Peer-review started: September 13, 2022
First decision: October 3, 2022
Revised: October 15, 2022
Accepted: December 1, 2022
Article in press: December 1, 2022
Published online: January 25, 2023
Processing time: 126 Days and 16.1 Hours
Empirical use of potentially hepatotoxic drugs in the management of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection is considered as one of the major etiopathogenetic factors for liver injury. Recent evidence has shown that an underlying genetic factor may also occur. Hence, it is important to understand the host genetics and iatrogenic-based mechanisms for liver dysfunction to make timely remedial measures.
To investigate drug-induced and genetic perspectives for the development of coronavirus disease 2019 (COVID-19)-related liver injury.
Reference Citation Analysis, PubMed, Google Scholar and China National Knowledge Infrastructure were searched by employing the relevant MeSH keywords and pertaining data of the duration, site and type of study, sample size with any subgroups and drug-induced liver injury outcome. Genetic aspects were extracted from the most current pertinent publications.
In all studies, the hepatic specific aminotransferase and other biochemical indices were more than their prescribed upper normal limit in COVID-19 patients and were found to be significantly related with the gravity of disease, hospital stay, number of COVID-19 treatment drugs and worse clinical outcomes. In addition, membrane bound O-acyltransferase domain containing 7 rs641738, rs11385942 G>GA at chromosome 3 gene cluster and rs657152 C>A at ABO blood locus was significantly associated with severity of livery injury in admitted SARS-CoV-2 patients.
Hepatic dysfunction in SARS-CoV-2 infection could be the result of individual drugs or due to drug-drug interactions and may be in a subset of patients with a genetic propensity. Thus, serial estimation of hepatic indices in hospitalized SARS-CoV-2 patients should be done to make timely corrective actions for iatrogenic causes to avoid clinical deterioration. Additional molecular and translational research is warranted in this regard.
Core Tip: Evidence highlights the multisystemic nature of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection. Hepatic dysfunction is the primary extrapulmonary manifestation. In addition to the direct cytopathic effect of the virus, iatrogenic causes and genetic susceptibility are also postulated in the pathogenesis of hepatic damage in SARS-CoV-2 infection. Degree of liver toxicity in terms of altered biochemical indices were consistent with severity of coronavirus disease 2019 (COVID-19) illness and hospital stay. Hence, serial monitoring of hepatic indices in COVID-19 hospitalized patients may provide useful prognostic value to make timely corrective actions to avoid clinical deterioration.
- Citation: Parchwani D, Sonagra AD, Dholariya S, Motiani A, Singh R. COVID-19-related liver injury: Focus on genetic and drug-induced perspectives. World J Virol 2023; 12(1): 53-67
- URL: https://www.wjgnet.com/2220-3249/full/v12/i1/53.htm
- DOI: https://dx.doi.org/10.5501/wjv.v12.i1.53
Since the index case of coronavirus disease 2019 (COVID-19) infection was confirmed in the month of December 2019 in China[1], the upsurge of COVID-19 has led to devastating effects on global health[2]. Considering the incessant evolution of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and its impact on public health, SARS-CoV-2 variants were labelled as “variants of concern” (e.g., Alpha, Beta, Gamma, Delta and Omicron) and “variants of interest” (e.g., Eta, Iota, Kappa and Lambda) based on their attributes[3,4].
Each of the variants penetrates human cells by binding the cell surface receptor angiotensin converting enzyme (ACE) 2 via the spike protein subunit 1, while spike protein subunit 2 permits entry of the virus by enabling fusion of the virus envelope with the host cell membrane. This virus-cell fusion is facilitated by S protein priming by host cell proteases viz transmembrane protease serine 2 at a cleavage site (spike protein subunit 1/spike protein subunit 2), which is a polybasic furin cleft[3,4]. Fusion of the viral and cell membrane is followed by the entry of the virus inside the host cell to release the genetic material, i.e., positive sense RNA. This RNA genome is the template for synthesis of new negative sense RNA with the help of RNA-dependent RNA polymerase. Newly synthesized RNA in turn facilitate the synthesis of positive sense RNA, which is responsible for the production of new cytoplasmic proteins, namely nucleocapsid protein and membrane protein. Nucleocapsid protein binds to freshly synthesized positive sense RNA, and membrane protein facilitates its assimilation into the endoplasmic reticulum (ER) to form nucleocapsids. These nucleocapsids are finally transferred to the cell membrane via the ER lumen and Golgi vesicle to the extracellular space via exocytosis[5,6].
These newly released virions infect the neighboring healthy cells and manifest COVID-19 with a diverse spectrum of symptoms, ranging from asymptomatic disease to severe symptoms, primarily associated with the respiratory system. However, emerging scientific evidence highlights the multisystemic nature of the disease, i.e., involving extrapulmonary clinical manifestations such as myocardial infarction, neurological, ocular, dermatologic, gastrointestinal, kidney failure and liver dysfunction, owing to the tropism of the virus for ACE2 expressed in different human cells[7]. In fact, liver injury is the primary extrapulmonary manifestation, and the most common pattern is mild to moderate hepatocellular injury, observed in 14%-53% of the hospitalized patients with COVID-19. Furthermore, epidemiological studies have revealed that over one-half of infected patients with SARS-CoV-2 had deranged liver function tests characterized by abnormal levels of hepatic specific aminotransferases and other hepatic specific biochemical indices[8,9], while a small subset of patients was found with acute liver damage and fulminant hepatic failure[10,11]. Altered biochemical indices were more frequent in severely ill COVID-19 patients in contrast to patients presenting with mild to moderate illness[12,13].
In addition, certain therapeutic compounds can cause drug-induced liver injury (DILI)[14]. This was substantiated by the findings that toxicity was relieved after the cessation of these agents in in vitro and in vivo experiments[15,16]. These compounds include: (1) Antibiotics (azithromycin and ceftriaxone); (2) Antivirals [remdesivir (RDV), lopinavir (LPV)/ritonavir (RTV), favipiravir, umifenovir and triazavirin]; (3) Antimalarial [hydroxychloroquine (HCQ)]; (4) Adjuncts; (5) Steroids (dexamethasone); and (6) Immunomodulators [tocilizumab (TCZ)]. Overall, available data suggest that the spectrum of hepatic damage in SARS-CoV-2 infection may be accredited to the direct cytopathic effect of the virus through the ACE2 receptor, indirect involvement by systemic immune-mediated inflammation and by iatrogenic causes, i.e., drug-induced[17,18].
Moreover, underlying genetic factors could also contribute to COVID-19-related liver abnormalities due to the occurrence in a subset of the patient population. In accordance with this, a substantial number of genetic-based and or association studies have addressed the genetic makeup of the host in regards to the predisposition to the development and progression of COVID-19-related liver injury to recognize the patient cohort for high clinical priority in terms of early or novel therapeutic interventions albeit with equivocal results.
Since there are limited data on the individual genetic susceptibility to SARS-CoV-2 infection-related liver abnormalities, a detailed understanding of the influence of specific genotypes will be crucial for clinical outcomes. In addition, substantial evidence from the scientific literature indicate that the degree of liver toxicity is due to a certain therapeutic regime employed in the treatment of SARS-CoV-2. Only a handful of researchers methodically and comprehensively explored the complete array of DILI in COVID-19 patients. Hence, it is worth reviewing the genetic and drug-induced perspectives on COVID-19-related liver injury. This review emphasized DILI in COVID-19 patients along with genetic insight into the development of SARS-CoV-2 infection-related liver injury by providing evidence from the most current pertinent publications using relevant keywords from online databases.
Using various electronic databases, namely Reference Citation Analysis, PubMed, China National Knowledge Infrastructure and Web of Science, our team carried out the relevant literature search using the following MeSH keywords: DRUG INDUCED LIVER INJURY AND COVID-19 OR DRUG INDUCED LIVER INJURY AND SARS-COV-2 OR DRUG INDUCED LIVER INJURY AND 2019 nCOV OR DRUG INDUCED LIVER INJURY AND CORONAVIRUS DISEASE with regards to drug-induced perspectives and COVID-19 AND LIVER INJURY AND POLYMORPHISM OR SARS-COV-2 AND LIVER INJURY AND POLYMORPHISM OR COVID-19 AND LIVER INJURY AND GENETIC INSIGHT OR SARS-COV-2 AND LIVER INJURY AND GENETIC INSIGHT for genetic insight of hepatic damage.
The criteria for inclusion were: Original articles; case series or reports; brief communication; or letters to the editor. However, articles were in English and published during between December 1, 2020 and April 30, 2022. The references of the articles from the initial search were screened to add any plausible relevant literature. However, studies with animal or cellular models were not included. Other criteria for exclusion were: Injury due to SARS-CoV-2 infection itself; and hepatic injury from herbal or dietary supplements. Finally, after eliminating duplicate articles, 31 (2 and 29 relating to genetic insight and DILI, respectively) out of 727 (14 and 713 articles, respectively, for genetic insight and DILI) articles were selected for review.
By means of the aforementioned key words and in line with inclusion and exclusion criteria of the study, Sonagra AD, Dholariya S and Motiani A reviewed articles to ensure the fulfilment of inclusion criteria. Thereafter, Parchwani D and Singh R chose the articles to be finally included in the study. The authors then extracted the data: Author, site and sample size of study, stages of COVID-19, severity of disease, medication, outcome and/or DILI. All the mentioned information were extracted by standardized data extraction tables in duplicate.
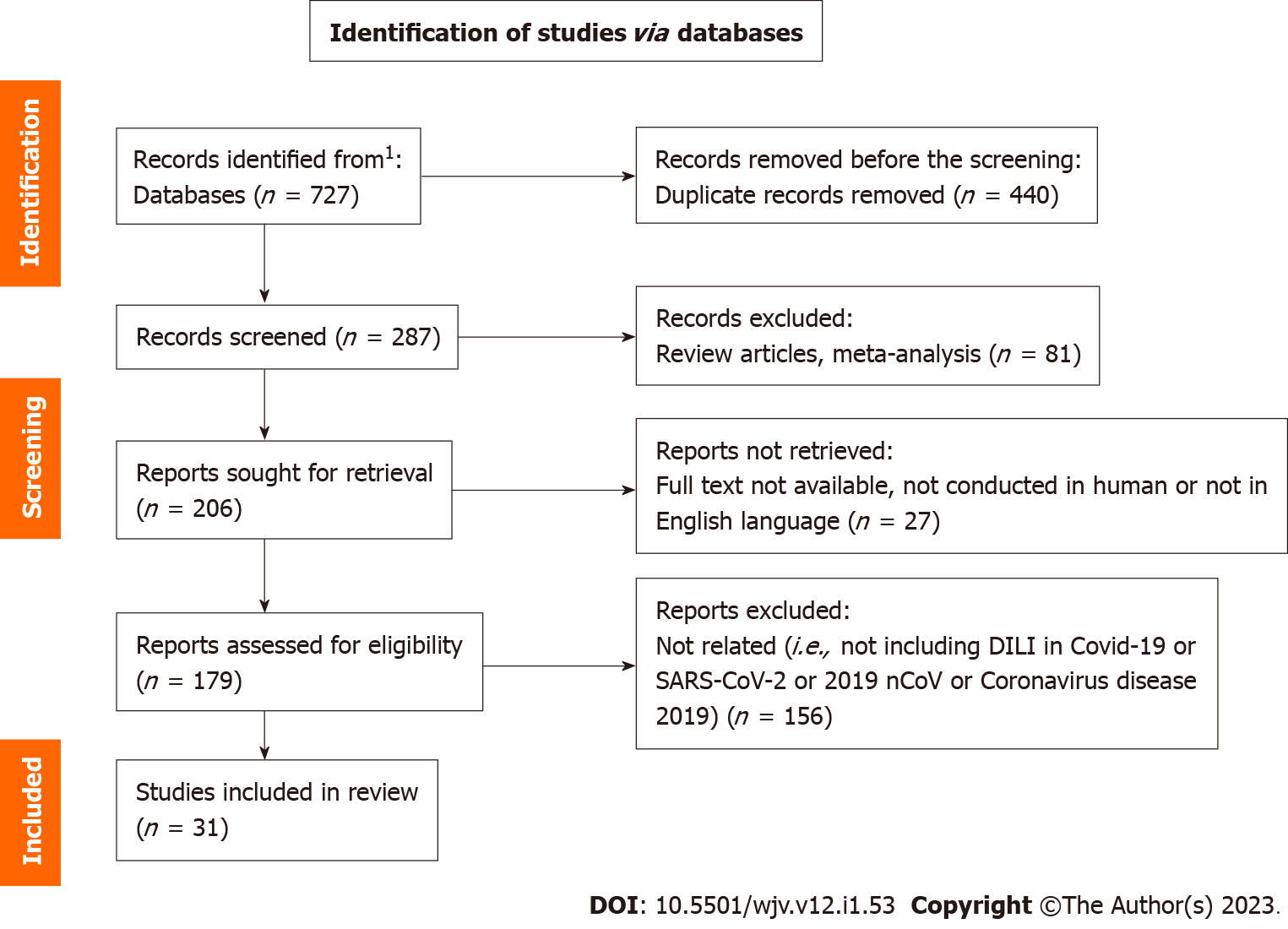
A total of 8 studies on genetic insight and 279 studies concerning DILI were screened after removing the duplicate publications. Among the included studies, a total of 31 studies were considered suitable for the qualitative synthesis comprising 2 studies regarding genetic insight and 29 studies regarding DILI. Extraction of research articles (Figure 1) were performed as per the guideline prescribed in PRISMA statement 2020 and was done according to the published protocol (PROSPERO ID: CRD42022311838).
Medications and or therapies employed in COVID-19 management, such as RDV, LPV/RTV, ribavirin, TCZ, hydroxyl chloroquine sulfate, etc are potentially hepatotoxic, specifically in high doses[19], and their administration in the form of polypharmacy exponentially increases the risk of DILI. The hepatotoxicity information as described across the included studies are compiled herewith for most frequently used drugs in the therapeutic regime of COVID-19. Table 1 depicts the relevant information per study. The most frequently associated drugs with DILI were RDV, LPV/RTV, TCZ, HCQ (+/-) azithromycin, ceftriaxone, paracetamol and enoxaparin.
| Sr. No. | Ref. | Type of study | Location | Number of study participants | Medication | Outcome |
| 1 | Grein et al[20], 2020 | Cross sectional/follow-up study | United States, Canada, Europe, Japan | 53 | Remdesivir | Elevation of liver enzymes in 12/53 patients |
| 2 | Goldman et al[76], 2020 | Randomized, open-label, phase 3 trial | United States, Italy, Spain, Germany, Hong Kong, Singapore, South Korea, Taiwan | 397 | Remdesivir | Elevated ALT in 26; elevated AST in 23; elevated bilirubin in 5 |
| 3 | Wang et al[1], 2020 | Randomized, double-blind, placebo-controlled, multicenter trial | China | 158 | Remdesivir | Elevated ALT in 2; elevated AST in 7, elevated bilirubin in 16, decreased albumin in 20 |
| 4 | Antinori et al[77], 2020 | Prospective (compassionate), open-label study | Italy | 35 | Remdesivir | Elevated transaminase in 15; elevated bilirubin in 7 |
| 5 | Leegwater et al[23], 2021 | Case study | Netherlands | 1 | Remdesivir | Elevated ALT, AST, ALP and GGT |
| 6 | Lee et al[24], 2020 | Case series | South Korea | 10 | Remdesivir | Elevated ALT in 5; elevated AST in 5 |
| 7 | Zampino et al[25], 2020 | Case series | Italy | 5 | Remdesivir | Elevated ALT in 4; elevated AST in 4 |
| 8 | Carothers et al[26], 2020 | Case series | United States | 2 | Remdesivir | Elevated ALT, AST, bilirubin, INR, ammonia in both patients; elevated ALP in 1 |
| 9 | Sun et al[27], 2020 | Active monitoring study by hospital pharmacovigilance system | China | 217 | LPV/ritonavir, umifenovir | Elevated ALT in 30 |
| 10 | Fan et al[28], 2020 | Retrospective, single-center study | China | 148 | LPV/ritonavir | Elevated ALT in 27; elevated AST in 32; elevated GGT in 26; elevated ALP in 6; elevated bilirubin in 18 |
| 11 | Cai et al[9], 2020 | Cross-sectional study | China | 417 | LPV/ ritonavir, oseltamivir, interferon, NSAIDs, ribavirin | Elevated ALT in 167; elevated AST in 137; elevated GGT in 143; elevated ALP in 71; elevated bilirubin in 196 |
| 12 | Jiang et al[29], 2020 | Multicenter, retrospective, observational study | China | 131 | LPV/ritonavir | Elevated ALT in 45; elevated AST in 4; elevated bilirubin in 43 |
| 13 | Serviddio et al[30], 2020 | Case series | Italy | 7 | LPV/ritonavir, HCQ, azithromycin | Elevated ALT, AST, GGT in all patients |
| 14 | Guaraldi et al[32], 2020 | Retrospective cohort study | Italy | 179 | Tocilizumab | No elevation of ALT or bilirubin was noted |
| 15 | Muhović et al[33], 2020 | Case report | Montenegro | 1 | Tocilizumab | Elevated ALT and AST |
| 16 | Hundt et al[34], 2020 | Retrospective observational cohort study | United States | 1827 | LPV/ritonavir, HCQ, remdesivir, tocilizumab | Elevated ALT in 1080 out of 1753; elevated AST in 1465 out of 1756; elevated ALP in 399 out of 1754; elevated total bilirubin in 284 out of 1747 |
| 17 | Kelly et al[35], 2021 | Retrospective analysis of hospitalized patients | Ireland | 82 | HCQ, azithromycin | Elevation of LFTs of more than ULN after 5 d therapy in 51/85 patients |
| 18 | Falcão et al[36], 2020 | Case report | Brazil | 1 | HCQ | Elevated ALT and AST with normal bilirubin and GGT |
| 19 | Yamazaki et al[38], 2021 | Case report | Japan | 1 | Favipiravir | Elevated ALT, AST, ALP, total bilirubin and LDH |
| 20 | Aiswarya et al[49], 2021 | Observational prospective study | India | 48 | Remdesivir | No significant change in serum transaminases and LDH |
| 22 | Kaur et al[78], 2022 | Case study | India | 1 | Remdesivir | Elevated ALT, AST and total bilirubin |
| 23 | Gao et al[79], 2022 | Retrospective study | China | 4010 | Oseltamivir, arbidol, interferon, ribavirin, LPV/ritonavir, HCQ/CQ, antibiotics, antifungals, corticosteroids | 395 out of 4010 developed DILI. 293 out of 395 received antibiotics, 25 out of 395 received antifungal, 42 out of 395 received oseltamivir, 52 out of 395 received ribavirin, 51 out of 395 received LPV/ritonavir, 47 out of 395 received interferon, 200 out of 395 received corticosteroid, 226 out of 395 received arbidol, 18 out of 395 received HCQ/CQ |
| 24 | Naseralallah et al[80], 2022 | Retrospective study | Qatar | 72 | Azithromycin, HCQ, LPV | Elevated ALT and AST was implicated in 24 patients due to azithromycin, in 11 patients due to HCQ and in 11 patients due to LPV |
| 25 | Chew et al[21], 2021 | Retrospective study | United States | 834 | Tocilizumab, remdesivir | 105 out of 834 (12.6%) had elevated AST |
| 26 | Delgado et al[22], 2021 | Retrospective observational study | Spain | 8719 | Remdesivir, hydroxychloroquine, azithromycin, tocilizumab and ceftriaxone | 4.9% of 8719 patients developed DILI. Out of which remdesivir had the highest incidence of DILI per 10000 defined daily doses |
| 27 | Durante-Mangoni et al[81], 2020 | Case report | Italy | 4 | Remdesivir | 3 out of 4 patients had elevated AST and ALT |
| 28 | Wong et al[82], 2022 | Self-controlled case series study | China | 860 | Remdesivir | 334 (38.8%) out of 860 had acute liver injury |
| 29 | Montastruc et al[83], 2020 | Multicenter study | France | 387 | Remdesivir | 130 (34%) out of 387 developed liver injury |
RDV: A broad-spectrum nucleotide analogue prodrug, primarily used for hospitalized patients with COVID-19 is known to inhibit viral RNA polymerases. RDV had the maximum DILI rate/administration. Screening of the World Health Organization safety reports database revealed a total of 387 adverse drug reactions (ADRs) reports of RDV by late 2021. Out of which the majority were hepatobiliary (61%) followed by 34% hepatic. The most common documented adverse liver outcome in different studies were elevated hepatic specific aminotransferase in the range of 15%-50%, hypoalbuminemia and hyperbilirubinemia[20]. Levels of aminotransferase elevation were more prominent in seriously ill patients, suggesting a possibility of occurrence of various adverse events due to severity and gravity of SARS-CoV-2 infection. Furthermore, studies also stated that in a subset of patients RDV treatment was discontinued on account of the abnormally high liver aminotransferase levels[20]. Chew et al[21] conducted a study in a sample of 834 COVID-19 hospitalized patients and reported that 12.6% (n = 105) of patients showed a > 5 upper limit of normal (ULN) of serum aspartate aminotransferase. Among the adverse lung events, TCZ and RDV were significantly associated with DILI on univariate analysis. Further, Delgado et al[22] conducted a retrospective observational study for the assessment of DILI by a pharmacovigilance program using laboratory signals. Out of 8719 patients admitted for COVID-19, 4.9% of patients developed DILI. The drugs commonly associated with DILI were HCQ, azithromycin, TCZ and ceftriaxone. Out of these, RDV had the highest incidence rate of 992.7 DILI per 10000 defined daily doses.
These adverse events were further corroborated by individual case reports/series. In one such report[23], after 2-d administration of RDV, a sharp elevation in the level of alanine aminotransferase (ALT) was observed, which was instantly corrected after discontinuing RDV. Correspondingly, in other reported cases[24,25], increased levels of liver enzymes were found in patients on RDV +/- HCQ, who were initially treated with LPV/RTV. In a case series reported in the United States, Carothers et al[26] suggested that administering acetyl cysteine had a positive impact on the overall health of the patient and reversed acute liver failure due to RDV with hepatic specific transaminase (ALT and aspartate aminotransferase) levels > 5000 IU/L and increased total bilirubin levels (3.1 mg/dL), serum ammonia (161 μmol/L) and international normalized ratio of 2.3.
LPV/RTV: Among the therapeutic regimes of COVID-19, LPV/RTV is one of the most common contributors of the liver ADRs. A study of 217 patients reported that LPV/RTV was found to be associated with 63% of total ADRs, while other drugs (umifenovir, chloroquine and antibacterial drugs) contributed to 47% of ADRs[27]. Correspondingly, a study of 148 patients reported that 48% developed hepatic function abnormality after admission to the hospital[28]. They emphasized that among such patients, 57.8% of the patients were treated with LPV/RTV. In another study, the authors reported an abnormal liver function in 12.1% of patients with the addition of each collateral medication[29]. Moreover, combined use of LPV/RTV with arbidol (umifenovir) in patients who were not terminally ill had an elevated risk of liver injury up to 3.58 times in comparison to those patient cohorts whose treatment regimen did not include LPV/RTV. It was postulated that RTV, being an inhibitor of chromosome 3 gene cluster A4, could promote hepatic toxicity from azithromycin via drug-drug interactions (polypharmacy)[26]. Similarly, metabolic interactions between the two medications [LPV/RTV and arbidol (umifenovir)] were studied in vitro using human liver microsomes by Serviddio et al[30] and concluded that LPV/RTV significantly impedes arbidol metabolism (P < 0.005), which may be the cause of DILI.
In a study with 163 mild and 29 severe patients with COVID-19, the multivariate analysis suggested RTV as one of the independent risk factors (odds ratio = 4.75, 95% confidence interval: 1.89-16.55, P < 0.001) in COVID-19 patients with liver injury[31]. In contrast to most of the studies that reported moderate-to-severe elevations in serum aminotransferase levels in patients under LPV/RTV treatment, Cai et al[9] reported increased odds of liver injury by four-fold in the LPV/RTV treated group among the enrolled 417 COVID-19 patients in China. The most significant increases were gamma-glutamyl transferase activity and total bilirubin. Because the drug was not efficacious, it was discontinued from the COVID-19 treatment regimen.
TCZ: TCZ, an interleukin-6 receptor antagonist monoclonal antibody, is primarily used for severely ill COVID-19 patients to arrest the cytokine storm. Guaraldi et al[32] did not find any adverse effects on the liver function test in a retrospective study involving 1351 COVID-19 patients treated with TCZ. In the following line of evidence, Serviddio et al[30] published a case series from Italy and displayed substantially altered hepatic and lung function tests after administration of LPV/RTV, HCQ and azithromycin for 5-7 d. These patients showed an improvement in both liver and lung function after the use of TCZ within 3 wk. However, the first case reported of DILI due to the use of TCZ did not deny the possibility of serious hepatotoxicity when used with other hepatotoxic drugs. In this case, 1 d after TCZ administration, the levels of serum transaminase increased up to 40-fold (aspartate aminotransferase of 1076 IU/L and ALT of 1541 IU/L)[33].
Another retrospective cohort study with 1827 patients conveyed that a positive correlation exists with usage of RDV, LPV/RTV, HCQ and TCZ and hepatotoxicity[34]. They observed peak hospitalization liver transaminase elevations more than 5 times the ULN.
HCQ with or without azithromycin: Most cases of DILI were reported for HCQ after its emergency use authorization for COVID-19 infection. Kelly et al[35] conducted an analysis in two groups of 134 patients. One group was treated with HCQ/azithromycin, while the other group was devoid of this targeted therapy. They reported no significant difference in the liver function tests between the two groups. On the contrary, a 10-fold elevation in levels of liver transaminases in the serum after HCQ administration was reported by Falcão et al[36]. They revealed that serum levels of hepatic enzymes rapidly declined after the withdrawal of HCQ from the treatment regimen.
Corticosteroids: Systemic corticosteroids, mainly dexamethasone, are widely used in patients with SARS-CoV-2 infections. However, an independent basis of hepatoxicity is uncommon. They are associated with minor, self-limiting elevations in serum aminotransferase. A study (n = 1040 COVID-19 patients) reported that the administration of corticosteroids was found to be correlated (adjusted odds ratio = 3.9) with development of acute liver injury on an independent basis (95% confidence interval: 2.1-7.2)[16].
Enoxaparin: Enoxaparin is associated with minor, self-limiting elevations in serum aminotransferase, but values > 5 ULN are uncommon. Sporadic cases of mild increases in serum bilirubin and alkaline phosphatase have been reported[37].
Favipiravir: A single study observed adverse liver events after favipiravir administration[38]. Authors of the study reported a COVID-19 patient who manifested bacterial pneumonia as a complication of COVID-19 during his hospital stay. The therapeutic regime of the patient was LPV/RTV combined with interferon β-1b. Following administration of favipiravir, liver transaminases and total bilirubin increased suggesting a cholestatic liver injury. The liver injury, in this case, may have been triggered by antibacterial treatment, which may have further deteriorated by treatment with a high dose of favipiravir.
Tang et al[39] found 17.31% (n = 3425) of patients exhibited DILI in a cohort of 19782 COVID-19 patients. The odds ratio for DILI was 2.99 (2.59-3.46), 5.39 (4.63-6.26) and 3.16 (2.68-3.73) when comparing LPV-RTV with all other drugs, RDV and HCQ/chloroquine, respectively. A single-center, open-label, parallel-arm, stratified randomized controlled trial completed by Panda et al[40] observed DILI in the form of elevated liver enzymes in 2 out of 67 participants who received a high dose of ribavirin.
In the matter pertaining to a genetic propensity towards SARS-CoV-2 induced liver injury, in the United Kingdom Biobank cohort[41] an elevated risk score of genetic fatty liver disease (FLD) based on glucokinase regulator, membrane bound O-acyltransferase domain containing 7, patatin like phospholipase domain containing 3 (PNPLA3) and transmembrane 6 superfamily 2 human gene genetic variants was not found to be associated with a higher probability of developing severe SARS-CoV-2. Hence, this finding challenges the causal role for metabolic-associated FLD in COVID-19 and implies that genetic susceptibility to hepatic fat deposition does not, in and of itself, increase the risk of developing a severe form of the disease[41]. However, contrary to this, membrane bound O-acyltransferase domain containing 7 rs641738[42], rs11385942 G>GA at chromosome 3 gene cluster and rs657152 C>A at the ABO blood locus were significantly associated with the severity of livery injury in admitted SARS-CoV-2 patients[43,44]. Thus, the genetic basis of SARS-CoV-2-induced liver injury is not yet fully understood, and additional research is required to validate the involvement of any specific variant form. Table 2 depicts the commonly employed therapeutic drugs for COVID-19, with its hepatic side effects and ‘Likelihood Score’ by the LiverTox database[45].
| Drug | Hepatotoxicity | Likelihood score |
| Remdesivir | A duration of 7-14 d of administration caused elevation of serum aminotransferases up to > 5 times of ULN. Elevation of > 5 times ULN were reported in 9% of patients but returned to normal after discontinuation. Prolonged and more severe effects were seen in critically ill patients with multiorgan involvement, pre-existing comorbidities and who had received combination therapy with other hepatotoxic agents like amiodarone | D |
| Lopinavir/ritonavir | A greater degree of rise in serum aminotransferase levels (> 5 times ULN) is mostly seen in association with immunodeficiency states. The pattern varies from hepatocellular to cholestatic or mixed type. Discontinuation leads to the normalization of enzyme levels. However, severe cases of acute liver failure or end stage liver disease are also reported with re-exposure of the drug | D |
| Tocilizumab | Reported to cause mild elevation of aminotransferases commonly, that is usually transient and asymptomatic, but rare instances of liver injury manifesting as jaundice and reactivation of hepatitis B are seen. ALT elevation (1-3 times ULN) was seen in 10%-50% of patients, which returned to baseline within 8 wk after stopping treatment. No effect on bilirubin or ALP levels were seen | C |
| Hydroxychloroquine | Clinically apparent liver injury is rare. In clinical trials for COVID-19 prevention and treatment, there were no reports of hepatotoxicity, and serum enzyme elevation was also low | C |
| Corticosteroids | Long-term use and high doses can result in hepatomegaly and steatosis. Can also trigger or exacerbate pre-existing or co-existing conditions like NASH, viral hepatitis or autoimmune hepatitis. Serum aminotransferase levels can rise up to 10-40 times ULN | A |
| Enoxaparin | 4%-13% of patients showed mild elevation in serum aminotransferase levels. Rapid onset of liver injury symptoms after starting the drug (within 3-5 d) but rapid recovery (1-4 wk) after discontinuation of therapy is seen | E |
| Favipiravir | Pretreatment with other hepatotoxic drugs like lopinavir/ritonavir and IF-β 1B lead to an increase in liver transaminase and bilirubin levels by manifold suggesting cholestatic injury. Isolated use is not known to cause any severe liver injury | D |
The primary analysis of this review revealed that DILI is due to the large-scale use of drugs/off-label drugs in the prophylactic and therapeutic regimen of COVID-19, and the causal relationship of genetic susceptibility with hepatic damage in SARS-CoV-2 infected patients is incomprehensible. Hepatic damage may arise either through intrinsic or idiosyncratic mechanisms. The intrinsic pathway is predictable and has a short latency period. However, COVID-19-related liver injury (drug-induced and/or genetic-based) predominantly follows the idiosyncratic mechanism, i.e., it is unpredictable with a variable latency period[19].
Drugs like LPV/RTV were associated with moderate to severe elevation (> 5 ULN) of hepatic specific aminotransferases in serum and exhibited a significantly (4 times) higher chances of liver injury[9]. The degree of hepatic damage varies widely, i.e., from injury of hepatocytes to complete stagnation of bile acid secretion (cholestatic injury) or may be both in certain cases[45]. Correspondingly, medication of COVID-19 patients with LPV/RTV might exaggerate dysfunction of hepatic cells in particular to hepatitis B virus and hepatitis C virus infection cases[46]. However, the efficacy of LPV/RTV in SARS-CoV-2 patients is not fully understood and requires further evaluation[47,48]. In contrast, a study in China suggested out that administration of antibiotics, ribavirin and nonsteroidal anti-inflammatory drugs is not associated with a statistically significant risk of hepatic damage[9].
Likewise, studies evaluating the ADRs of RDV reported that it could lead to liver injury, barring one[49]. Liver injury caused by RDV manifested only after the 3rd d of its administration as elevated hepatic specific transaminases, coagulopathy and hepatic encephalopathy. N-acetyl cysteine is recommended for the management of acute liver failure induced by RDV and discontinuation of drug for progression to acute liver failure[26]. It was suggested that the following criteria should be considered for an immediate cessation of RDV treatment: Elevation of ALT > 5 times ULN or elevation of alkaline phosphatase > 2 times ULN; increased level of total bilirubin more than > 2 times ULN; immediate incidence of coagulopathy; or in cases where the patient’s condition is deteriorating[50]. Thus, to diminish RDV-induced toxic effects, assessment of liver status must be completed before drug initiation, and continuous monitoring of the liver function test should be performed during the course of treatment.
However, the most contentious reports were of TCZ. A retrospective cohort study[32] and meta-analysis[47,48] reported that TCZ by itself is not associated with liver injury in COVID-19 patients. One study reported that TCZ had a positive effect on clinical and laboratory parameters caused by the use of LPV/RTV[30]. ALT levels fall within the normal range from > 5 times ULN after administration of TCZ. On the other hand, a study conducted by Muhović et al[33] reported that the hepatotoxic effects of TCZ were increased in cases of prior administration of antiviral drugs (LPV/RTV).
DILI in COVID-19 patients is often dependent on numerous factors. For instance, co-existing medical conditions (porphyria cutanea tarda, viral hepatitis and rheumatologic diseases) could increase the risk of developing toxicity due to recommended drugs[36]. Further, drug-drug interaction (e.g., chloroquine and its derivatives combined with anti-rejection immunosuppressants[51]) can lead to detrimental effects. For example, the prevalence of liver damage was 15.2% in a sample size of 208 COVID-19 patients on RDV only, whereas it was 37.2% among 775 patients treated with RDV and LPV/RTV[52], substantiating the concept of polypharmacy.
Among included studies, one study reported that the grade of hepatotoxicity was not statistically different between the controls and cases, who were treated with HCQ and azithromycin[35]. Nevertheless, divergent findings are also frequently reported for HCQ and has been hypothesized that the presence of pre-existing inflammation (mild to moderate) might increase the risk of liver damage by HCQ (+/- azithromycin) in the doses that are not hepatoxic due to the production of cytotoxic metabolites from drug metabolism by inflammatory cells with the help of myeloperoxidase enzyme. Ivermectin (anti-parasite medication) and colchicine (anti-inflammatory agent) are well-tolerated and have been reported to reduce the severity, length of hospital stay and prevention of a cytokine storm[47,53,54], but efficacy of these drugs in the management of SARS-CoV-2 infected patients is still not fully understood[55,56]. Hepatotoxic effects are not well documented.
Drugs employed in the management of SARS-CoV-2 infection (RDV, LPV/RTV, ribavirin, TCZ, HCQ or any other drugs) are metabolized by the hepatic cells. Liver damage with an associated increase of hepatic specific indices is predictable and has been corroborated and cited in the scientific literature[57-61] (Table 3).
| Proposed main mechanism | Explanation |
| Direct cytotoxicity | Active SARS-CoV-2 replication in hepatic cells, which further binds to hepatic and biliary epithelial cells by angiotensin-converting enzyme 2 and damage them by direct infection[57] |
| Immunological damage | Severe inflammatory response generated by SARS-CoV-2 further damages hepatic cells by immune mediated pathogenesis[58] |
| Drug-induced | Antiviral drugs such as remdesivir, chloroquine and ritonavir are possibly hepatotoxic[59] |
| Reactivation of pre-existing liver illness | Increased risk to develop hepatotoxicity in the presence of pre-existing liver diseases. In addition, baricitinib also causes reactivation of hepatitis B virus infection[60] |
| Anoxia | Anoxia or hypoxia leads to respiratory failure in SARS-CoV-2, which further leads to hypoxic hepatitis[61] |
Critical biochemical properties of anti-COVID-19 drugs that might lead to hepatotoxicity in susceptible hosts are lipophilicity, mitochondrial liability, generation of cytotoxic metabolites, their metabolic pathway in the liver and the ability to inhibit hepatic transporters[62]. Patients who died with severe COVID-19 had moderate microvesicular steatosis, a condition characterized by a variant form of hepatic fat accumulation and modest lobular and portal activity in their liver biopsies, suggesting that the liver injury may have been due to either viral- or drug-induced mechanisms. Steatosis in lieu of drugs occurs due to interference with β-oxidation of fatty acids, oxidative phosphorylation or both by certain drugs[63], resulting in the accumulation of free fatty acids, which are converted to triglycerides[64].
Clinical and murine studies have provided evidence that pre-existing medical conditions, e.g., inflammatory diseases, increased blood pressure and diabetes mellitus, augment SARS-CoV-2 hepatic injury, possibly because of ACE inhibitors or angiotensin receptor blockers, which results in ACE2 upregulation[65,66]. The presence of pre-existing nonalcoholic FLD sensitizes hepatocytes to antipyretic agents containing acetaminophen[67].
Reduced and/or suppressed activity of the CYP family or cytochrome P450 (enzyme responsible for metabolism of xenobiotics) is also a plausible mechanism to alter the activity of liver cells[68]. CYPs are downregulated due to repressive effects exerted by interleukins and cytokines, which are upregulated during COVID-19 infection, leading to toxicity of several COVID-19 drugs[68]. Drug-drug interactions also play an important role in the development and progression of DILI, as exemplified by the clearance of umifenovir, which is compromised by concomitant use of LPV/RTV due to its inhibitory effect on cytochrome P3A[29].
A precedent of hepatic transporter inhibition by COVID-19 drugs to manifest the liver injury is reported by many studies with regard to LPV, a prominent blocker of multidrug resistance-associated protein-2. A study performed on rats reported the accumulation of taurocholic acid inside the liver cells following the 10-min exposure of rat liver cells to protease inhibitor (PIs) drugs, LPV and RTV, indicating that the Pis inhibit the efflux of bile salts from liver cells[69]. Experimental studies showed the inhibitory effect of Pis on multidrug resistance-associated protein-2[70,71]. Holmstock et al[70] also reached a similar conclusion using 5(6)-carboxy-2’,7’-dichlorofluorescein through confocal imaging. Another recent study by Khalatbari et al[72] focused on oxidative stress damage leading to hepatotoxicity and FLD due to Pis, LPV and RTV. They reported the interference of these drugs by ER-Golgi trafficking through inhibition of Ras converting CAAX endopeptidase-1 and any of its substrate, which in turn leads to development of fatty liver and cellular stress.
Additionally, exhaustion of P450 activity to metabolize large and multiple amounts of COVID-19 drugs as a treatment regimen can also be the cause of hepatoxicity. Simultaneously, studies reported that administration of certain drugs (LPV/RTV) assists in the reactivation of hepatitis B and C viruses and results in hepatoxicity[73]; administration of HCQ in patients with porphyria cutanea tarda leads to significant liver damage[73] due to the interaction of reactive metabolites of HCQ and the inflammatory response due to SARS-CoV-2 infection[74].
However, there is a deficit of uniformity and standardization of DILI due to a lack of reliable and exclusive evidence pointing towards the drugs used in the treatment of COVID-19. Moreover, there is considerable overlap and commonality in the presenting symptoms of hepatic damage due to COVID-19 infection per se and due to drugs given for its treatment. Increased vigilance on the part of the clinicians is warranted so that cases of severe liver damage suspected to be caused by the drugs can be reported and entered into the National/International database. The R value can be considered as a diagnostic approach for the pattern of liver injury (i.e., R > 5 is considered hepatocellular DILI, R < 2 is considered cholestatic DILI, and R = 2-5 is considered mixed DILI; R value = ALT value/ULN divided by alkaline phosphatase value/ULN).
Irrespective of the aforementioned drugs in the treatment regime, Machill et al[42] reported that patients with carrier genotypes of membrane bound O-acyltransferase domain containing 7 rs641738 polymorphism had significantly elevated bilirubin, ALT and alkaline phosphatase levels and decreased serum albumin levels during hospitalization. This points towards genetic susceptibility.
Dongiovanni et al[75] explored the polygenic risk score of hepatic fat content and genetic markers of liver fibrosis (PNPLA3 I148M variant). Both polygenic risk score of hepatic fat content and PNPLA3 I148M were found to be inherited independently of dysmetabolism at conception to gain a better understanding into the relationship between FLD, liver damage and COVID-19[23]. They reported that rs11385942 G>A at chromosome 3 gene cluster and rs657152 C>A at the ABO blood locus were significantly associated with the severity of liver injury in admitted SARS-CoV-2 patients[75]. In fact, although a greater ALT was related to a genetic propensity to FLDs during SARS-CoV-2, this was accompanied by reduced systemic inflammation or C-reactive protein levels and maintenance of hepatic production or circulating serum albumin levels in carriers with the PNPLA3 I148M variant. The protective impact of the non-secretor ABO phenotype against SARS-CoV-2 infection has yet to be explained; it depends on whether or not differences in membrane glycan shedding underlie differential tissue susceptibility such as the liver and lungs. Finally, it was observed that the risk of severe COVID-19 in hospitalized patients was not elevated by the use of genetics-based assessment, which is a reliable unconfounded all-time proxy of a tendency to and progression of FLD. Therefore, genetic propensity to obtain liver fat, despite aiding in liver injury, may unexpectedly defend against inflammation throughout SARS-CoV-2, suggesting that FLD predilection does not automatically lead to increased inflammation[43].
To summarize, the available evidence outlines that the degree of lipophilicity of drugs, inflammatory response to the antivirals, metabolization by CYP3A4 in the liver, interference of drugs with various transporters in the liver and molecules/proteins accountable for protection against the xenobiotics (e.g., organic anion transporting polypeptide 1B1, p-glycoprotein, multidrug resistance-associated protein-2, breast cancer resistance proteins and ER-Golgi trafficking primarily by inhibiting Ras converting CAAX endopeptidase-1) are the underlying factors responsible for drug hepatotoxicity in SARS-CoV-2 infection treatment. Concurrently, drugs have a detrimental effect on bile salt export pump activity (an outflow transporter system responsible for excretion of waste and foreign substances from the hepatic cells) and thus becomes a central factor in the cholestasis process. In addition, robust shreds of evidence are lacking regarding the genetic predisposition to hepatic dysfunction in SARS-CoV-2 infection. Larger prospective studies are warranted in this regard.
Hepatic dysfunction in SARS-CoV-2 infection could be the result of an individual drug or due to interactions among more than one drug and may include a subset of the patient population that has a genetic propensity. Thus, serial estimation of hepatic indices in SARS-CoV-2 hospitalized patients, especially patients on treatment with drugs like RDV, LPV/RTV, favipiravir, HCQ and TCZ should be performed to take corrective actions for iatrogenic causes to avoid clinical deterioration.
Nonetheless, our findings described here are only an assortment of studies and do not imply causation. Other limitations include the sample sizes that were comparatively small. The methodology adopted in the included studies had a wide variation (as few studies only raised the probability of DILI rather than confirming the role of drugs with certainty). The therapeutic regimen, duration difference in the samples collected after hospital admission, the failure to record or the variation of the onset of disease/degree of liver injury and discrepancy in correcting different clinic and biochemical indices (sex, co-existing morbidities and age) varied between studies. The discrepancy in the measurement of liver indices and sub-classification of the cases was another limitation. Finally, only articles published in English were considered for analysis, which may have a local literature bias. In spite of all the aforementioned confines, the present review detailed important systematic data on the genetic susceptibility of liver damage and DILI in SARS-CoV-2 infection.
Available data advocate that the spectrum of hepatic damage in severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection may be accredited to the direct cytopathic effect of the virus, indirect involvement by systemic immune-mediated inflammation and by iatrogenic causes, i.e., drug induced. Empirical use of potentially hepatotoxic drugs in the management of SARS-CoV-2 infection is considered as one of the major etiopathogenetic factor for liver injury. Moreover, experimental and clinical evidence has shown that an underlying genetic factor may also be present. Hence, it is important to understand the genetics and iatrogenic-based mechanisms for liver dysfunction to make timely remedial measures.
To identify drug-induced liver injury in coronavirus disease 2019 (COVID-19) patients along with a genetic insight for the development of SARS-CoV-2 infection related liver injury to provide better care and timely management of critical patients.
To explore drug-induced and genetic perspectives in the development of SARS-CoV-2 infection related liver injury.
A systematic literature search was carried out in multiple electronic databases: PubMed, Reference Citation Analysis, China National Knowledge Infrastructure and Goggle Scholar. The literature was screened using related MeSH keywords and relevant data. The inclusion criteria were English language articles published between December 1, 2020 and April 30, 2022. Reference lists from the articles in the initial search were screened to identify additional literature. There was no exclusion based on the study outcome and stage or severity of SARS-CoV-2 infection. However, studies with animal or cellular models were not included. Other criteria for exclusion were: injury due to SARS-CoV-2 infection itself; and hepatic injury from herbal or dietary supplements.
The primary analysis of this review revealed that DILI was due to the large-scale use of drugs/off-label drugs in the prophylactic and therapeutic regimen of COVID-19, and the genetic susceptibility underlying liver damage in COVID-19 patients is not yet fully understood. COVID-19-related liver injury (drug-induced and/or genetic-based) predominantly follows the idiosyncratic mechanism, i.e., it is unpredictable with a variable latency period. In most commonly used drugs, the hepatic specific aminotransferases and other biochemical indices were elevated and were significantly associated with severity, hospital stay, number of COVID-19 treatment drugs and worse clinical outcomes.
Hepatic dysfunction in SARS-CoV-2 infection could be the result of individual drugs or due to drug-drug interactions and may include a subset of the patient population with a genetic propensity. Thus, serial estimation of hepatic indices in SARS-CoV-2 infection hospitalized patients should be performed to make timely corrective actions for iatrogenic causes to avoid clinical deterioration.
Additional prospective studies are warranted in this regard to justify drug-induced liver injury due to COVID-19 treatment along with the genetic predisposition, which should provide optimization of disease status.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Virology
Country/Territory of origin: India
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C, C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Bellini MI, Italy; Teixeira KN, Brazil; Yu F, China S-Editor: Wang JJ L-Editor: Filipodia P-Editor: Wang JJ
| 1. | Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470-473. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4848] [Cited by in RCA: 4388] [Article Influence: 877.6] [Reference Citation Analysis (1)] |
| 2. | Nicola M, Alsafi Z, Sohrabi C, Kerwan A, Al-Jabir A, Iosifidis C, Agha M, Agha R. The socio-economic implications of the coronavirus pandemic (COVID-19): A review. Int J Surg. 2020;78:185-193. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2872] [Cited by in RCA: 2681] [Article Influence: 536.2] [Reference Citation Analysis (0)] |
| 3. | Chan JF, Kok KH, Zhu Z, Chu H, To KK, Yuan S, Yuen KY. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg Microbes Infect. 2020;9:221-236. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1813] [Cited by in RCA: 1953] [Article Influence: 390.6] [Reference Citation Analysis (0)] |
| 4. | Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z. Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020;11:1620. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2003] [Cited by in RCA: 2291] [Article Influence: 458.2] [Reference Citation Analysis (0)] |
| 5. | Cascarina SM, Ross ED. A proposed role for the SARS-CoV-2 nucleocapsid protein in the formation and regulation of biomolecular condensates. FASEB J. 2020;34:9832-9842. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 103] [Cited by in RCA: 102] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 6. | Hartenian E, Nandakumar D, Lari A, Ly M, Tucker JM, Glaunsinger BA. The molecular virology of coronaviruses. J Biol Chem. 2020;295:12910-12934. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 326] [Cited by in RCA: 343] [Article Influence: 68.6] [Reference Citation Analysis (0)] |
| 7. | Canatan D, Vives Corrons JL, De Sanctis V. The Multifacets of COVID-19 in Adult Patients: A Concise Clinical Review on Pulmonary and Extrapulmonary Manifestations for Healthcare Physicians. Acta Biomed. 2020;91:e2020173. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 8. | Nkeih C, Sisti G, Schiattarella A. Elevated transaminases in a COVID-19 positive patient at term of gestation: a case report. Acta Biomed. 2020;91:e2020002. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 2] [Reference Citation Analysis (0)] |
| 9. | Cai Q, Huang D, Yu H, Zhu Z, Xia Z, Su Y, Li Z, Zhou G, Gou J, Qu J, Sun Y, Liu Y, He Q, Chen J, Liu L, Xu L. COVID-19: Abnormal liver function tests. J Hepatol. 2020;73:566-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 623] [Cited by in RCA: 661] [Article Influence: 132.2] [Reference Citation Analysis (0)] |
| 10. | Melquist S, Estepp K, Aleksandrovich Y, Lee A, Beiseker A, Hamedani FS, Bassett J. COVID-19 presenting as fulminant hepatic failure: A case report. Medicine (Baltimore). 2020;99:e22818. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 35] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 11. | Cichoż-Lach H, Michalak A. Liver injury in the era of COVID-19. World J Gastroenterol. 2021;27:377-390. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 53] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (2)] |
| 12. | Ghoda A, Ghoda M. Liver Injury in COVID-19 Infection: A Systematic Review. Cureus. 2020;12:e9487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 13. | Anirvan P, Bharali P, Gogoi M, Thuluvath PJ, Singh SP, Satapathy SK. Liver injury in COVID-19: The hepatic aspect of the respiratory syndrome - what we know so far. World J Hepatol. 2020;12:1182-1197. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (1)] |
| 14. | Abubakar AR, Sani IH, Godman B, Kumar S, Islam S, Jahan I, Haque M. Systematic Review on the Therapeutic Options for COVID-19: Clinical Evidence of Drug Efficacy and Implications. Infect Drug Resist. 2020;13:4673-4695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 15. | Wu J, Song S, Cao HC, Li LJ. Liver diseases in COVID-19: Etiology, treatment and prognosis. World J Gastroenterol. 2020;26:2286-2293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 105] [Cited by in RCA: 97] [Article Influence: 19.4] [Reference Citation Analysis (1)] |
| 16. | Yip TC, Lui GC, Wong VW, Chow VC, Ho TH, Li TC, Tse YK, Hui DS, Chan HL, Wong GL. Liver injury is independently associated with adverse clinical outcomes in patients with COVID-19. Gut. 2021;70:733-742. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 123] [Article Influence: 30.8] [Reference Citation Analysis (0)] |
| 17. | Taneva G, Dimitrov D, Velikova T. Liver dysfunction as a cytokine storm manifestation and prognostic factor for severe COVID-19. World J Hepatol. 2021;13:2005-2012. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 18. | Jothimani D, Venugopal R, Abedin MF, Kaliamoorthy I, Rela M. COVID-19 and the liver. J Hepatol. 2020;73:1231-1240. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 364] [Cited by in RCA: 358] [Article Influence: 71.6] [Reference Citation Analysis (1)] |
| 19. | Sandhu N, Navarro V. Drug-Induced Liver Injury in GI Practice. Hepatol Commun. 2020;4:631-645. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 41] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 20. | Grein J, Ohmagari N, Shin D, Diaz G, Asperges E, Castagna A, Feldt T, Green G, Green ML, Lescure FX, Nicastri E, Oda R, Yo K, Quiros-Roldan E, Studemeister A, Redinski J, Ahmed S, Bernett J, Chelliah D, Chen D, Chihara S, Cohen SH, Cunningham J, D'Arminio Monforte A, Ismail S, Kato H, Lapadula G, L'Her E, Maeno T, Majumder S, Massari M, Mora-Rillo M, Mutoh Y, Nguyen D, Verweij E, Zoufaly A, Osinusi AO, DeZure A, Zhao Y, Zhong L, Chokkalingam A, Elboudwarej E, Telep L, Timbs L, Henne I, Sellers S, Cao H, Tan SK, Winterbourne L, Desai P, Mera R, Gaggar A, Myers RP, Brainard DM, Childs R, Flanigan T. Compassionate Use of Remdesivir for Patients with Severe Covid-19. N Engl J Med. 2020;382:2327-2336. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1926] [Cited by in RCA: 1884] [Article Influence: 376.8] [Reference Citation Analysis (0)] |
| 21. | Chew M, Tang Z, Radcliffe C, Caruana D, Doilicho N, Ciarleglio MM, Deng Y, Garcia-Tsao G. Significant Liver Injury During Hospitalization for COVID-19 Is Not Associated With Liver Insufficiency or Death. Clin Gastroenterol Hepatol. 2021;19:2182-2191.e7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 22. | Delgado A, Stewart S, Urroz M, Rodríguez A, Borobia AM, Akatbach-Bousaid I, González-Muñoz M, Ramírez E. Characterisation of Drug-Induced Liver Injury in Patients with COVID-19 Detected by a Proactive Pharmacovigilance Program from Laboratory Signals. J Clin Med. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 23. | Leegwater E, Strik A, Wilms EB, Bosma LBE, Burger DM, Ottens TH, van Nieuwkoop C. Drug-induced Liver Injury in a Patient With Coronavirus Disease 2019: Potential Interaction of Remdesivir With P-Glycoprotein Inhibitors. Clin Infect Dis. 2021;72:1256-1258. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 24. | Lee C, Ahn MY, Byeon K, Choi JP, Hahm C, Kim H, Kim S, Kim TH, Oh J, Oh DH. Clinical Experience with Use of Remdesivir in the Treatment of Severe Acute Respiratory Syndrome Coronavirus 2: a Case Series. Infect Chemother. 2020;52:369-380. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 25. | Zampino R, Mele F, Florio LL, Bertolino L, Andini R, Galdo M, De Rosa R, Corcione A, Durante-Mangoni E. Liver injury in remdesivir-treated COVID-19 patients. Hepatol Int. 2020;14:881-883. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 91] [Article Influence: 18.2] [Reference Citation Analysis (0)] |
| 26. | Carothers C, Birrer K, Vo M. Acetylcysteine for the Treatment of Suspected Remdesivir-Associated Acute Liver Failure in COVID-19: A Case Series. Pharmacotherapy. 2020;40:1166-1171. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 50] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 27. | Sun J, Deng X, Chen X, Huang J, Huang S, Li Y, Feng J, Liu J, He G. Incidence of Adverse Drug Reactions in COVID-19 Patients in China: An Active Monitoring Study by Hospital Pharmacovigilance System. Clin Pharmacol Ther. 2020;108:791-797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 14.8] [Reference Citation Analysis (0)] |
| 28. | Fan Z, Chen L, Li J, Cheng X, Yang J, Tian C, Zhang Y, Huang S, Liu Z, Cheng J. Clinical Features of COVID-19-Related Liver Functional Abnormality. Clin Gastroenterol Hepatol. 2020;18:1561-1566. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 559] [Cited by in RCA: 556] [Article Influence: 111.2] [Reference Citation Analysis (0)] |
| 29. | Jiang S, Wang R, Li L, Hong D, Ru R, Rao Y, Miao J, Chen N, Wu X, Ye Z, Hu Y, Xie M, Zuo M, Lu X, Qiu Y, Liang T. Liver Injury in Critically Ill and Non-critically Ill COVID-19 Patients: A Multicenter, Retrospective, Observational Study. Front Med (Lausanne). 2020;7:347. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 29] [Article Influence: 5.8] [Reference Citation Analysis (1)] |
| 30. | Serviddio G, Villani R, Stallone G, Scioscia G, Foschino-Barbaro MP, Lacedonia D. Tocilizumab and liver injury in patients with COVID-19. Therap Adv Gastroenterol. 2020;13:1756284820959183. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 31. | Liao S, Zhan K, Gan L, Bai Y, Li J, Yuan G, Cai Y, Zhang A, He S, Mei Z. Inflammatory cytokines, T lymphocyte subsets, and ritonavir involved in liver injury of COVID-19 patients. Signal Transduct Target Ther. 2020;5:255. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 32. | Guaraldi G, Meschiari M, Cozzi-Lepri A, Milic J, Tonelli R, Menozzi M, Franceschini E, Cuomo G, Orlando G, Borghi V, Santoro A, Di Gaetano M, Puzzolante C, Carli F, Bedini A, Corradi L, Fantini R, Castaniere I, Tabbì L, Girardis M, Tedeschi S, Giannella M, Bartoletti M, Pascale R, Dolci G, Brugioni L, Pietrangelo A, Cossarizza A, Pea F, Clini E, Salvarani C, Massari M, Viale PL, Mussini C. Tocilizumab in patients with severe COVID-19: a retrospective cohort study. Lancet Rheumatol. 2020;2:e474-e484. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 604] [Cited by in RCA: 688] [Article Influence: 137.6] [Reference Citation Analysis (0)] |
| 33. | Muhović D, Bojović J, Bulatović A, Vukčević B, Ratković M, Lazović R, Smolović B. First case of drug-induced liver injury associated with the use of tocilizumab in a patient with COVID-19. Liver Int. 2020;40:1901-1905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 94] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 34. | Hundt MA, Deng Y, Ciarleglio MM, Nathanson MH, Lim JK. Abnormal Liver Tests in COVID-19: A Retrospective Observational Cohort Study of 1,827 Patients in a Major U.S. Hospital Network. Hepatology. 2020;72:1169-1176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 35. | Kelly M, O'Connor R, Townsend L, Coghlan M, Relihan E, Moriarty M, Carr B, Melanophy G, Doyle C, Bannan C, O'Riordan R, Merry C, Clarke S, Bergin C. Clinical outcomes and adverse events in patients hospitalised with COVID-19, treated with off-label hydroxychloroquine and azithromycin. Br J Clin Pharmacol. 2021;87:1150-1154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 36. | Falcão MB, Pamplona de Góes Cavalcanti L, Filgueiras Filho NM, Antunes de Brito CA. Case Report: Hepatotoxicity Associated with the Use of Hydroxychloroquine in a Patient with COVID-19. Am J Trop Med Hyg. 2020;102:1214-1216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 49] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 37. | Gabrielli M, Franza L, Esperide A, Gasparrini I, Gasbarrini A, Franceschi F. On Behalf Of Gemelli Against Covid. Liver Injury in Patients Hospitalized for COVID-19: Possible Role of Therapy. Vaccines (Basel). 2022;10:192. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 38. | Yamazaki S, Suzuki T, Sayama M, Nakada TA, Igari H, Ishii I. Suspected cholestatic liver injury induced by favipiravir in a patient with COVID-19. J Infect Chemother. 2021;27:390-392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 39. | Tang H, Zhou L, Li X, Kinlaw AC, Yang JY, Moon AM, Barnes EL, Wang T. Drug-induced liver injury associated with lopinavir-ritonavir in patients with COVID-19: a disproportionality analysis of U.S. food and drug administration adverse event reporting system (FAERS) data. Int J Clin Pharm. 2021;43:1116-1122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 9] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Panda PK, Singh BO, Moirangthem B, Bahurupi YA, Saha S, Saini G, Dhar M, Bairwa M, Pai VS, Agarwal A, Sindhwani G, Handu S, Kant R. Antiviral Combination Clinically Better Than Standard Therapy in Severe but Not in Non-Severe COVID-19. Clin Pharmacol. 2021;13:185-195. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 41. | Valenti L, Jamialahmadi O, Romeo S. Lack of genetic evidence that fatty liver disease predisposes to COVID-19. J Hepatol. 2020;73:709-711. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 26] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 42. | Machill A, Bals R, Lammert F, Krawczyk M. Genetic insight into COVID-19 related liver injury: A note on MBOAT7. Liver Int. 2021;41:1157-1159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 43. | Bianco C, Baselli G, Malvestiti F, Santoro L, Pelusi S, Manunta M, Grasselli G, Bandera A, Scudeller L, Prati D, Valenti L. Genetic insight into COVID-19-related liver injury. Liver Int. 2021;41:227-229. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 44. | Valenti L, Griffini S, Lamorte G, Grovetti E, Uceda Renteria SC, Malvestiti F, Scudeller L, Bandera A, Peyvandi F, Prati D, Meroni P, Cugno M. Chromosome 3 cluster rs11385942 variant links complement activation with severe COVID-19. J Autoimmun. 2021;117:102595. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 45. | LiverTox: Clinical and Research Information on Drug-Induced Liver Injury [Internet]. Bethesda (MD): National Institute of Diabetes and Digestive and Kidney Diseases; 2012- . [PubMed] |
| 46. | Olry A, Meunier L, Délire B, Larrey D, Horsmans Y, Le Louët H. Drug-Induced Liver Injury and COVID-19 Infection: The Rules Remain the Same. Drug Saf. 2020;43:615-617. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 68] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 47. | Hariyanto TI, Kristine E, Jillian Hardi C, Kurniawan A. Efficacy of Lopinavir/Ritonavir Compared With Standard Care for Treatment of Coronavirus Disease 2019 (COVID-19): A Systematic Review. Infect Disord Drug Targets. 2021;21:e270421187364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 48. | Patel TK, Patel PB, Barvaliya M, Saurabh MK, Bhalla HL, Khosla PP. Efficacy and safety of lopinavir-ritonavir in COVID-19: A systematic review of randomized controlled trials. J Infect Public Health. 2021;14:740-748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 53] [Cited by in RCA: 46] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 49. | Aiswarya D, Arumugam V, Dineshkumar T, Gopalakrishnan N, Lamech TM, Nithya G, Sastry BVRH, Vathsalyan P, Dhanapriya J, Sakthirajan R. Use of Remdesivir in Patients With COVID-19 on Hemodialysis: A Study of Safety and Tolerance. Kidney Int Rep. 2021;6:586-593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 58] [Cited by in RCA: 52] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 50. | Wong GL, Wong VW, Thompson A, Jia J, Hou J, Lesmana CRA, Susilo A, Tanaka Y, Chan WK, Gane E, Ong-Go AK, Lim SG, Ahn SH, Yu ML, Piratvisuth T, Chan HL; Asia-Pacific Working Group for Liver Derangement during the COVID-19 Pandemic. Management of patients with liver derangement during the COVID-19 pandemic: an Asia-Pacific position statement. Lancet Gastroenterol Hepatol. 2020;5:776-787. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 70] [Article Influence: 14.0] [Reference Citation Analysis (1)] |
| 51. | Ridruejo E, Soza A. The liver in times of COVID-19: What hepatologists should know. Ann Hepatol. 2020;19:353-358. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 52. | Kulkarni AV, Kumar P, Tevethia HV, Premkumar M, Arab JP, Candia R, Talukdar R, Sharma M, Qi X, Rao PN, Reddy DN. Systematic review with meta-analysis: liver manifestations and outcomes in COVID-19. Aliment Pharmacol Ther. 2020;52:584-599. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 183] [Article Influence: 36.6] [Reference Citation Analysis (0)] |
| 53. | Mareev VY, Orlova YA, Plisyk AG, Pavlikova EP, Akopyan ZA, Matskeplishvili ST, Malakhov PS, Krasnova TN, Seredenina EM, Potapenko AV, Agapov MA, Asratyan DA, Dyachuk LI, Samokhodskaya LM, Mershina ЕА, Sinitsyn VE, Pakhomov PV, Zhdanova EA, Mareev YV, Begrambekova YL, Kamalov АА. Proactive anti-inflammatory therapy with colchicine in the treatment of advanced stages of new coronavirus infection. The first results of the COLORIT study. Kardiologiia. 2021;61:15-27. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 54. | Vitiello A, La Porta R, D'Aiuto V, Ferrara F. The risks of liver injury in COVID-19 patients and pharmacological management to reduce or prevent the damage induced. Egypt Liver J. 2021;11:11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 55. | Bryant A, Lawrie TA, Dowswell T, Fordham EJ, Mitchell S, Hill SR, Tham TC. Ivermectin for Prevention and Treatment of COVID-19 Infection: A Systematic Review, Meta-analysis, and Trial Sequential Analysis to Inform Clinical Guidelines. Am J Ther. 2021;28:e434-e460. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 145] [Cited by in RCA: 142] [Article Influence: 35.5] [Reference Citation Analysis (0)] |
| 56. | Okumuş N, Demirtürk N, Çetinkaya RA, Güner R, Avcı İY, Orhan S, Konya P, Şaylan B, Karalezli A, Yamanel L, Kayaaslan B, Yılmaz G, Savaşçı Ü, Eser F, Taşkın G. Evaluation of the effectiveness and safety of adding ivermectin to treatment in severe COVID-19 patients. BMC Infect Dis. 2021;21:411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 57. | Chai X, Hu L, Zhang Y, Han W, Lu Z, Ke A, Zhou J, Shi G, Fang N, Fan J, Cai J, Lan F. Specific ACE2 Expression in Cholangiocytes May Cause Liver Damage After 2019-nCoV Infection. 2020 Preprint. Available from: bioRxiv: 2020.02.03.931766. [DOI] [Full Text] |
| 58. | Liu J, Li S, Liu J, Liang B, Wang X, Wang H, Li W, Tong Q, Yi J, Zhao L, Xiong L, Guo C, Tian J, Luo J, Yao J, Pang R, Shen H, Peng C, Liu T, Zhang Q, Wu J, Xu L, Lu S, Wang B, Weng Z, Han C, Zhu H, Zhou R, Zhou H, Chen X, Ye P, Zhu B, Wang L, Zhou W, He S, He Y, Jie S, Wei P, Zhang J, Lu Y, Wang W, Zhang L, Li L, Zhou F, Wang J, Dittmer U, Lu M, Hu Y, Yang D, Zheng X. Longitudinal characteristics of lymphocyte responses and cytokine profiles in the peripheral blood of SARS-CoV-2 infected patients. EBioMedicine. 2020;55:102763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1171] [Cited by in RCA: 1207] [Article Influence: 241.4] [Reference Citation Analysis (0)] |
| 59. | Teschke R, Méndez-Sánchez N, Eickhoff A. Liver Injury in COVID-19 Patients with Drugs as Causatives: A Systematic Review of 996 DILI Cases Published 2020/2021 Based on RUCAM as Causality Assessment Method. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 60. | Mantovani A, Beatrice G, Dalbeni A. Coronavirus disease 2019 and prevalence of chronic liver disease: A meta-analysis. Liver Int. 2020;40:1316-1320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 120] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 61. | Huang H, Li H, Chen S, Zhou X, Dai X, Wu J, Zhang J, Shao L, Yan R, Wang M, Wang J, Tu Y, Ge M. Prevalence and Characteristics of Hypoxic Hepatitis in COVID-19 Patients in the Intensive Care Unit: A First Retrospective Study. Front Med (Lausanne). 2020;7:607206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 18] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 62. | Raschi E, Caraceni P, Poluzzi E, De Ponti F. Baricitinib, JAK inhibitors and liver injury: a cause for concern in COVID-19? Expert Opin Drug Saf. 2020;19:1367-1369. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 11] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 63. | Fromenty B, Pessayre D. Inhibition of mitochondrial beta-oxidation as a mechanism of hepatotoxicity. Pharmacol Ther. 1995;67:101-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 475] [Cited by in RCA: 455] [Article Influence: 15.2] [Reference Citation Analysis (1)] |
| 64. | Pessayre D, Fromenty B, Berson A, Robin MA, Lettéron P, Moreau R, Mansouri A. Central role of mitochondria in drug-induced liver injury. Drug Metab Rev. 2012;44:34-87. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 187] [Cited by in RCA: 195] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 65. | FitzGerald GA. Misguided drug advice for COVID-19. Science. 2020;367:1434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 98] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 66. | Fang L, Karakiulakis G, Roth M. Are patients with hypertension and diabetes mellitus at increased risk for COVID-19 infection? Lancet Respir Med. 2020;8:e21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1758] [Cited by in RCA: 1951] [Article Influence: 390.2] [Reference Citation Analysis (0)] |
| 67. | Massart J, Begriche K, Moreau C, Fromenty B. Role of nonalcoholic fatty liver disease as risk factor for drug-induced hepatotoxicity. J Clin Transl Res. 2017;3:212-232. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 51] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 68. | El-Ghiaty MA, Shoieb SM, El-Kadi AOS. Cytochrome P450-mediated drug interactions in COVID-19 patients: Current findings and possible mechanisms. Med Hypotheses. 2020;144:110033. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 69. | Griffin LM, Watkins PB, Perry CH, St Claire RL 3rd, Brouwer KL. Combination lopinavir and ritonavir alter exogenous and endogenous bile acid disposition in sandwich-cultured rat hepatocytes. Drug Metab Dispos. 2013;41:188-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Holmstock N, Oorts M, Snoeys J, Annaert P. MRP2 Inhibition by HIV Protease Inhibitors in Rat and Human Hepatocytes: A Quantitative Confocal Microscopy Study. Drug Metab Dispos. 2018;46:697-703. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 71. | Ye ZW, Camus S, Augustijns P, Annaert P. Interaction of eight HIV protease inhibitors with the canalicular efflux transporter ABCC2 (MRP2) in sandwich-cultured rat and human hepatocytes. Biopharm Drug Dispos. 2010;31:178-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 72. | Khalatbari A, Mishra P, Han H, He Y, MacVeigh-Aloni M, Ji C. Ritonavir and Lopinavir Suppress RCE1 and CAAX Rab Proteins Sensitizing the Liver to Organelle Stress and Injury. Hepatol Commun. 2020;4:932-944. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 73. | Kudaravalli P, Saleem SA, Ibeche B, John S. Case series and review of liver dysfunction in COVID-19 patients. Eur J Gastroenterol Hepatol. 2020;32:1244-1250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 74. | Rismanbaf A, Zarei S. Liver and Kidney Injuries in COVID-19 and Their Effects on Drug Therapy; a Letter to Editor. Arch Acad Emerg Med. 2020;8:e17. [PubMed] |
| 75. | Dongiovanni P, Stender S, Pietrelli A, Mancina RM, Cespiati A, Petta S, Pelusi S, Pingitore P, Badiali S, Maggioni M, Mannisto V, Grimaudo S, Pipitone RM, Pihlajamaki J, Craxi A, Taube M, Carlsson LMS, Fargion S, Romeo S, Kozlitina J, Valenti L. Causal relationship of hepatic fat with liver damage and insulin resistance in nonalcoholic fatty liver. J Intern Med. 2018;283:356-370. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 198] [Cited by in RCA: 284] [Article Influence: 40.6] [Reference Citation Analysis (0)] |
| 76. | Goldman JD, Lye DCB, Hui DS, Marks KM, Bruno R, Montejano R, Spinner CD, Galli M, Ahn MY, Nahass RG, Chen YS, SenGupta D, Hyland RH, Osinusi AO, Cao H, Blair C, Wei X, Gaggar A, Brainard DM, Towner WJ, Muñoz J, Mullane KM, Marty FM, Tashima KT, Diaz G, Subramanian A; GS-US-540-5773 Investigators. Remdesivir for 5 or 10 Days in Patients with Severe Covid-19. N Engl J Med. 2020;383:1827-1837. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 987] [Cited by in RCA: 987] [Article Influence: 197.4] [Reference Citation Analysis (0)] |
| 77. | Antinori S, Cossu MV, Ridolfo AL, Rech R, Bonazzetti C, Pagani G, Gubertini G, Coen M, Magni C, Castelli A, Borghi B, Colombo R, Giorgi R, Angeli E, Mileto D, Milazzo L, Vimercati S, Pellicciotta M, Corbellino M, Torre A, Rusconi S, Oreni L, Gismondo MR, Giacomelli A, Meroni L, Rizzardini G, Galli M. Compassionate remdesivir treatment of severe Covid-19 pneumonia in intensive care unit (ICU) and Non-ICU patients: Clinical outcome and differences in post-treatment hospitalisation status. Pharmacol Res. 2020;158:104899. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 153] [Cited by in RCA: 143] [Article Influence: 28.6] [Reference Citation Analysis (1)] |
| 78. | Kaur M, Tiwari D, Sidana V, Mukhopadhyay K. Remdesivir-Induced Liver Injury in a COVID-Positive Newborn. Indian J Pediatr. 2022;89:826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 79. | Gao S, Yang Q, Wang X, Hu W, Lu Y, Yang K, Jiang Q, Li W, Song H, Sun F, Cheng H. Association Between Drug Treatments and the Incidence of Liver Injury in Hospitalized Patients With COVID-19. Front Pharmacol. 2022;13:799338. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Reference Citation Analysis (0)] |
| 80. | Naseralallah LM, Aboujabal BA, Geryo NM, Al Boinin A, Al Hattab F, Akbar R, Umer W, Abdul Jabbar L, Danjuma MI. The determination of causality of drug induced liver injury in patients with COVID-19 clinical syndrome. PLoS One. 2022;17:e0268705. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 9] [Reference Citation Analysis (0)] |
| 81. | Durante-Mangoni E, Andini R, Bertolino L, Mele F, Florio LL, Murino P, Corcione A, Zampino R. Early experience with remdesivir in SARS-CoV-2 pneumonia. Infection. 2020;48:779-782. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 82. | Wong CKH, Au ICH, Cheng WY, Man KKC, Lau KTK, Mak LY, Lui SL, Chung MSH, Xiong X, Lau EHY, Cowling BJ. Remdesivir use and risks of acute kidney injury and acute liver injury among patients hospitalised with COVID-19: a self-controlled case series study. Aliment Pharmacol Ther. 2022;56:121-130. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 83. | Montastruc F, Thuriot S, Durrieu G. Hepatic Disorders With the Use of Remdesivir for Coronavirus 2019. Clin Gastroenterol Hepatol. 2020;18:2835-2836. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 60] [Article Influence: 12.0] [Reference Citation Analysis (0)] |