Published online Sep 25, 2023. doi: 10.5501/wjv.v12.i4.233
Peer-review started: June 21, 2023
First decision: July 5, 2023
Revised: July 19, 2023
Accepted: August 7, 2023
Article in press: August 7, 2023
Published online: September 25, 2023
Processing time: 90 Days and 1 Hours
Hepatitis B virus (HBV) and hepatitis C virus (HCV) viral load (VL) estimation is essential for the management of both HBV and HCV infections. Due to a longer turnaround time for VL estimation, many patients drop out from the cascade of care. To achieve the global goals of reducing morbidity and mortality due to HBV/HCV and moving towards their elimination by 2030, molecular diagnostic platforms with faster and random (i.e. single sample) access are needed.
To evaluate the performance of the recently launched NeuMoDx 96 random access system with the conventional COBAS®AmpliPrep/COBAS TaqMan system for HBV and HCV VL estimation.
Archived once-thawed plasma samples were retrieved and tested on both platforms. Correlation between the assays was determined by linear regression and Bland-Altman analysis. The study included samples from 186 patients, 99 for HBV of which 49 were true infected HBV cases (hepatitis B surface antigen, anti-hepatitis B core antibody, and HBV DNA-positive) and 87 for HCV assay in which 39 were true positives for HCV infection (anti-HCV and HCV RNA-positive).
The median VL detected by NeuMoDx for HBV was 2.9 (interquartile range [IQR]: 2.0-4.3) log10 IU/mL and by COBAS it was 3.70 (IQR: 2.28-4.56) log10 IU/mL, with excellent correlation (R2 = 0.98). In HCV, the median VL detected by NeuMoDx was 4.9 (IQR: 4.2-5.4) log10 IU/mL and by COBAS it was 5.10 (IQR: 4.07-5.80) log10 IU/mL with good correlation (R2 = 0.96).
The overall concordance between both the systems was 100% for both HBV and HCV VL estimation. Moreover, no genotype-specific bias for HBV/HCV VL quantification was seen in both the systems. Our findings reveal that NeuMoDx HBV and HCV quantitative assays have shown overall good clinical performance and provide faster results with 100% sensitivity and specificity compared to the COBAS AmpliPrep/COBAS TaqMan system.
Core Tip: In this study, the clinical performance of the NeuMoDx 96 random access system for hepatitis B virus (HBV) and hepatitis C virus (HCV) viral load (VL) quantification was evaluated and compared to the routinely used COBAS AmpliPrep/COBAS TaqMan system. Good concordance was observed between both systems for the HBV and HCV VL assays. Due to its random access (i.e. single sample access) characteristics and shorter turnaround, the NeuMoDx 96 can be considered a faster and effortless molecular testing system.
- Citation: Chooramani G, Samal J, Rani N, Singh G, Agarwal R, Bajpai M, Kumar M, Prasad M, Gupta E. Performance evaluation of NeuMoDx 96 system for hepatitis B and C viral load. World J Virol 2023; 12(4): 233-241
- URL: https://www.wjgnet.com/2220-3249/full/v12/i4/233.htm
- DOI: https://dx.doi.org/10.5501/wjv.v12.i4.233
Hepatitis B virus (HBV) and hepatitis C virus (HCV) are among the most common causes of chronic liver diseases worldwide. This underscores its public health significance, with more than 250 million HBV carriers and about 70 million people with chronic HCV infection[1]. As a result of HBV vaccination programs[2,3] and direct-acting antiviral treatment against HCV[4], the prevalence of these infections has significantly decreased but both are still major risk factors for the development of cirrhosis and hepatocellular carcinoma. In resource-poor nations, gaps in HBV and HCV screening, treatment, and monitoring, as well as a lack of efficient therapeutic management choices for HBV/HCV-related liver cancer, have resulted in a low survival rate among infected persons. In response to this significant worldwide public health burden, the World Health Organization launched a worldwide Health Sector Strategy on Hepatitis 2016-2021 in 2016, with the objective of eliminating viral hepatitis as a public health issue by 2030[5]. Moreover, HBV and HCV account for about 96% of all viral hepatitis deaths in low- and middle-income countries[1]. The complete burden of disease includes not just death but also decreased quality of life for patients (due to cirrhosis and associated comor-bidities), financial expenses of care for people and health care systems alike, and social economic costs[6]. Serological assays to detect antigens/antibodies to HBV/HCV do not differentiate between an active and past infection; thus, an active ongoing infection must be confirmed by the detection of viral DNA/RNA[7]. This two-step process for diagnosing active HBV & HCV infection, i.e. first a serological test to screen for exposure, followed by a nucleic acid test (NAT) to confirm viremia often takes a longer time and leads to patient loss to follow-up[6].
Therefore, to prevent loss to follow-up of a patient from the cascade of care, it is important to have molecular assays that are fully automated with high throughput and short turnaround (TAT), particularly in low-income countries[8].
At present, batch-based closed molecular testing platforms are routinely being used but recently fully automated random access (i.e. single sample) closed systems are coming up and have demonstrated good potential[9,10]. The NeuMoDx 96 molecular system (QIAGEN Sciences, Waltham, MA, United States) is one such recent fully automated random access closed system. Therefore, the present study compared and evaluated the clinical performance of the NeuMoDx 96 molecular system (referred to as assay 1) for HBV and HCV viral load (VL) testing with the routinely used COBAS®AmpliPrep/COBAS® TaqMan®, v2.0 (Roche Diagnostics, GmbH, Mannheim, Germany) system (referred to as assay 2).
This was a retrospective study conducted in a tertiary care liver center in Delhi, India. A total of 186 archived once-thawed plasma samples with pre-existing test results from initial routine lab testing were used. Among these, 99 samples for the HBV assay and 87 samples for the HCV assay were simultaneously tested on both platforms from the same freeze-thaw cycle. Of the 99 samples, 49 were HBV true positive (hepatitis B surface antigen [HBsAg], anti-hepatitis B core antibody [anti-HBc] total, and HBV DNA-positive; VL range: 1 to > 106 log10 IU/mL), and the remaining 50 were confirmed negatives for HBV (HBsAg, anti-HBc and HBV DNA-negative). In the HCV assay, of the 87 samples, 39 were true positives for HCV infection (anti-HCV antibody and HCV RNA-positive; VL range: 1 to > 106 log10 IU/mL), and the remaining 48 were confirmed negatives for HCV infection (anti-HCV antibody and HCV RNA negative).
The clinical details along with other virological parameters were obtained from our Hospital Information System. Samples from patients with other co-infections, such as human immunodeficiency virus and hepatitis delta virus, on immunosuppressants and samples with insufficient available volume were excluded. This study was approved by the Institutional Ethics Board (Approval No. IEC/2023/102/MA06) and conducted as per the principles of the Declaration of Helsinki. The study was performed on deidentified, anonymous, archived once-thawed clinical plasma samples. Therefore, the requirement of individual patient consent was waived.
In the NeuMoDx 96, 550 µL plasma was used in the instrument as per the manufacturer’s instructions. The results of the test were automatically displayed on the system approximately within 1 h for HBV DNA and approximately within 1.25 h for HCV RNA. The results are expressed as IU/mL. The lower limit of detection (LOD) and lower limit of quantification (LLOQ) for both HBV and HCV assays provided by the manufacturer are 8.0 IU/mL and 0.9 log10 IU/mL, respectively (Table 1).
| Parameter | HBV | HCV | ||
| Assay 1 | Assay 2 | Assay 1 | Assay 2 | |
| Sample volume | 550 µL plasma | 650 µL plasma | 550 µL plasma | 650 µL plasma |
| Turnaround time | Approximately 1.00 h | Approximately 5.00 h | Approximately 1.25 h | Approximately 5.45 h |
| LOD in IU/mL | 8.0 | 20 | 8.0 | ≥ 15 |
| LLOQ as log10 IU/mL | 0.9 | 1.3 | 0.9 | 1.5 |
| Linear range quantification as log10 IU/mL | 1-9 | 1.3-8.2 | 1-9 | 1.5 |
In the COBAS®AmpliPrep/COBAS® TaqMan®, v2.0, 650 µL plasma sample was used in the instrument. The results of the test were automatically displayed on the system within 5 h for HBV DNA and about 5 h 45 min for HCV RNA. For the HBV assay, the LOD and LLOQ claimed by the manufacturer are 20 IU/mL and 1.3 log10 IU/mL, respectively. For the HCV assay, the LOD and LLOQ are ≥ 15 IU/mL and 1.5 log10 IU/mL, respectively (Table 1).
The HBV and HCV VLs are expressed in log10 format. Continuous variables were summarized using the mean ± SD or median with interquartile range (IQR), as applicable. Categorical variables are presented as percentages. Linear regression and Pearson’s correlation coefficient were determined for VL results between assay 1 and assay 2. The Bland-Altman plot was used to determine the level of agreement between the assays, in which the difference in HBV VLs measured by the two assays was plotted against the mean of the assays. All analyses were done using Statistical Package for the Social Sciences software version 22 (SPSS Inc., Chicago, IL, United States).
The baseline demographic and clinical characteristics of the study population are described in Table 2. The male:female ratio was 2.5:1.0. The overall mean age was 45 years ± 15 years. The median VL detected for the HBV DNA-positive samples was 2.9 (IQR 2.0-4.3) log10 IU/mL and 3.7 (IQR: 2.28-4.56) log10 IU/mL for assay 1 and assay 2, respectively. For HCV RNA, the median VL detected was 4.9 (IQR: 4.2-5.4) log10 IU/mL and 5.1 (IQR: 4.07-5.80) log10 IU/mL for assay 1 and assay 2, respectively. Further, for the HBV assay, genotyping data were available for 24 plasma samples with the following information: GTD (n = 10 as D1 and n = 8 as D2), GTA (n = 4, all as A1) and GTC (n = 2, all as C1). For the HCV assay, genotype results were available for 34 plasma samples with the following distribution: GT3 (n = 20 as 3a and n = 4 as 3b), GT1 (n = 6, all were GT1a), and GT4 (n = 4, all were GT4c).
| Characteristic | HBV patient group | HCV patient group |
| Patients, n | 99 | 87 |
| Sex as M/F ratio | 3.34:1.00 | 1.71:1.00 |
| Age in yr, mean ± SD | 44.0 ± 14.6 | 46.0 ± 14.9 |
| Total bilirubin in mg/dL, median (IQR) | 0.83 (0.59-0.83) | 0.95 (0.58-2.15) |
| Direct bilirubin in mg/dL, median (IQR) | 0.18 (0.10-0.47) | 0.25 (0.10-0.62) |
| ALT values in IU/mL, median (IQR) | 32.5 (23.2-60.1) | 33 (17.0-59.7) |
| AST values in IU/mL, median (IQR) | 36 (28.00-62.75) | 46 (27.0-99.2) |
| NeuMoDx viral load as log10 IU/mL, median (IQR) | 2.9 (2.0-4.3) | 4.9 (4.2-5.4) |
| COBAS viral load as log10 IU/mL, median (IQR) | 3.7 (2.28-4.56) | 5.1 (4.07-5.80) |
A total of 99 samples were analyzed, of which 49 were true HBV positives and 50 were true negatives for HBV infection. A 100% concordance was observed for both positive and negative samples. Therefore, the sensitivity (95% confidence interval [CI]: 92.75-100.00) and the specificity (95%CI: 92.89-100.00) were found to be 100%. Assay 1 showed no difference in quantification of HBV VL across different genotypes compared to assay 2.
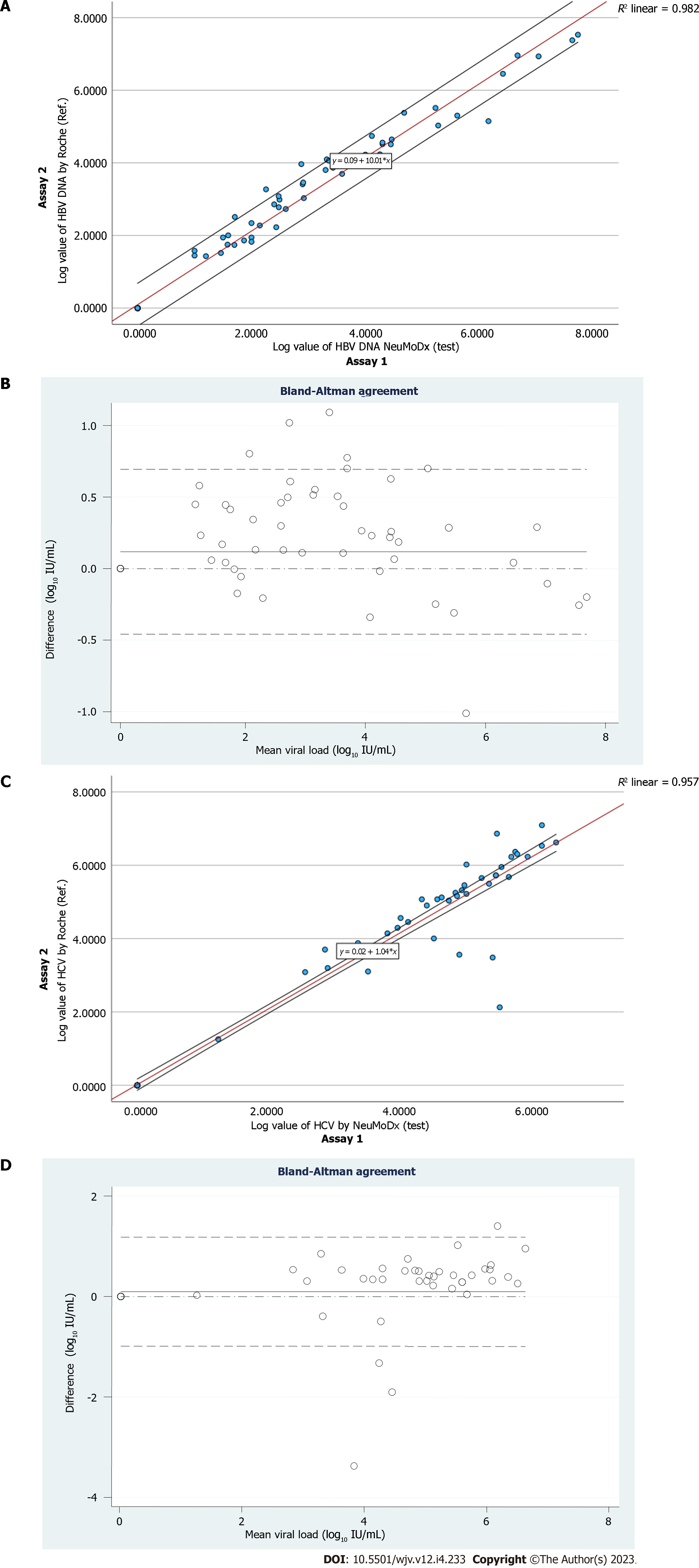
A total of 49 confirmed HBV DNA-positive samples were used with VL ranging from 1.0 to 7.7 log10 IU/mL (Table 3). Linear regression analysis of the quantifiable VLs obtained by both assays demonstrated a good correlation (Pearson’s correlation coefficient; r2 = 0.991) (Figure 1A). The Bland-Altman plot was used to determine the level of agreement between the assays (Figure 1B). The mean difference between assay 1 and assay 2 was 0.05 log10 IU/mL, with agreement limits ranging from -0.458 to 0.694 log10 IU/mL. With an intraclass correlation coefficient (ICC) of 0.995 (95%CI: 0.990-0.997; P = 0.001), the two systems agreed well (Figure 1B).
| Range of viral load as log10 IU/mL | HBV | |
| Assay 1, n = 49 | Assay 2, n = 49 | |
| 101-103 | n = 25 (51.0%) | n = 19 (38.7%) |
| 103-106 | n = 18 (36.7%) | n = 25 (51.0%) |
| > 106 | n = 6 (12.2%) | n = 5 (10.2%) |
A total of 87 samples were analyzed, of which 39 were true HCV positives and 48 were confirmed negatives for HCV infection. A 100% concordance was observed for both the positive and negative samples. Therefore, the sensitivity (95%CI: 90.97-100.00) and specificity (95%CI: 92.6-100.0) were found to be 100% compared to assay 2. No difference was seen in VL quantification among the different HCV genotypes between assay 1 and assay 2.
A total of 39 confirmed HCV RNA-positive samples were used with VL ranging from 1.26 to 7.09 log10 IU/mL (Table 4). Linear regression analysis of the quantifiable VLs obtained by both assays demonstrated a good correlation (Pearson’s correlation coefficient; r2 = 0.978) (Figure 1C). The Bland-Altman plot was used to determine the level of agreement between the assays (Figure 1C). The mean difference between assay 1 and assay 2 was 0.06 log10 IU/mL, with agreement limits ranging from -0.987 to 1.182 log10 IU/mL. With an ICC of 0.988 (95%CI: 0.981-0.992; P = 0.001), the two systems agreed well (Figure 1D).
| Range of viral load (log10 IU/mL) | HCV | |
| Assay 1 (n = 39) | Assay 2 (n = 39) | |
| 101-103 | n = 4 (10.2%) | n = 2 (5.1%) |
| 103-106 | n = 32 (82.0%) | n = 28 (71.7%) |
| > 106 | n = 3 (7.69%) | n = 9 (23.07%) |
In this study, the performance of a random access system (NeuMoDx 96) was compared with the currently used batch system (COBAS AmpliPrep/COBAS TaqMan) for the detection of HBV DNA and HCV RNA from clinical samples. A concordance of 100% was observed between both systems for both HBV DNA and HCV RNA analyses. Moreover, both assays demonstrated a good correlation with respect to the VL estimation. Such systems with random access to samples are the need of the hour, so that results are provided in a shorter TAT to the clinicians[11]. In the cascade of care for both HBV DNA and HCV RNA, patients screened initially through serological tests often are lost to follow-up for VL testing due to either non-availability of the assays or a high TAT. The use of existing batch testing platforms across most of the diagnostic labs results in longer “sample-to-result” time[12-15].
At present, for conventional real-time PCR, closed systems are commonly used[16]. Though the analytical sensitivity or LOD of these assays is good and lies between 5 and 15 IU/mL, these available assays typically run on batch testing to justify and minimize the cost of the test. This further leads to an increase in TAT, which eventually delays the test results, and augments the total number of visits of the patients to the health care facility, and dropouts from the care process[17]. Another important aspect is that if viremic patients are not identified earlier, they pose a significant risk of transmission of active infection to their family members and close contacts due to the nature of transmission of both HBV and HCV infections.
Almost near point-of-care molecular testing platforms that are available at present for HBV/HCV VL are very limited (GeneXpert, TrueNAT)[9,18]. These systems provide random access to samples but are useful in diagnostic sites with low sample throughput such as in peripheral settings or remote areas, whereas systems such as NeuMoDx can cater to large numbers of samples and be placed easily in a high-throughput laboratory. Molecular testing platforms such as NeuMoDx can provide faster results with a very short TAT in a cost-effective manner. Limited studies regarding the evaluation of the NeuMoDx platform are available. Our study focused on its real-life evaluation on clinical samples[19-21].
We found good correlation of HBV DNA and HCV RNA levels, especially when the VL in the sample was higher (> 106 log10 IU/mL). However, in VL 101-103 log10 IU/mL, despite good concordance, only satisfactory correlation was seen. This could be because very low VLs in clinical samples are often difficult to quantitate in terms of their exact values.
This system includes various functions including specimen preparation, nucleic acid extraction, real-time PCR set-up, amplification, and detection, all integrated within a single platform. Notably, this system provides numerous advantages, including high-throughput capacity, complete traceability, minimal risk of cross-contamination, and stringent safety standards for end users. The assay holds all of the reagents required for up to 20 different assays on board and works at room temperature. Therefore, this automated assay can be used in resource-limited settings, particularly in areas where refrigeration could be a problem. This can be used just like chemiluminescence platforms used for serological testing and would not require specialized molecular testing facilities or separate infra-structure requirements[22], which are extremely expensive to design and require specialized trained manpower to perform such assays. The expertise needed to perform testing on this platform and subsequent manual steps are greatly reduced and a single person can operate on the machine and perform multiple assays together.
An important limitation of our study is that it was a retrospective, single-center study done on already archived tested samples; thus, large-scale prospective studies in real time are needed to further evaluate their reproducibility, stability, and cost-effectiveness.
The findings of the study demonstrated excellent clinical performance (100% concordance) of the random access system compared to the conventional routinely used batch system. With a short TAT and user-friendly operation, it is a reliable assay for HBV and HCV VL assessment. These promising preliminary results indicate that the NeuMoDx 96 system holds great potential as a novel VL measurement solution in effective patient care and management.
Large-scale prospective studies are needed to evaluate the overall analytical, clinical performance, and reproducibility of the NeuMoDx 96 system.
The findings of this study demonstrated excellent clinical performance (100% concordance) of the random access system compared to the conventional commercially available batch system. With a short turnaround time (TAT) and user-friendly operation, it is a reliable assay for hepatitis B virus (HBV) and hepatitis C virus (HCV) viral load (VL) ass-essment.
In this study, overall a good correlation (100% concordance) and agreement were found between the two systems for HBV DNA and HCV RNA VL quantification. The sensitivity and specificity of the NeuMoDx 96 for both HBV and HCV assays were found to be 100%. Moreover, no difference was found in the quantification of HBV/HCV VL across different genotypes.
A total of 186 archived once-thawed plasma samples with pre-existing test results from initial routine lab testing were used. The overall concordance, correlation, and agreement were evaluated among all samples for HBV and HCV VL estimation using both systems.
The objectives of the study were: (1) Comparison of a random access system with conventional routinely used real-time PCR for quantifying HBV and HCV VL in plasma samples; (2) to estimate the overall concordance and agreement of VL quantification of clinical samples between the two systems; and (3) to evaluate genotype-based comparison of HBV and HCV VL between both systems.
To date, most routinely used assays for HBV/HCV VL estimation typically run on batch testing. The use of such platforms across most of the diagnostic lab results in longer “sample-to-result” time, leading to loss to follow-up. Therefore, a reliable random access system with a shorter TAT is the need of the hour for all clinicians.
HBV and HCV pose a significant health burden in low- and middle-income countries. The TAT for VL quantification (from receiving samples to giving out results) is longer for the conventional routinely used batch system-based real-time PCR assays. Therefore, to prevent loss to follow-up of a patient from the cascade of care, it is important to have molecular assays that are fully automated with high throughput and short TAT.
We acknowledge QIAGEN Sciences (Waltham, MA, United States) for providing the NeuMoDx 96 system and the reagents for the study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Virology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Emran TB, Bangladesh; Gerlich W, Germany S-Editor: Chen YL L-Editor: Filipodia P-Editor: Chen YX
| 1. | World Health Organization. Global hepatitis report 2017. [cited 20 July 2023]. Available from: https://www.who.int/publications/i/item/9789241565455. |
| 2. | Chen CL, Yang JY, Lin SF, Sun CA, Bai CH, You SL, Chen CJ, Kao JH, Chen PJ, Chen DS. Slow decline of hepatitis B burden in general population: Results from a population-based survey and longitudinal follow-up study in Taiwan. J Hepatol. 2015;63:354-363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 61] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 3. | Goyal A, Murray JM. The impact of vaccination and antiviral therapy on hepatitis B and hepatitis D epidemiology. PLoS One. 2014;9:e110143. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 41] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 4. | Falade-Nwulia O, Suarez-Cuervo C, Nelson DR, Fried MW, Segal JB, Sulkowski MS. Oral Direct-Acting Agent Therapy for Hepatitis C Virus Infection: A Systematic Review. Ann Intern Med. 2017;166:637-648. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 497] [Cited by in RCA: 549] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 5. | World Health Organization. Progress report on HIV, viral hepatitis and sexually transmitted infections 2019. [cited 20 July 2023]. Available from:https://www.who.int/publications/i/item/progress-report-on-hiv-viral-hepatitis-and-sexually-transmitted-infections-2019. |
| 6. | Heffernan A, Barber E, Cook NA, Gomaa AI, Harley YX, Jones CR, Lim AG, Mohamed Z, Nayagam S, Ndow G, Shah R, Sonderup MW, Spearman CW, Waked I, Wilkinson RJ, Taylor-Robinson SD. Aiming at the Global Elimination of Viral Hepatitis: Challenges Along the Care Continuum. Open Forum Infect Dis. 2018;5:ofx252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 18] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Chevaliez S, Pawlotsky JM. Diagnosis and management of chronic viral hepatitis: antigens, antibodies and viral genomes. Best Pract Res Clin Gastroenterol. 2008;22:1031-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 49] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 8. | AASLD-IDSA HCV Guidance Panel. Hepatitis C Guidance 2018 Update: AASLD-IDSA Recommendations for Testing, Managing, and Treating Hepatitis C Virus Infection. Clin Infect Dis. 2018;67:1477-1492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 477] [Cited by in RCA: 477] [Article Influence: 68.1] [Reference Citation Analysis (0)] |
| 9. | Gupta E, Agarwala P, Kumar G, Maiwall R, Sarin SK. Point -of -care testing (POCT) in molecular diagnostics: Performance evaluation of GeneXpert HCV RNA test in diagnosing and monitoring of HCV infection. J Clin Virol. 2017;88:46-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 10. | Fourati S, Challine D, Poveda JD, Laperche S, Rallier S, Pawlotsky JM, Chevaliez S. Evaluation of a new random-access HBV DNA molecular assay: The VERIS HBV assay. J Clin Virol. 2017;92:69-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 11. | Cooke GS, Andrieux-Meyer I, Applegate TL, Atun R, Burry JR, Cheinquer H, Dusheiko G, Feld JJ, Gore C, Griswold MG, Hamid S, Hellard ME, Hou J, Howell J, Jia J, Kravchenko N, Lazarus JV, Lemoine M, Lesi OA, Maistat L, McMahon BJ, Razavi H, Roberts T, Simmons B, Sonderup MW, Spearman CW, Taylor BE, Thomas DL, Waked I, Ward JW, Wiktor SZ; Lancet Gastroenterology & Hepatology Commissioners. Accelerating the elimination of viral hepatitis: a Lancet Gastroenterology & Hepatology Commission. Lancet Gastroenterol Hepatol. 2019;4:135-184. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 395] [Article Influence: 65.8] [Reference Citation Analysis (0)] |
| 12. | Sauné K, Abravanel F, Haslé C, Boineau J, Mengelle C, Izopet J. Analytical performance of the VERIS MDx system HCV assay for detecting and quantifying HCV RNA. J Clin Virol. 2016;84:7-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 13. | Han MS, Park Y, Nah H, Kim HS. Comparison of the QIAGEN artus HBV QS-RGQ Assay With the Roche COBAS AmpliPrep/COBAS TaqMan HBV Assay for Quantifying Viral DNA in Sera of Chronic Hepatitis B Patients. Ann Lab Med. 2017;37:248-253. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 14. | Lô G, Ndiaye AJS, Sow A, Fall A, Barry O, Diop NM, Ndiaye HD, Kâne NCT, Mboup S. Comparison of Amplix® hepatitis B virus and Roche COBAS AmpliPrep/COBAS Taqman hepatitis B virus assay in quantifying HBV DNA in plasma of chronic hepatitis B in Senegal. Pan Afr Med J. 2021;38:294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 15. | Abravanel F, Lhomme S, Trémeaux P, Migueres M, Harter A, Haslé C, Bruel P, Alric L, Métivier S, Raymond S, Izopet J. Performance of the Xpert HBV Viral Load assay vs the Aptima Quant assay for quantifying hepatitis B virus DNA. DiagnMicrobiol Infect Dis. 2020;96:114946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 16. | Adler M, Goubau P, Leroux-Roels G, Sprengers D, Pawlotsky JM. Practical use of hepatitis C and B molecular tools: Belgian guidelines. Acta Gastroenterol Belg. 2005;68:308-313. [PubMed] |
| 17. | Hans R, Marwaha N. Nucleic acid testing-benefits and constraints. Asian J Transfus Sci. 2014;8:2-3. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 18. | McHugh MP, Wu AHB, Chevaliez S, Pawlotsky JM, Hallin M, Templeton KE. Multicenter Evaluation of the Cepheid Xpert Hepatitis C Virus Viral Load Assay. J Clin Microbiol. 2017;55:1550-1556. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 41] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 19. | Mourik K, Boers SA, van Rijn AL, Thijssen JCP, Wessels E, Claas ECJ. Clinical performance of two new, fully integrated molecular platforms used for HIV-1, HBV and HCV viral load analysis, the NeuMoDx 288 and the Alinity m. J Clin Virol. 2023;160:105376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 20. | Arikan A, Sayan M, Doluca O. NeuMoDx random access molecular diagnostic system for detection and quantification of hepatitis B virus in clinical samples. J Infect Dev Ctries. 2020;14:1197-1203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (1)] |
| 21. | Besombes J, Pronier C, Lefevre C, Lagathu G, Maillard A, Grolhier C, Thibault V. Performances of NeuMoDx™, a random-access system for hepatitis B virus DNA and hepatitis C virus RNA quantification. Clin Microbiol Infect. 2021;27:1693.e9-1693.e15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 22. | World Health Organization. Establishment of PCR laboratory in developing countries, 2nd edition. 2016. [cited 20 July 2023]. Available from: https://apps.who.int/iris/handle/10665/249549. |