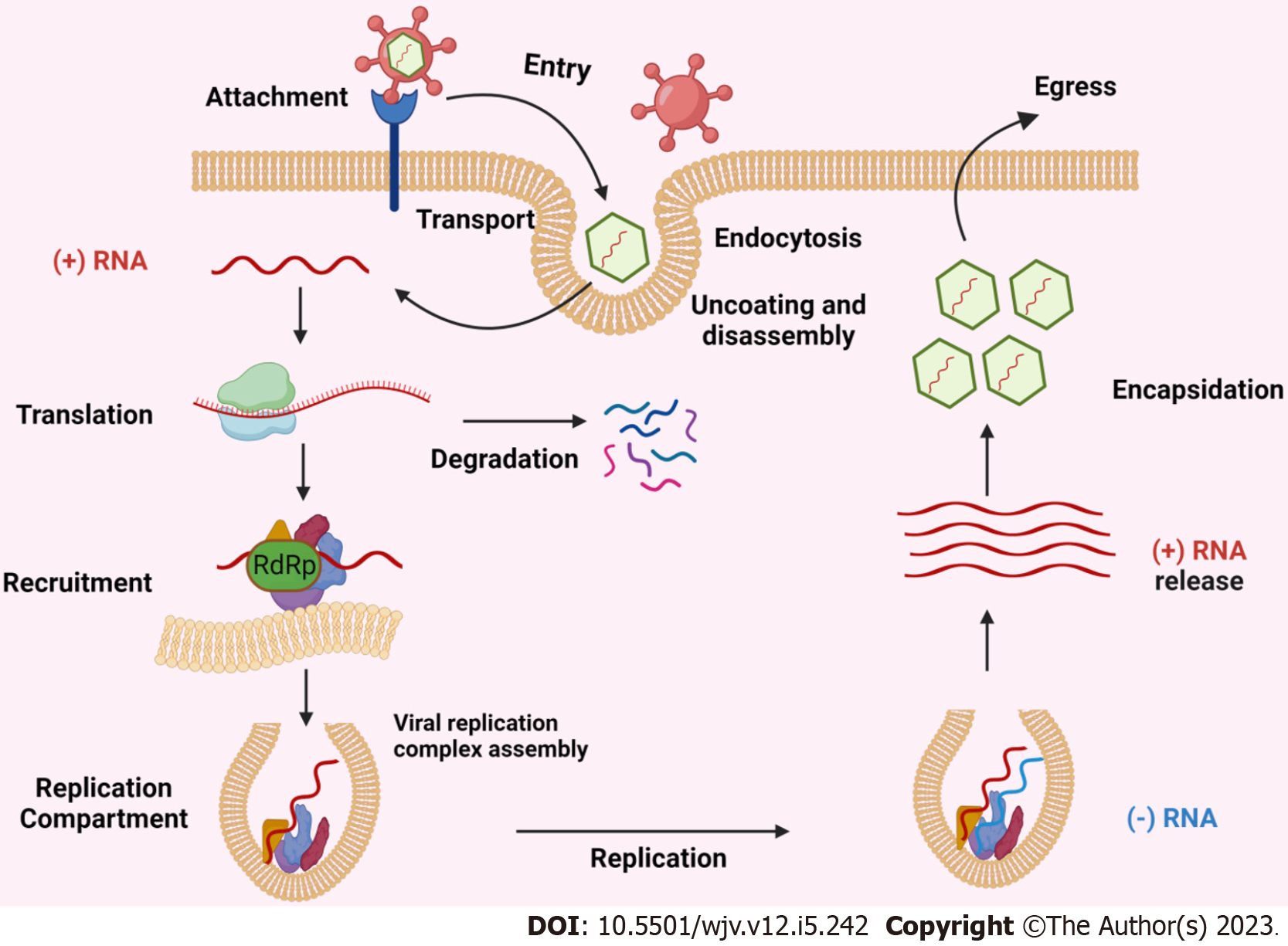
The primary processes and entrance points for both enveloped and non-enveloped viruses are attachment to cell-surface receptors and transport of the viral genome to the host cell's cytoplasm. After attaching to receptors, which might be proteins, carbohydrates, or lipids, viruses enter the cell by one of two routes: endocytic or non-endocytic. After attaching to receptors, which might be proteins, carbohydrates, or lipids, viruses enter the cell by one of two routes: endocytic or non-endocytic[14]. Alphaviruses, Coronaviruses, Picornaviridae enteroviruses, and Flaviviruses are examples of positive-sense RNA (+RNA) viruses that significantly alter cellular membranes to act as platforms for replication and the assembly of new virions[15]. Since viral genomic RNA replication occurs in the cytosol of host cells, viruses must be able to distinguish between their own genome and many cellular RNAs that are present in cells, so that they amplify only their own genome. The co-optation of host RNA-binding proteins by RNA viruses to speed up replication or dodge host RNA breakdown mechanisms is expected[16].
All (+) RNA viruses can sequester host intracellular membranes to produce replication compartments (RCs). These RCs contain recruited host proteins and lipids as well as viral RNA and proteins, which together produce an environment that is favourable for RNA replication[17]. Capsids have not been found in RCs, suggesting that viral RNA is duplicated within RCs and then (+) RNAs are transferred outside to virion assembly sites. Figure 1 depicted the schematic representation of replication and transmission of positive-sense RNA viruses.
Figure 1 Schematic representation of replication and transmission of positive-sense RNA viruses.
RdRp: RNA-dependent RNA polymerase.
FACTORS CONTRIBUTING TO VIRAL ADAPTABILITY AND EVOLUTION
Similar to animal RNA viruses, plant RNA viruses may have evolved through three different pathways, including transfer of genes horizontally from hosts, parallel evolution with similar genetic components and coevolution or codivergence with hosts[18,19]. Three temporal phases of emergence were identified by Elena et al[20-22], these were: The host moves to a different species or the same species but in a different ecological condition, acclimatization to the new environment or host, and epidemiology in the recently arrived host population, often by adjusting to a new vector species or mechanism of transmission. The evolution and host adaptability of animal RNA viruses have piqued the interest of many researchers. The majority of animal RNA viruses have A-rich coding sequences, as reported by Kustin et al[23] They also proposed possible explanations such as codon usage bias, weakened RNA secondary structures, and selection for a particular composition of amino acids, concluding that host immunological forces may be the cause of similar biases in the makeup of coding sequences among animal RNA viruses.
ROLE OF HOST FACTORS IN VIRAL REPLICATION AND DISSEMINATION
Various strategies have evolved in (+) RNA viruses to utilize host cell resources. For replication to occur, the viral genomic RNA, along with viral and host components, must be actively attracted to the relevant subcellular membrane surfaces[13]. In the case of human poliovirus (PV), the host poly(rC)-binding protein 2 (PCBP2) plays a crucial role in recruiting RNA templates. PCBP2, an RNA-binding protein, facilitates cap-independent translation by binding to the internal ribosome entry site (IRES) in PV (+)RNA and stabilizing mRNA[20]. Upon binding to viral RNA, PCBP2 is cleaved by the PV-encoded RdRp precursor (3CD) or protease (3C) protein. Studies have demonstrated that the viral proteinase 3CD cleaves PCBP2, thereby suppressing viral translation[24]. Although cleaved, PCBP2 retains two RNA-binding sites, allowing it to attach to the cloverleaf structure at the PV (+) RNA’s 5′ untranslated region (UTR). This interaction is vital for PV RNA replication as it brings together the 3′ and 5′ ends of the viral RNA through interaction with another host protein family, the poly(A)-binding proteins[25].
In the context of tomato mosaic virus (ToMV) replication, two Arabidopsis thaliana membrane proteins, TOM1 and TOM3, are essential. They interact with the helicase-like ToMV replication protein 130K, facilitating ToMV replication[26]. Additionally, the SNARE-like protein, human vesicle-associated membrane protein-associated protein A (VAPA), serves as a membrane anchor for HCV replication proteins[27]. HCV replication proteins NS5A and NS5B66 interact with VAPA, a contact crucial for the association of NS5A and NS5B with intracellular ER-derived membranes, which serve as the site of HCV replication[28]. Moreover, LSM1 protein aids in RNA recruitment in Brome mosaic virus and HCV. In PV, HCV, and coronaviruses, host heterogeneous nuclear ribonucleoproteins promote RNA recruitment and (+) RNA and (-) RNA synthesis[16,20].
VIRAL-HOST INTERACTIONS AND IMMUNE RESPONSE
During a viral infection, the host's pattern recognition receptors (PRRs) detect viral pathogen-associated molecular patterns (PAMPs). These PAMPs represent distinct molecular attributes found in viruses that are absent in the host cell, enabling cells to differentiate between self and non-self entities, thereby initiating an immune response against infection[29,30]. The activation of these PRRs initiates intracellular signaling cascades involving adaptor proteins like MAVS and STING[6]. Subsequently, these adaptor proteins trigger the activation of kinases and transcription factors, which, in turn, promote the transcriptional upregulation of type I and type III interferons (IFNs) and the synthesis of antiviral proteins[31].
Efforts have been made to understand how virus infections affect host cell protein synthesis, but inhibiting host protein synthesis is not always necessary for successful virus replication[32]. Many viruses like paramyxoviruses, papovaviruses, and retroviruses typically do not block host protein synthesis during their replication. Moreover, mutant viruses that cannot effectively halt host protein synthesis are not necessarily impaired in their ability to replicate[13]. For instance, in the case of VSV mutants selected during persistent infections, they may have a reduced capacity to interfere with the host's translational processes, yet they can achieve higher virus titers during a lytic growth cycle compared to the wild-type virus[33,34]. In many instances, this inhibition is accompanied by an overall reduction in the rate of protein synthesis within the infected cell. This broad inhibition likely occurs at the initiation stage of protein synthesis, as observed by a decrease in the average size of polysomes in infected cells where examined[35].
Before the initiation of targeted cellular or humoral immune responses against a specific virus, the activation of apoptosis can serve as an initial defensive mechanism within host cells. This process aids in the removal of cells that have been infected by the virus, thereby restricting viral replication[36]. Molluscum contagiosum virus, equine herpesvirus 2, bovine herpesvirus 4, human herpesvirus 8, and herpesvirus saimiri encode FLICE-inhibitory proteins (FLIPs) that exhibit structural similarities to FLICE (caspase-8)[37]. These FLIPs engage with the Fas-associated death domain protein adapter within the host cell[20].
Studies on candidate genes and genome-wide associations have yielded important information on the genetic foundations of many infectious illnesses. The loss-of-function mutation known as CCR5Δ32, which results in the absence of CCR5 expression on the surface of host cells, confers resistance to HIV infection in homozygous individuals[31]. Polymorphic variations in HLA genes have been associated with a wide range of infectious diseases, including RNA viruses such as SARS, influenza, HIV, hepatitis C, rabies, West Nile fever, rubella, mumps, and measles, among others. Genetic association studies of this nature are pivotal for pinpointing HLA alleles that might be correlated with immune responses offering protection. Variations in HLA alleles have prompted inquiries into their potential role in the distinct immune responses observed between mild and severe coronavirus disease 2019 (COVID-19) cases, such as delayed immunoglobulin M (IgM) responses and elevated S protein immunoglobulin G titers in non-intensive care unit patients[38,39].
ANTIVIRAL DRUGS
RNA virus-mediated infections include HCV, SARS, influenza, Ebola, polio, measles, HIV, HTLV-1, Respiratory syncytial virus (RSV), and others. Most RNA viruses have single-stranded or double-stranded RNA as their genetic material. Viruses primarily possess RNA-dependent RNA polymerase for genome replication or have reverse transcriptase for genome replication. Idoxuridine was the first antiviral drug approved in 1963. Since then, 90 antiviral drugs categorized into 13 functional groups have been approved for the treatment of 9 infectious diseases. Antiviral drugs approved for RNA virus-mediated infections include trifluridine, vidarabine, entecavir, zidovudine, didanosine, lamivudine, abacavir, nevirapine, efavirenz, rilpivirine, ritonavir, indinavir, lopinavir, simeprevir, paritaprevir, raltegravir, elvitegravir, palivizumab, tenofovir disoproxil fumarate, sofosbuvir with ribavirin, amantadine, zanamivir, rimantadine, laninamivir octanoate, and favipiravir[40].
In the realm of influenza treatment, antiviral drugs primarily include adamantanes (M2 ion channel blockers) which can block an ion channel formed of M2 protein encoded by the M gene in influenza A virus. This category comprises two classes of drugs: Amantadine and rimantadine. Additionally, neuraminidase inhibitors, approved for use against both influenza A and influenza B viruses, include major drugs such as zanamivir, oseltamivir, Peramivir, and Laninamivir[41]. Another class, the RNA-Dependent RNA Polymerase Inhibitors, includes favipiravir, which hampers viral RNA synthesis. Notably, the absence of favipiravir-resistant viruses is a remarkable property, indicating its effectiveness as an antiviral medication. A novel class, the Polymerase Acidic Endonuclease Inhibitor, encompasses baloxavir marboxil, which inhibits the cap-dependent endonuclease of the viral RdRp complex. This disruption restricts mRNA production and prevents subsequent viral protein synthesis. Baloxavir marboxil is one of the most recently developed anti-influenza drugs[42].
Challenges in developing anti-influenza drugs stem from the virus's antigenic evolution mechanisms: Antigenic shifts and drifts in surface glycoproteins. Immunization against these processes is challenging due to their natural immunity combat mechanisms. Major limitations in antiviral treatment include antiviral resistance of most human influenza A virus strains to M2 inhibitors and the need for in vivo disease models for influenza-related research. Evaluating antiviral treatment efficacy poses challenges from the standpoint of drug resistance. One of the main hurdles in treating influenza is the emergence of drug-resistant influenza viruses due to current antiviral regimens. Alternative treatments include drug combination therapies that synergistically minimize drug resistance and reduce drug toxicity. Commonly used antiviral combination therapies include Oseltamivir + zanamivir (targeting the same viral protein), Baloxavir + favipiravir (targeting Cap-Dependent Endonuclease & RNA-Dependent RNA Polymerase), Baloxavir + oseltamivir (targeting Cap-Dependent Endonuclease & Neuraminidase), and Oseltamivir + amantadine + ribavirin (Triple-combination antiviral drug treatment targeting M2 Ion Channel, Neuraminidase & RNA-Dependent RNA Polymerase)[43]. Additionally, next-generation influenza virus inhibitor candidates in early-stage development include EIDD-2801 and Pimodivir, a cyclohexyl carboxylic acid analogue targeting the polymerase PB2 subunit to hinder influenza virus replication[44].
Moving to HCV treatment, initial antiviral efforts utilized IFNα monotherapy followed by ribavirin (RBV), a synthetic triazole guanosine analogue active against both DNA and RNA viruses. Pegylated IFN, a modified interferon with a prolonged pharmacokinetic profile, showed favorable results. Subsequently, direct-acting antivirals (DAAs) emerged to directly interfere with viral proteins, marking a significant breakthrough. Protease inhibitors (PIs) like boceprevir and telaprevir were among the first DAAs, substantially increasing sustained virologic response (SVR) rates. However, these drugs accelerated the generation of resistance-associated substitutions, leading to virological breakthroughs in almost all treated individuals. Compared to the prior standard regimen of Peg-IFN–RBV, Peg-IFN–RBV–triple therapy raised SVR rates in treatment-naive patients by about 30%[45].
The COVID-19 pandemic spurred progress in antiviral medication development, introducing both DAAs and host-based antivirals. Commonly used DAAs include remdesivir and molnupiravir, inhibiting RNA replication through interaction with RdRp. Nirmatrelvir targets the main protease or 3-chymotrypsin-like protease, while favipiravir and ritonavir are used for mild to moderate COVID-19 cases[46,47]. However, these drugs have limitations. Molnupiravir, in particular, has mutagenic potential for both the virus and the host. Host-based antiviral drugs like camostat and ivermectin target transmembrane protease serine 2. Other options include fluvoxamine, thapsigargin, and plitidepsin[48].
For HIV, various antiretroviral drugs have been developed, including Non-nucleoside reverse transcriptase inhibitors, Nucleoside reverse transcriptase inhibitors, PIs, Fusion inhibitors, CCR5 antagonists, Integrase strand transfer inhibitors, and Post-attachment inhibitors. Combination antiretroviral therapy or highly active antiretroviral therapy (HAART) uses these drugs in combination. Food and Drug Administration (FDA)-approved HAART treatments include bictegravir, emtricitabine, and tenofovir alafenamide, cabotegravir, and rilpivirine, doravirine, lamivudine, and tenofovir disoproxil fumarate, among others. Direct-Acting Anti-HIV Agents such as cabotegravir, doravirine, and islatravir, along with host-based antivirals like the monoclonal antibody ibalizumab, are being used or are in the developmental stage[47,49]. Despite their efficacy, these drugs pose various risks and side effects, including elevated liver enzyme levels, gastrointestinal toxicity, rashes, benign hyperbilirubinemia, nausea, headache, anemia, leukopenia, reversible peripheral neuropathy, lactic acid elevation, low phosphate levels, and CNS toxicity[50].
In the fight against polio, large-scale oral vaccination programs have been implemented through the Global Polio Eradication Initiative. However, no specific antipicornavirus drugs have been FDA approved. Promising candidates include pirodavir (capsid binders), rupintrivir (protease inhibitors), Enviroxime (protein 3A inhibitors), ribavirin (nucleoside analogues), and compounds like MDL-860, discovered as a broad-spectrum inhibitor of viruses, although its exact mechanism of action remains unknown[51,52].
Ebola virus, a highly lethal pathogen, lacks FDA-approved drugs or vaccines. However, several potential drugs are under investigation, including favipiravir, amiodarone, amodiaquine, chloroquine, clomiphene, toremifene, Brincidofovir, sertraline, and BCX4430[53].
RSV is a negative-sense, ssRNA virus that primarily causes acute lower respiratory tract infections in infants, children, adults, and immunocompromised individuals. Currently, the FDA has approved two drugs for RSV treatment: ribavirin (a guanosine analogue) and palivizumab (a monoclonal antibody)[54]. Several antiviral candidates against RSV are under clinical research and trial stages, including REGN2222, MEDI8897, and Motavizumab. Additionally, various fusion inhibitors, nucleoprotein inhibitors, nucleoside analogues, and non-nucleoside inhibitors are under development for RSV therapy[54].
HTLV-1 is an RNA virus mainly responsible for HTLV-1-associated diseases such as Adult T-cell leukemia (ATLL) and neurological disorders like HTLV-1 Associated Myelopathy (HAM). It is a retrovirus that affects CD4+ cells and, to some extent, CD8+ cells and dendritic cells. The treatment approach for ATLL diseases mainly involves a combination of drugs such as interferon α and zidovudine (IFN-α/AZT). However, there is no established treatment approach for HAM disorder[55]. These are some of the most commonly encountered RNA virus infections and the commonly used antiviral agents/drugs for their treatment.
RNA-BASED THERAPEUTICS
RNA holds significant potential for therapeutic applications. Growing understanding and recent advancements in RNA studies have paved the way for various innovative RNA-based therapeutic approaches. Several RNA-based therapeutic methods are gaining popularity and receiving clinical approval for use. These approaches offer certain advantages over antiviral drugs, conventional protein targeting, and DNA-based medicines. The key advantage lies in targeting the RNA of the virus, providing a broader and more efficient target. Small molecular drugs, in contrast, target about 0.05% of the human genome. Moreover, many targets of disease lack clearly defined active regions for binding of small molecule.
RNA-based treatments face significant challenges in terms of intracellular trafficking and metabolic stability. However, researchers have explored a variety of strategies to overcome these obstacles[56].
TYPES OF RNA-BASED THERAPEUTICS
RNA interference
RNA interference (RNAi) is an in-vivo cellular process which leads to silencing of RNA expression by using ds RNAs, this provides an intrinsic defensive mechanism against invading viruses and transposable elements. miRNAs and siRNAs are small oligonucleotide sequences of 20-22 nucleotides with definite structures composed of 5’-phosphate and 3’-hydroxyl endings and two 3’-overhang ribonucleotides on each duplex strand. Within the RNA-induced silencing complex, the endoribonuclease Dicer isolates the guide and passenger strands by cutting dsRNAs. While the passenger siRNA strand is broken down by the argonaute2 (AGO2) protein, the guide siRNA strand attaches itself directly to the target RNA and initiates AGO2-mediated cleavage[57]. When siRNAs bind to the promoter regions, they can also cause chromatin remodelling and histone changes in the nucleus, which silences transcription in addition to destroying cytoplasmic RNAs. This therapeutic approach has a lot of potential to be use in HIV, Influenza, SARS-CoV treatment[58].
Antisense oligonucleotides
In antisense oligonucleotides (ASOs)-mediated gene regulation, short single-stranded oligonucleotides (12-24 nt) are utilized. These oligonucleotides are complementary to specific RNA sequences through Watson-Crick base pairing, enabling them to alter the expression of proteins, reduce, or restore their expression[59]. There are two types of mechanisms used to modify expression: One is Occupancy-Mediated Degradation: In this mechanism, ASOs induce target mRNA cleavage by RNase H1 or ribozymes. The ASOs lead to the degradation of the target RNA, and 2nd is Occupancy-Only Mechanism in this mechanism, the target RNA is not directly degraded. Instead, various mechanisms are employed to modify expression. These include altering RNA splicing using splice switching, blocking miRNA binding to the target RNA, inhibiting or activating translation, and triggering nonsense-mediated mRNA decay[60].
CRISPR-based genome editing
Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) is a prokaryotic defense system widely applied in genome editing. The CRISPR-associated protein (Cas) system enables precise genome sequence editing in mammalian cells and organisms, leading to the target gene's irreversible knockout or knockin. This system relies on guide RNA and Cas nucleases. The complex recognizes the protospacer adjacent motif sequence in the target RNA, initiating its activity. For effective genome editing, the Cas nuclease cleaves either the double-stranded DNA or a single-stranded RNA at the designated spot[61]. The most commonly used Cas systems are Cas9 and Cas13. The Cas9 system can target both double-stranded DNA and single-stranded RNA. This technique finds applications in detecting SARS-CoV-2, with CRISPR-Cas13-based assay designs used for detecting 67 diseases, including SARS-CoV-2, Zika virus, and dengue fever, among others. The CRISPR/Cas9 system aids in studying the regulation pathway of the influenza virus, representing an emerging field in developing antiviral therapies against diseases like HIV, Hepatitis, and SARS[62].
Aptamer: It is synthetically designed chemical antibodies, are single-stranded oligonucleotide sequences that specifically bind to and inhibit protein expressions. Aptamer selection is based on the methodical evolution of ligands by exponential enrichment. Aptamers function primarily by interfering with interactions between disease-related targets, such as those between proteins or between receptors and ligands[63]. Aptamers can also deliver therapeutic agents to specific cells. They have applications in controlling SAR-CoV-2 infection; slow off-rate modified aptamers (SOMAmers) are DNA-based aptamers that bind to specific S protein fragments of SARS-CoV-2, preventing virus interaction with ACE-2 receptors[64]. Aptamers like anti-CCR5 are designed to prevent the interaction of HIV with the T-cell GPCR receptor. They serve as prospective anti-HIV/AIDS drugs, offering targeted delivery of various therapeutic options through CCR5-targeted aptamers and aptamer-siRNA conjugates[65]. Aptamers are also used in viral disease diagnosis; aptasensors, electrochemical diagnostic tools, demonstrate advantages such as low cost, specificity and early detection for influenza A and HA glycoprotein virus particles[66].
mRNA-based therapeutics
mRNA is generally single-stranded and is transcribed from the antisense strand of DNA, carrying information about the expression of functional proteins. mRNA-based therapeutics represent the future of treating various refractory diseases, including infectious diseases, metabolic genetic disorders, cancer, cardiovascular diseases, and others. In this therapy, exogenous mRNA is introduced with the help of a carrier, acting as a vaccine or therapeutic agent that expresses the necessary functional protein. This approach offers several advantages over conventional therapies, including higher efficiency, faster design and production, adaptability, and lower costs. These benefits are possible due to a planned manufacturing method developed for in vitro transcribed mRNA[67].
However, there are challenges associated with mRNA-based therapies. The anionic nature of cell membranes makes it difficult for mRNA to translate functional proteins in the cytoplasm. Additionally, mRNA has a median intracellular half-life of approximately 7 hours, and efficient carriers are crucial for overcoming cellular barriers, improving immunogenicity, and addressing stability issues. Moreover, mRNA may trigger immunological reactions and related toxicities, hindering the development of mRNA-based treatments[68,69].
The basic steps for designing and manufacturing mRNA-based therapeutic agents include mRNA design and synthesis, mRNA entrapment, pharmacodynamics and safety evaluation, manufacturing, and clinical trials[70]. Quality control measures are essential, such as codon optimization for mRNA encoding the antigen, ensuring mRNA sequence identity and integrity, assessing nucleic acid quantity, 5’ capping, poly A tail length, optimizing 5’-UTRs and 3’-UTRs, and ensuring mRNA purity. During the drug delivery process, mass spectrometry analysis, nuclear magnetic resonance analysis, evaluation of lipid electric charges and ratios, assessment of lipid impurities, and transfection efficiency are crucial. For mRNA-lipid nanoparticle drugs, encapsulation efficiency, particle size, storage conditions, and zeta potential must be carefully considered[67,70].
Apart from the conventional linear mRNA form, there are other structural forms such as self-amplifying RNA derived from alphaviruses, circular RNAs, noncoding RNAs, and competitive endogenous RNAs. These diverse forms of mRNA can be utilized for therapeutics[71,72]. Correct delivery of mRNA inside living systems is pivotal, and various delivery systems like lipid nanoparticles (LNP), polymeric nanoparticles, cationic nanoemulsions, protamine-condensed mRNA, exosomes, extracellular vesicles, and mesoporous silica are used[73]. These delivery systems utilize electrostatic interactions, hydrogen bonds, or coordination interactions through methods like thin-film hydration, nanoprecipitation, or microfluidic mixing. Nanoparticle-based delivery systems enhance cell uptake, facilitate lysosomal escape, and accelerate translation, maximizing mRNA availability. Achieving effective in vivo distribution of mRNA necessitates tissue-targeted delivery of mRNA-based therapies[74]. Precision nanoparticle engineering has been developed to cross biological barriers, expanding its applications in various therapeutic areas for mRNA-based drug delivery[75].
LNPs are a popular mRNA-based delivery method targeting the liver. Current research focuses on improving LNP platforms for administration to additional tissues. Through the exact and predictable customization of LNPs to transport mRNA, Cas9 mRNA/single-guide RNA, and Cas9 ribonucleoprotein complexes to target organs via intravenous injection into the liver and lungs, selective organ targeting has emerged as a therapeutic method. Another critical aspect of drug delivery is the route of administration. While the majority of disorders can be treated with intravenous administration specific administration routes are tailored to the targeted organ or organ system[76,77].
Over the past three to four years, mRNA-based therapeutics have gained significant popularity due to their role in designing treatments for COVID-19 and are now extensively explored for their applications in other viral infections, specifically RNA virus infections such as influenza, SARS, HIV, HCV, RSV, and various cancer therapies. A growing number of well-funded biotechnology companies, including Moderna, CureVac, BioNTech, Argos Therapeutics, RaNA, Translate Bio, Ethris, Arcturus, and Acuitas, are investing billions of dollars in mRNA therapy. Clearly, one of the most compelling topics in medication research is mRNA, which is worth investigating in the long run[67].
MICRORNA (MIRNA) PATTERN AND ITS ROLE IN GLIOMAS
Circulating microRNAs, have gained significant attention in the field of cancer research, as potential non-invasive diagnostic biomarkers for various types of tumors, including gliomas (a type of brain tumor)[78]. miRNAs can be found in different body fluids, including blood (serum and plasma), and cerebrospinal fluid and can be transported between cells, including tumor cells and neighboring normal cells, through exosomes so these can be used as reliable biomarkers[79]. miRNAs interfere the protein translation through complementary base-pairing, or degrade mRNA, thus dysregulation of miRNAs lead to tumor progression[80]. The miR-21 has the potential to predict the radiation necrosis compared to tumor progression[78]. The potential utility of miR-128 and miR-342-3p as biomarkers for assessing glioma grades and monitoring treatment response has been advocated[81]. Indeed, the profiling of these miRNAs can provide valuable insights into the presence, type, and stage of the tumour. Consequently, early detection and accurate diagnosis are crucial for patient prognosis and treatment planning, and survival.
For rapid and early detection of miRNAs high sensitive methods such as a toehold-mediated strand displacement reaction, the enzyme-free surface plasmon resonance imaging biosensing method, and the ultrasensitive electrochemical method, should be integrated with existing diagnostic modalities, such as imaging and molecular profiling (genetic and epigenetic markers), to enhance diagnostic accuracy and guide treatment decisions. But challenges are the heterogeneity nature of gliomas, the varying level of sensitivity and specificity of miRNA, small number of studies, lack of standardized protocols for miRNA isolation, quantification that makes miRNA to accurately detect all glioma types and stages[82]. With the advancement of glioma biology and miRNA function, possibility of miRNA-based tests may eventually become a valuable part of the diagnostic and screening toolkit for glioma patients.
DEVELOPMENT AND APPLICATION OF MRNA-BASED VACCINATION
Vaccination plays a important role in dealing with communicable diseases, remarkably contributing to global public health. In simple terms, vaccination aims to generate immunity against specific diseases by using vaccines. Conventional vaccine candidates mainly include whole organism vaccines (live attenuated or inactivated pathogens), subunit vaccines, viral vectors, etc., which have been crucial in disease prevention. However, the scalability, speed of development, and ability to respond to newly emerging pathogens of these conventional vaccination platforms are often limited.
Recently, mRNA-based vaccination has emerged as the most advanced technology offering various benefits. It provides a flexible framework for the quick and focused development of vaccines against infectious illnesses, such as viral outbreaks and new infections. mRNA vaccines also have the advantage of being developed and produced more quickly than traditional vaccinations, which frequently need expensive and time-consuming manufacturing procedures. When it comes to responding to emerging infections or developing variants, mRNA vaccines offer unparalleled flexibility and speed because they can be designed and manufactured in a few of weeks[83].
During recent times, mRNA-based vaccination against SARS-CoV-2 has been a groundbreaking discovery in tackling diseases. The design of mRNA vaccines is adaptable to various diseases by simply changing the mRNA sequence encoding the required antigen. However, their efficient delivery poses a substantial challenge due to their susceptibility to degradation, poor stability, and obstacles in reaching the targeted areas of action[84].
The delivery of mRNA-based vaccines is a major challenge due to the internal environment inside the cytoplasm and the need to pass through the cell membrane. Various delivery systems have been developed, among which nanoparticles have emerged as promising tools in mRNA vaccine delivery, overcoming the inherent limitations of naked mRNA molecules. These nanoscale delivery and protection systems offer effective cellular absorption, defense against enzymatic breakdown and controlled mRNA payload release. Furthermore, nanoparticles can be developed to increase balance, extend their duration of circulation, and enable targeted administration to immune cells or organs, enhancing the immunogenicity and effectiveness of mRNA vaccines[85].
Various types of nanoparticle-based carrier systems include LNPs, polymeric nanoparticles, peptides, and protamine-based delivery systems, as well as cationic nanoparticles. Among them, for the administration of mRNA vaccines, LNPs have become the most popular class of nanoparticles. Due to their hydrophobic core, LNPs can enclose and safeguard mRNA, facilitating effective cellular absorption and intracellular release. This strategy is highly effective in designing various mRNA vaccines against infectious diseases, including COVID-19[86,87]. LNPs are positioned as prospective tools for successful vaccination techniques due to their outstanding safety profiles, high transfection efficiency, and capacity to elicit robust immune responses. This nanoparticle-based delivery system for mRNA is also useful in addressing other issues like cancer immunotherapy, personalized medicine, and therapeutic interventions for genetic disorders[88].
TYPES AND MECHANISM OF ACTION OF MRNA VACCINES
Self-amplifying mRNA, trans-amplifying mRNA, and conventional mRNA are the three forms of mRNA vaccines that are now on the market. Conventional mRNA vaccines, also known as non-replicating or non-amplifying mRNA vaccines, mainly consist of untranslated regions (5′UTR, 3′UTR) and the coding part of mRNA, which by transcription produces one copy of the immunogenic protein[71]. Self-amplifying mRNA vaccines are genetically modified mRNA, incorporating engineered replicons from self-replicating RNA viruses. They possess 5′ and 3′ conserved sequence elements (CSE) that regulate viral RNA synthesis and facilitate attachment to viral or cellular proteins. Self-amplifying RNA contains non-structural proteins 1-4 (nsP 1-4) sequences[89]. Trans-amplifying mRNA is also genetically modified mRNA with 5′ and 3′ CSE. Trans-amplifying mRNA requires two RNA genes to be co-delivered: the mRNA without nsP 1-4 and the mRNA encoding nsP1-4 genes[90].
In mRNA vaccination technology, mRNA is synthesized outside the body, injected, and then transported across cell membranes for translation in the cytoplasm. Once in the cytoplasm, the mRNA is translated into the necessary protein by ribosomes. In the case of naturally occurring mRNA, this process occurs after the mRNA moves from the nucleus or cell membrane. The poly-A tail get attached to the poly-A-binding protein during translation, and the eIFs attach to the 5′UTR cap to start translation[70]. Ribosomes convert each codon, consisting of three nucleotides in the translated portion of the mRNA, into an amino acid. After injection, immune cells internalize mRNA-LNPs, leading to the release of mRNA from the LNPs. Ribosomes recognize the mRNA, translating it into antigenic proteins. These proteins are broken down and processed by proteasomes, resulting in small peptides presented on the cell surface by major histocompatibility complex class I (MHC I) molecules, and activates CD8+ T lymphocytes to eliminate infected cells. These produced antigen can be broken down further by lysosomes, loading small peptides on major histocompatibility complex class II (MHC II) molecules, recognized by CD4+ T lymphocytes. These cells stimulate B cells to stimulate humoral immune responses and inflammatory cytokine release to stimulate cellular immunological responses. Successful mRNA translation into the necessary antigen requires recognition of the 5′ cap, poly-A tail, 5′UTR, 3′UTR, and translated region of the synthesized mRNA vaccine by the ribosomes[67].
Optimizing mRNA vaccines is essential for enhancing their stability, safety, and efficiency. Modifications in 5′ cap of the 5′UTR and also in the poly(A) tail of 3′UTR can regulate translational efficiency of mRNA. For instance, converting the mRNA cap into phosphorothioate when the mRNA vaccine is transfected in immature dendritic cells may increase stability and expression. Furthermore, by including altered nucleosides into the mRNA, the Toll-like receptor activation may be decreased or eliminated, improving the vaccine's safety via nucleoside modification[91].
DELIVERY SYSTEMS
The most commonly used mRNA vaccine delivery systems include LNPs, Polymeric nanoparticles, peptides, protein nanoparticles, protamine nanoparticles, and other systems like cationic lipid amphiphiles. Because they may effectively transfer mRNA intracellularly by merging with the lipid bilayer of early endosomes, lipid-based nanoparticles are frequently preferred. Through this procedure, the mRNA is shielded from RNase breakdown during systemic circulation and is allowed to enter the cytoplasm. These LNPs primarily has three important components, i.e., 40%-50% ionizable lipids, 38%–45% cholesterol, and 10%–12% helper phospholipid. In certain cases, a fourth component, such as 1%-2% PEGylated lipid, is added. LNPs have been utilized for delivering mRNA vaccines and drugs against diseases like COVID-19 and influenza[92,93].
Polymeric nanoparticles involve the addition of low molecular weight polyethyleneimine with polyethylene glycol, which is then linked to cyclodextrin. Conjugation with cyclodextrin has been proven as a reliable and safe method for delivering mRNA. This approach has versatile applications and may lead to the development of specific antibodies[94].
Peptides and protein-based nanoparticles are extensively used due to their excellent biocompatibility and accessibility. Amphipathic peptides, with cationic or amphipathic amine groups (arginine), can facilitate delivery of mRNA into the cells. These peptides binds electrostatically to the mRNA, forming nano-complexes[95].
Protamine nanoparticles leverage the net positive charge of protamine, a cationic protein primarily composed of positively charged amino acids. Protamine can complex with nucleic acids, such as RNAs, enhancing their stability and shielding them from enzymatic degradation by nucleases. This property facilitates their delivery to specific tissues[96]. Protamine's cationic nature, attributed to an arginine-rich sequence, enables it to interact with negatively charged mRNA, making it valuable in the design of mRNA-based vaccines.
APPLICATION OF MRNA VACCINES
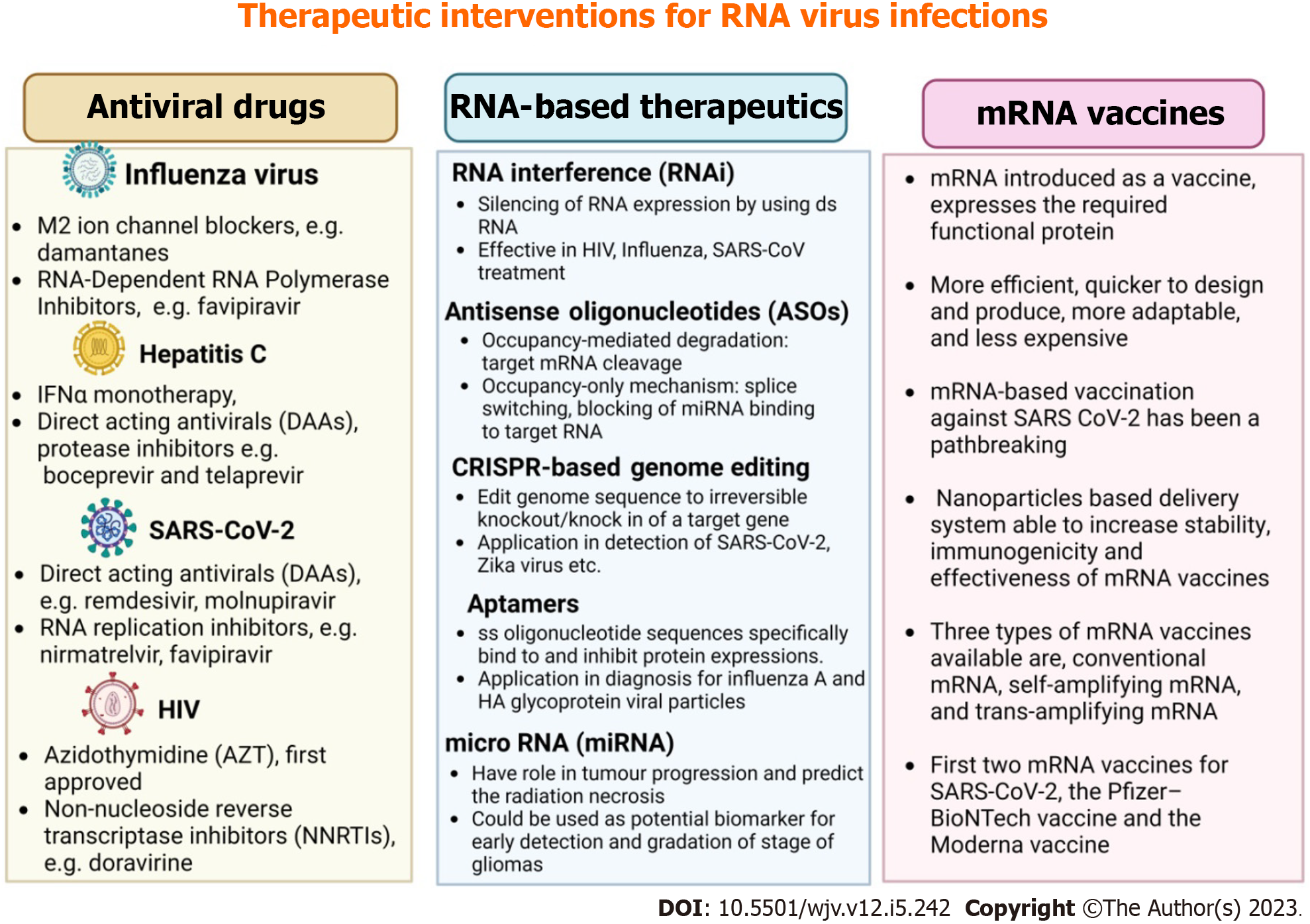
mRNA vaccines are primarily designed to generate immunity against infectious diseases and cancers. Due to the COVID-19 pandemic, these vaccines were developed more quickly, and as a result, the FDA approved the first two mRNA vaccines for SARS-CoV-2: The Moderna vaccine (mRNA-1273) and the Pfizer-BioNTech vaccine (BNT162b2)[97]. In addition to SARS virus mRNA vaccines, various mRNA vaccines for different viral infections such as influenza, HIV, Zika, and Rabies are under development. Numerous potential mRNA vaccine candidates are undergoing clinical trials, including mRNA-1345 against RSV (in phases two and three trials developed by Moderna), mRNA-1273 against SARS-CoV-2 variant B.1.351 (in phase two trials), and mRNA-1893 against Zika virus (in phase two trials). Apart from their application in preventing infectious diseases, mRNA vaccines also have broad applications in cancer treatment[98,99]. Figure 2 summarizes therapeutic interventions like antiviral drugs, RNA-based therapeutics, and mRNA vaccines for RNA virus infections.
Figure 2 Summary of therapeutic interventions for RNA virus infections has been summarized.
HIV: Human immunodeficiency virus; SARS-CoV-2: Severe acute respiratory syndrome coronavirus 2; IFN: Interferon.
FUTURE PERSPECTIVES AND CHALLENGES
The idea of personalized medicine may bring a great revolution in the field of medicine. Personalized medicine, an emerging practice of medicine uses a person's genetic profile to guide decisions which are made in regards to the prevention, diagnosis, and treatment of the disease[100]. The use of personalized medicine may be beneficial in many aspects, such as diagnostic accuracy improvement, better disease prevention, targeted therapy, reducing side effects, and the health care cost and promotion of research[101].
For instance, despite of rapid advancement in medical science, the modern medicine could not provide adequate treatment for COVID-19. In case of COVID-19 pandemic, it was established that observing the genetic background of each patient can contribute greatly to the drug effectiveness[102]. After the arrival of the challenges regarding treatment of coronavirus infection, the crucial roles of personalized medicine were realized by the physicians and healthcare workers. Hence, utilization of personalized medicine may stay as a potential therapeutic strategy for RNA virus mediated infections.
Previously, in late 2003, a coronavirus, i.e., SARS-CoV-1 caused an outbreak of severe acute respiratory illness (now called SARS), and acute respiratory distress syndrome type findings[103]. In 2012, another coronavirus witnessed to cause Middle East respiratory syndrome, having a case-fatality rate of more than 35%. An outbreak of a disastrous pandemic occurred in late 2019, which in due course spread all over the world. Out of all these three coronavirus outbreaks the SARS-CoV-2 infection was newer and the first two outbreaks helped us to prepare for this third one[104].
The absence of suitable animal models, an insufficient understanding of the correlates of immune protection, and limited pharmaceutical industry investment are obstacles that affect the development of an HIV vaccine[105].
If the viral pathogens are zoonotic, it must be needed a prevention barrier to reduce their chances of first introduction to human population. ‘One health’ approach reduces the prevalence of viral pathogens with high zoonotic potential in animals and which in turn reduces the viral introduction into human population[106]. It is quite easy to monitor, treat or vaccinate the domestic animals, but is complicated for the wild animals. An inclusive surveillance plan must be executed for the wild animals in order to recognize the pathogens having possibility of transmission to human[107]. Advanced sequencing methods can be used for the surveillance to find out the variations which could have enhanced the zoonotic potential or pathogenicity[108]. Surveillance of bush meat market may be important for detecting such zoonotic viral pathogens.
For the management of any future pandemics, we must be ready with some preventable approaches. These approaches may be the non-pharmacological approach (such as mask wearing and social distancing), vaccine anticipation, and anticipating therapies to reduce morbidity and mortality[107].
Orthomyxoviruses, HCV, Ebola illness, SARS, influenza, polio, measles, and retroviruses including adult HTLV-1 and HIV are among the human diseases caused by RNA viruses. The genetic material of RNA viruses is RNA, which can be single-stranded or double-stranded[109]. Reverse transcriptase produces viral DNA that can be integrated into host DNA through its integrase activity, and viruses can also use RNA-dependent RNA polymerases to make copies of their genomes. Retroviruses, on the other hand, have two copies of their single-strand RNA genomes. Due to the error-prone nature of RNA-dependent RNA polymerases of RNA viruses, they live as quasispecies with several variants within their populations[110].
CONCLUSION
In this review article, we have highlighted the pathogenesis and recent advances in the treatment of RNA virus-mediated infections. We discussed RNAi and various RNA-based antiviral drugs, as well as the development of RNA vaccines and the challenges associated with their administration. Different types of vaccines exhibit distinct efficacy, and we emphasized various strategies to enhance vaccine effectiveness. Additionally, our focus was on host-directed therapies, which represent an antiviral strategy. However, the development of these therapies poses significant challenges. Overcoming these challenges is crucial to transforming host-directed therapies into potent antiviral treatments.