Published online Nov 25, 2021. doi: 10.5501/wjv.v10.i6.288
Peer-review started: March 15, 2021
First decision: April 6, 2021
Revised: April 19, 2021
Accepted: July 30, 2021
Article in press: July 30, 2021
Published online: November 25, 2021
Processing time: 252 Days and 0.4 Hours
Almost all the cellular processes in a living system are controlled by proteins: They regulate gene expression, catalyze chemical reactions, transport small molecules across membranes, and transmit signal across membranes. Even, a viral infection is often initiated through virus-host protein interactions. Protein-protein interactions (PPIs) are the physical contacts between two or more proteins and they represent complex biological functions. Nowadays, PPIs have been used to construct PPI networks to study complex pathways for revealing the functions of unknown proteins. Scientists have used PPIs to find the molecular basis of certain diseases and also some potential drug targets. In this review, we will discuss how PPI networks are essential to understand the molecular basis of virus-host relationships and several databases which are dedicated to virus-host interaction studies. Here, we present a short but comprehensive review on PPIs, including the experimental and computational methods of finding PPIs, the databases dedicated to virus-host PPIs, and the associated various applications in protein interaction networks of some lethal viruses with their hosts.
Core Tip: This paper provides a comprehensive review on protein-protein interactions (PPIs), including the experimental and computational methods of finding PPIs, the databases dedicated to virus-host PPIs, and the associated applications in the studies of some lethal viruses with their hosts. PPIs can be mapped into networks and innumerable novel insights into the functional organization of proteomes can be gained by analyzing the networks. Many studies have used network biology to construct PPI networks of lethal pathogens with their host Homo sapiens to dig deep down into the molecular constitution of the disease pathways, and have successfully found multiple potential drug targets against the viruses.
- Citation: Farooq QUA, Shaukat Z, Aiman S, Li CH. Protein-protein interactions: Methods, databases, and applications in virus-host study. World J Virol 2021; 10(6): 288-300
- URL: https://www.wjgnet.com/2220-3249/full/v10/i6/288.htm
- DOI: https://dx.doi.org/10.5501/wjv.v10.i6.288
Proteins have been declared as the chief representative of biological function[1]. It has been reported that more than 80% of proteins do not function alone[2], but instead often interact with each other or with other molecules like DNA or RNA to perform distinct cellular functions. Protein-protein interactions (PPIs) are thought to execute many biological processes including complex metabolic pathways and signaling cascades, and hence it is crucial to understand the particular nature of these associations[1,3].
De Las Rivas and Fontanillo[4] defined PPIs as “physical contacts with molecular docking between the proteins that occur in a cell or in a living organism in vivo”. The physical contacts between the proteins should be specific and intentional, i.e., evolved for a particular function. Protein interacting with other proteins can be in any form, i.e., in binary, multi-protein complexes or in the form of long chains[1,4]. Proteins involved in a certain cellular pathway or biological process are often found to interact with each other repeatedly, suggesting that the proteins with associated functions are more likely to interact with each other[2,5]. Conversely, researchers can reveal the functions of unidentified or uncharacterized proteins if the proteins with which they are interacting are known[6,7]. The outcome of most of the cellular processes can be deciphered by protein interactions. The information about PPIs can help scientists find out potential drug targets by investigating the pathogen-host interaction network[8,9]. Therefore, it is significant to study PPIs for understanding the functions of proteins within a cell or a living organism.
PPIs can be determined by different high-throughput experimental and computational methods which yield different types of PPI data. The high-throughput experimental techniques either identify the interactions directly or infer them indirectly based on different approaches[1,4]. In the following, the two main experimental methods, yeast two-hybrid (Y2H) and tandem affinity purification-mass spectrometry (MS), will be introduced.
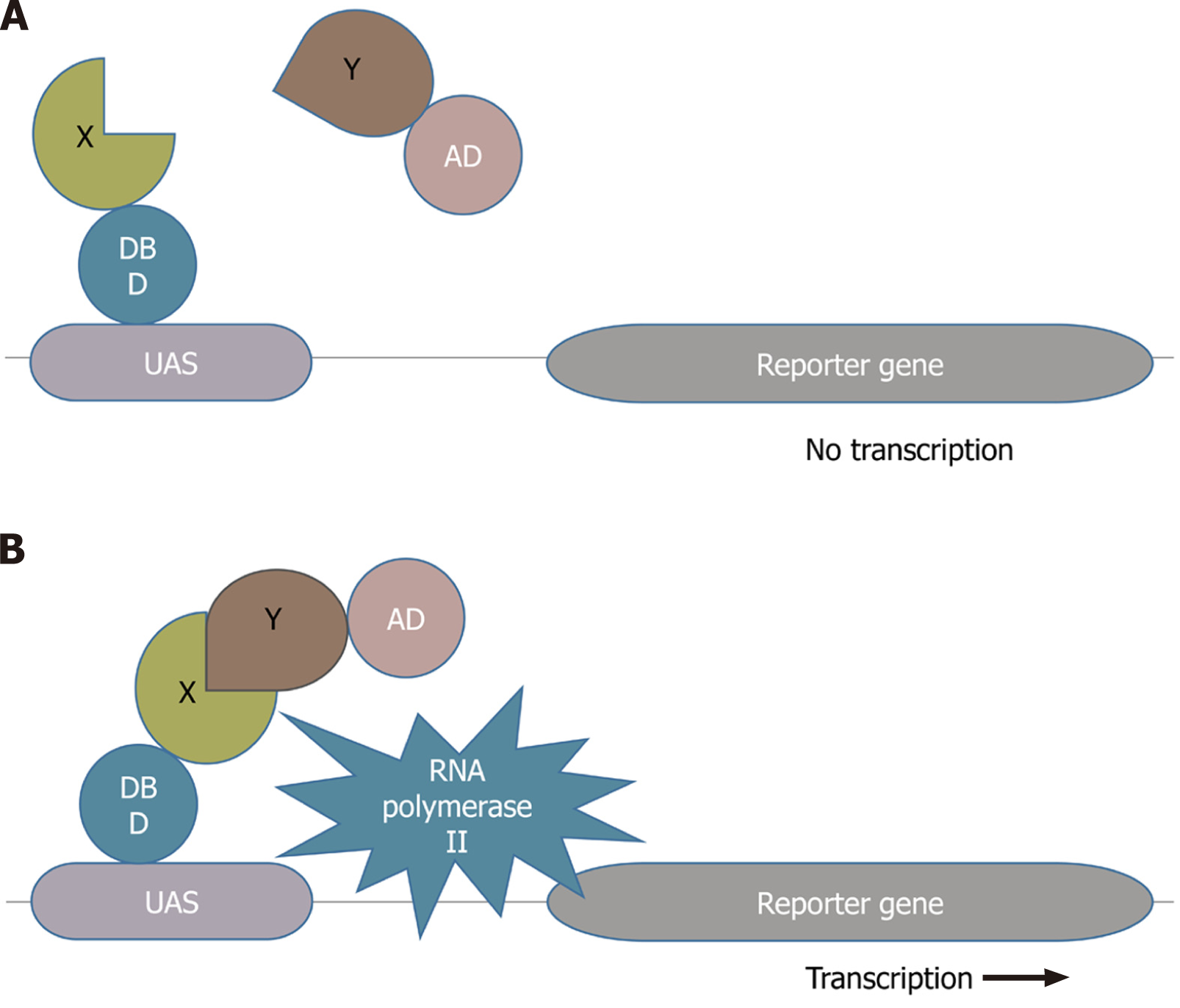
Y2H, also known as a binary method initially reported in 1989, is the most widely and commonly used interaction detection approach that identifies direct physical interactions between two proteins in vivo[10]. It detects the interactions between the query protein of interest and the protein library. In this approach, the former fused with the binding domain of a particular transcription factor is known as the bait and the latter fused with the activation domain of the transcription factor is referred to as the prey. If the bait and prey can interact with each other, they will bring together the two halves of the transcription factor to activate the transcription complex (shown in Figure 1), which transcribes the downstream reporter gene leading to the expression of the reporter gene[1,4,11]. The availability of many full genomes with the advancement of next-generation sequencing techniques allows us to use protein interactions to help understand the functions of their gene products. Y2H has outranked the other experimental techniques and has become the system of choice for researchers in large-scale, high-throughput, and comprehensive investigations of PPIs. The complete proteomes of several pathogens including hepatitis C virus (HCV), bacteriophage T7, and vaccinia have been analyzed using the Y2H screen[12-14]. Several scientists have performed the comprehensive two-hybrid analysis of the yeast protein interactome, including the construction and analysis of PPI map of all possible associations between the yeast proteins[15-17].
Y2H has been used massively by scientists to infer physical interactions between macromolecules. It is advantageous because of its simple organization and easy detection for the transient interactions. However, despite its importance, there are certain disadvantages[10,18] which will be discussed in the section of experimental errors in PPI detection.
MS is a powerful in vitro tool for the detection of macromolecular interactions. The principle of MS was explained extensively in one of our previous reviews[19]. MS allows us to identify polypeptide sequences by ionizing them and then detecting analyte ions based on their mass-to-charge ratios[20,21]. To interpret the mass spectra and detect PPIs, various MS-based methods have been developed so far. The MS-based detection of PPIs has become significant in the recent era especially for the large-scale investigations, through which high-throughput and high confidence PPIs can be identified[22,23]. These MS-based technologies include cross-linking MS (CLMS)[24], tandem affinity purification MS (TAP-MS)[25,26], and several others.
TAP-MS is a conventional MS-based qualitative method to study protein functions and interactions. Sinz[27] and Yugandhar et al[28] have extensively reviewed CLMS, which is a more recent and advanced MS technique for interpreting protein interaction networks. Many scientists have been working on the techniques using MS for finding potential interactors where true positives are segregated and prioritized from false positives. Gavin et al[29] and Collins et al[30] developed score-based methods to infer high-accuracy physical interactions.
According to the EMBL-EBI statistics (https://www.ebi.ac.uk/intact/about/statistics?conversationContext=2), TAP-MS has overtaken Y2H as a major source of generating PPI data.
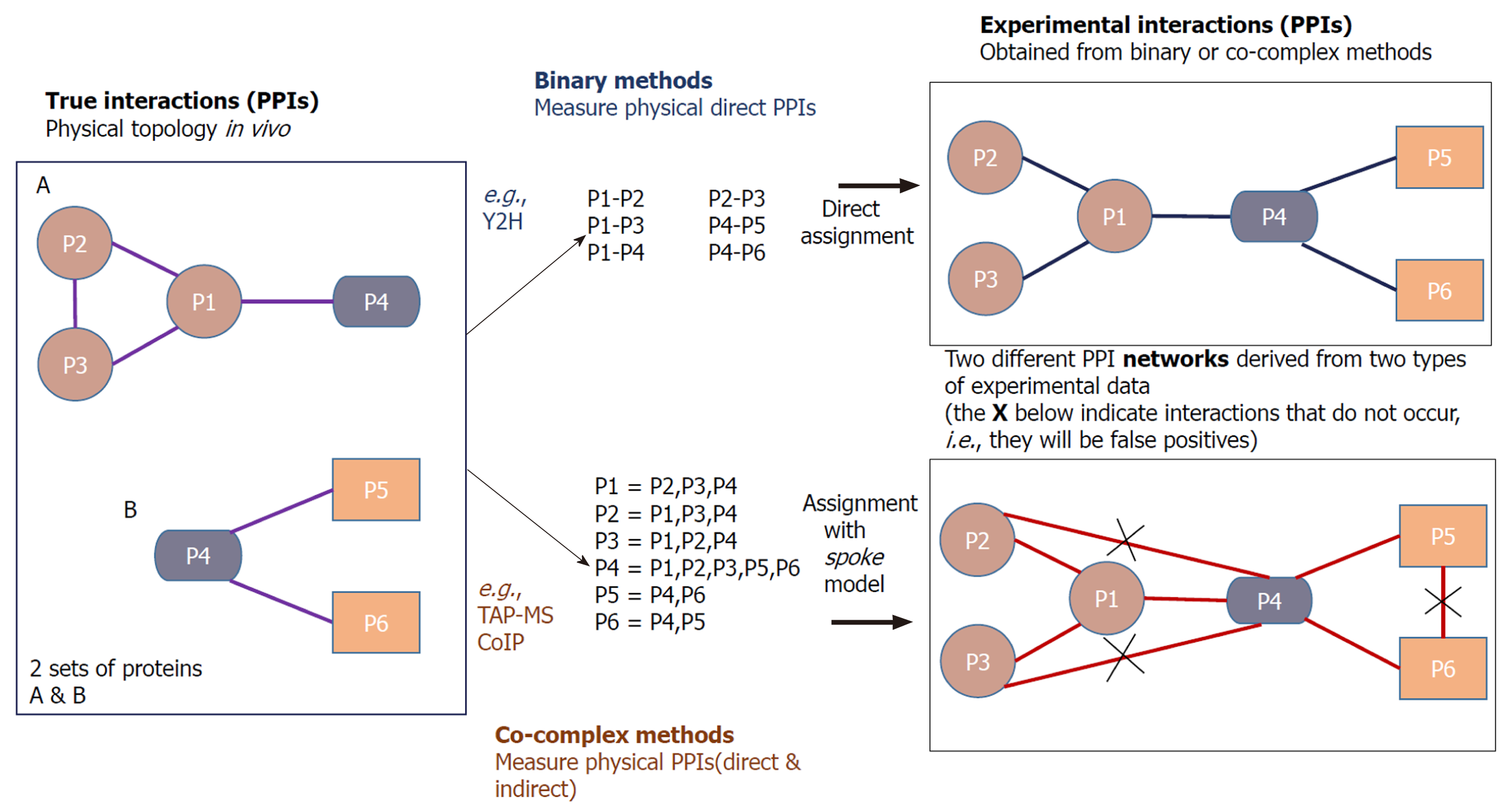
Compared with Y2H which detects only binary interactions, TAP-MS is a co-complex method which determines both direct and indirect associations between proteins in vitro. In this technique, a TAP tag is fused at the C- or N-terminus of a protein of interest (the bait), which has two independent binding regions, allowing two successive affinity purification steps. The most common TAP tag consists of two immunoglobulin G binding repeats of Protein A from Staphylococcus aureus (ProtA) and a calmodulin-binding peptide which are separated by a tobacco etch virus protease cleavage site. In TAP, a group of protein complexes can be caught by a tagged bait protein in a pull-down assay, which are called prey proteins[2,4,31]. The prey proteins interacting with the bait are separated via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and then identified by MS[18,32].
In addition to the tandem affinity purification, there is another co-complex method called co-immunoprecipitation (CoIP) for determining PPIs. The interaction data derived from co-complex methods cannot be used to infer binary interactions directly, and the related algorithms are needed to interpret the pairwise interactions from the experimental data[4].
High-throughput experimental approaches for determining PPIs are very efficient, but they also have some limitations. They have a high possibility of false negative and false positive errors. False positives in an experimental system are those interactions that do not occur in the system naturally. One reason for the false positives in Y2H can be the auto-activation of transcription by the bait protein itself or sometimes the transient interactions that are not always specific, i.e., the interactors can be the sticky prey proteins fused with the bait protein and chosen by Y2H analysis[4,10,33]. The precise percentage of the false-positive interactions in Y2H is not well known but the estimated rate of the inaccurate interactions is about 50%, which is quite a big percentage, yet still Y2H is one of the most powerful interaction determining methods[2,10]. Additionally, the experimental system for determining PPIs faces false negative errors too, i.e., some interactions cannot be identified due to the flaws in the experimental system. In Y2H, most of the interactions between membrane proteins are undetectable. Hence, it is important to choose the Y2H design thoughtfully based on the type of cellular proteome. Sometimes in Y2H, very weak transient interactions escape from being identified by the method[10].
Co-complex methods also encounter errors in their interaction detection mechanisms. There can be sticky prey proteins in the TAP pull down assay that are detected by the method as interacting partners of the bait protein. The TAP is an in vitro technique, which means that it is not sure whether the interactions that occur in vitro will surely exist in vivo. Additionally in TAP, the very transient interactions often vanish due to the series of purification levels[1,2]. Another drawback of co-complex methods is that they might analyze all the elements of a protein complex which certainly may not have direct interactions with each other[10] (crossed links in Figure 2).
PPI studies do not just rely on Y2H or affinity purification methods, and due to the false positives and false negatives, several other methods have also been made into practice by researchers for PPI detection. Some of these in vitro techniques are CoIP[18], protein microarrays[34], protein-fragment complementation[35], X-ray crystallography, and nuclear magnetic resonance spectroscopy[36].
As discussed in the previous section, experimental methods for PPI detection have many limitations including a high percentage of false positives, high cost, and being significantly laborious and time-consuming. Besides, due to the completion of various genome sequencing projects, it is necessary to speed up to find the functional linkages between proteins. Thus, the computational prediction of PPIs seems to be very crucial. Now, computational methods are being practiced successfully to evaluate and analyze the interaction data generated by high-throughput experimental approaches as well as to predict novel PPIs by gaining insights from the already known interactions.
The computational methods are a quick and low-cost alternative to the traditional experimental techniques to predict PPIs. An important advantage of computational methods over the experimental ones is that we can study proteins by mapping the pairwise associations into a comprehensive network according to their distinct functional level[1,37]. Table 1 lists some of the important in silico methods of PPI prediction.
| Method | Description | Ref. |
| In silico two-hybrid (I2H) | The I2H method is based on the detection of direct physical associations between the interacting proteins and it relies on the presumption that in order to maintain the protein function reliable, the interacting proteins should go through coevolution | Pazos and Valencia[38] |
| Ortholog-based approach | It is a sequence-based approach that uses a pairwise local search algorithm to obtain the similarities between the query protein pairs and the known interaction pairs. It is dependent upon the homologous nature of the target proteins | Lee et al[39] |
| Gene fusion | Also known as Rosetta stone method. According to this method, some of the proteins with single domains fuse together in one organism and form a multi-domain protein in another organism | Enright et al[40] |
| Domain-pairs-based approach | This method predicts the interactions between proteins by the domain-domain interactions | Wojcik and Schächter[41] |
| Gene expression | An indirect way to predict PPIs. Based on the concept that the proteins translated from the genes that belong to the common expression profiling clusters more likely interact with each other than the proteins translated from the genes that belong to different clusters | Grigoriev[42] |
| Structure-based approaches | It predicts protein-protein interactions based on the structural similarity | Zhang et al[43] |
| Phylogenetic tree | This method predicts protein-protein interactions based on the concept that the interacting proteins show similarity in their evolution history | Sato et al[44] |
The continuous increase in PPI data produced by high-throughput technologies needs the formation of biological repositories where these data should be stored in an effective and organized way. The data in the publicly available PPI databases makes it much easier to analyze different types of interactions according to our concerns[37]. There are more than 100 repositories accessible online related to PPI data[45]. Here we will discuss the most popular databases (see Table 2) of PPI information that have been used by most of the researchers worldwide and contain experimentally verified virus-host PPIs.
| PPI database | URL | Total interactions | Last updated | Ref. |
| STRING | http://string-db.org/ | > 2000 mio | 2020 | Szklarczyk et al[50] |
| BioGrid | http://thebiogrid.org/ | 1746922 | 2021 | Oughtred et al[47] |
| HPIDB | https://hpidb.igbb.msstate.edu/index.html | 69787 | 2019 | Ammari et al[51] |
| MINT | https://mint.bio.uniroma2.it/ | 131695 | 2012 | Zahiri et al[3] and Licata et al[55] |
| DIP | https://dip.doe-mbi.ucla.edu/dip/Main.cgi | 81923 | 2017 | Zahiri et al[3] and Salwinski et al[56] |
| IntAct | http://www.ebi.ac.uk/intact/ | 1130596 | 2020 | Orchard et al[52] |
| HPRD | http://www.hprd.org/ | 41327 | 2010 | Zahiri et al[3] and Keshava Prasad et al[57] |
The Biological General Repository for Interaction Datasets (BioGRID) is a publicly retrievable and comprehensive database which stores experimentally determined PPI data of almost all important model organisms[3,46]. It has constantly being updated and according to the February 2021 release, it carries 1740000 non-redundant protein and genetic interactions collected from 70000+ publications[47]. The current version of BioGrid (v 4.3.194) themed curation projects focuses on curated interactions of different diseases including coronavirus disease 2019 (COVID-19), ubiquitin-proteosome system, fanconi anemia, glioblastoma, and autophagy.
Search Tool for Retrieval of Interacting Genes (STRING) is equipped with the complete information about the functional relationships between proteins. The current version STRING v11.0 contains interaction data of 5090 organisms that is the highest number of organisms covered by any PPI database. The major assets of STRING database are its exhaustive coverage, confidence scoring of the interactions, and its intuitive user interface[48,49]. Currently, the database covers 3123056667 PPIs which are the sum of high-confidence and low-confidence interactions. An important new feature in the current version of STRING is that users can perform Gene Ontology and KEGG analysis of their input which has provided ease in gene-set enrichment analysis[50].
HPIDB is a curated database that contains host-pathogen interaction data. Developed in 2010, it is updated yearly and presents new versions. Currently, it contains protein interaction data between 66 hosts and 668 infectious pathogen species. The number of unique interactions is 69787 according to the last update (July 29, 2019). The pathogenic species that can be found superabundantly in HPIDB are influenza virus, herpes virus, papillomaviruses, Saccharomyces cerevisiae, and several others[51].
Developed in 2002, IntAct is a freely available molecular interaction data source and contains the data obtained from literature curation or deposited directly by the researchers. In 2013, IntAct and MINT joined their efforts and started the MINTACT project to maximize the coverage and curation output[52].
The International Molecular Exchange Consortium (IMEx) is an international consortium established by the joint efforts of prime public interaction databases including DIP, IntAct, HPIDB, MINT, BioGRID, MatrixDB, I2D, and some others. BIND and MPIDB which used to be large PPI databases are also members of IMEx but they no longer are active anymore. The data in IMEx is a comprehensive and integrated consortium of databases recording meta data for PPIs in a standard PSI-MS format and is available for all the researchers to re-use and re-analyze. Over the last two decades, there has been a massive increase in protein interaction data and out of all the resources, IMEx is the only source which is providing up to the minute information regarding protein interactions and annotations[45,53,54].
Some protein interaction databases are dedicated to a specific viral pathogen for example HCVPro[58] containing the data on PPIs between HCV and human. VirHostNet[59] covers an extensive range of human specific viruses and contains nearly 22000 virus-human PPIs.
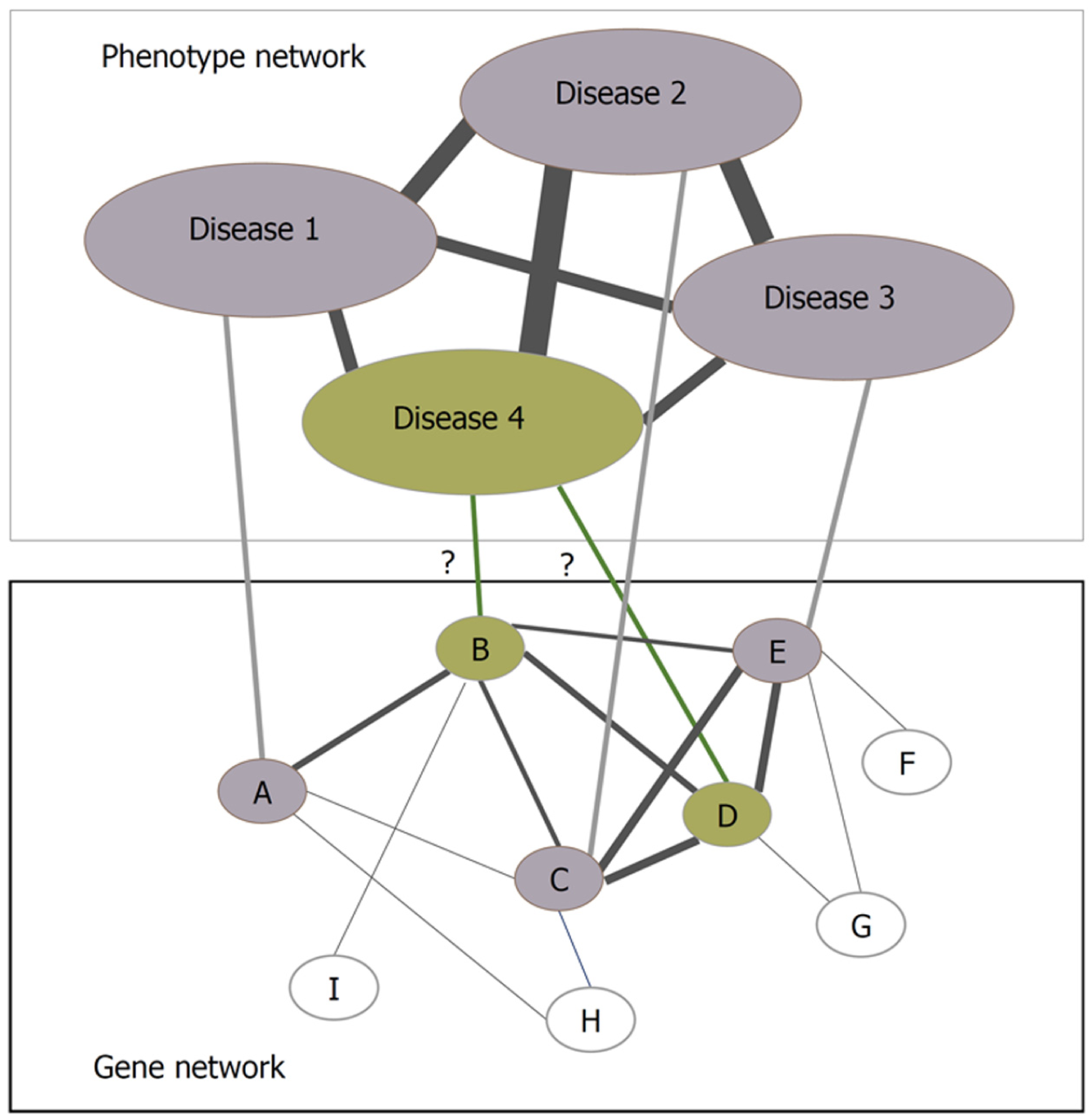
Bacteria and viruses are the major pathogens affecting humans on earth. Bacterial infections can be eradicated by using antibiotics, and viruses not easy to be eliminated can only be inhibited in their growth. Viruses depend entirely on their hosts and infect hosts often by virus-host protein interactions[54]. PPIs can be mapped into networks and innumerable novel insights into the functional organization of proteomes can be gained by analyzing the networks. Several protein interaction network construction and visualization tools are available, including Cytoscape[60], BioLayout[61], and VisANT[62]. Analyzed by these tools, PPI networks can provide the differences between normal and the diseased states, and thus the fundamental knowledge about the disease can be obtained based on the related pathways revealed through the analyses of PPI network, i.e., by looking into the subnetworks constructed by the proteins involved in the disease[1,63]. Protein interaction networks can help find new disease-related genes by the presumption that the neighboring genes of the disease-causing gene are expected to be causing the same disease or involved in causing some similar diseases (Figure 3)[64]. Various researchers have been using network biology to study pathogen-host relationship at the molecular level, which ultimately helps in identifying key viral proteins and their human targets and helps scientists in further biological investigations.
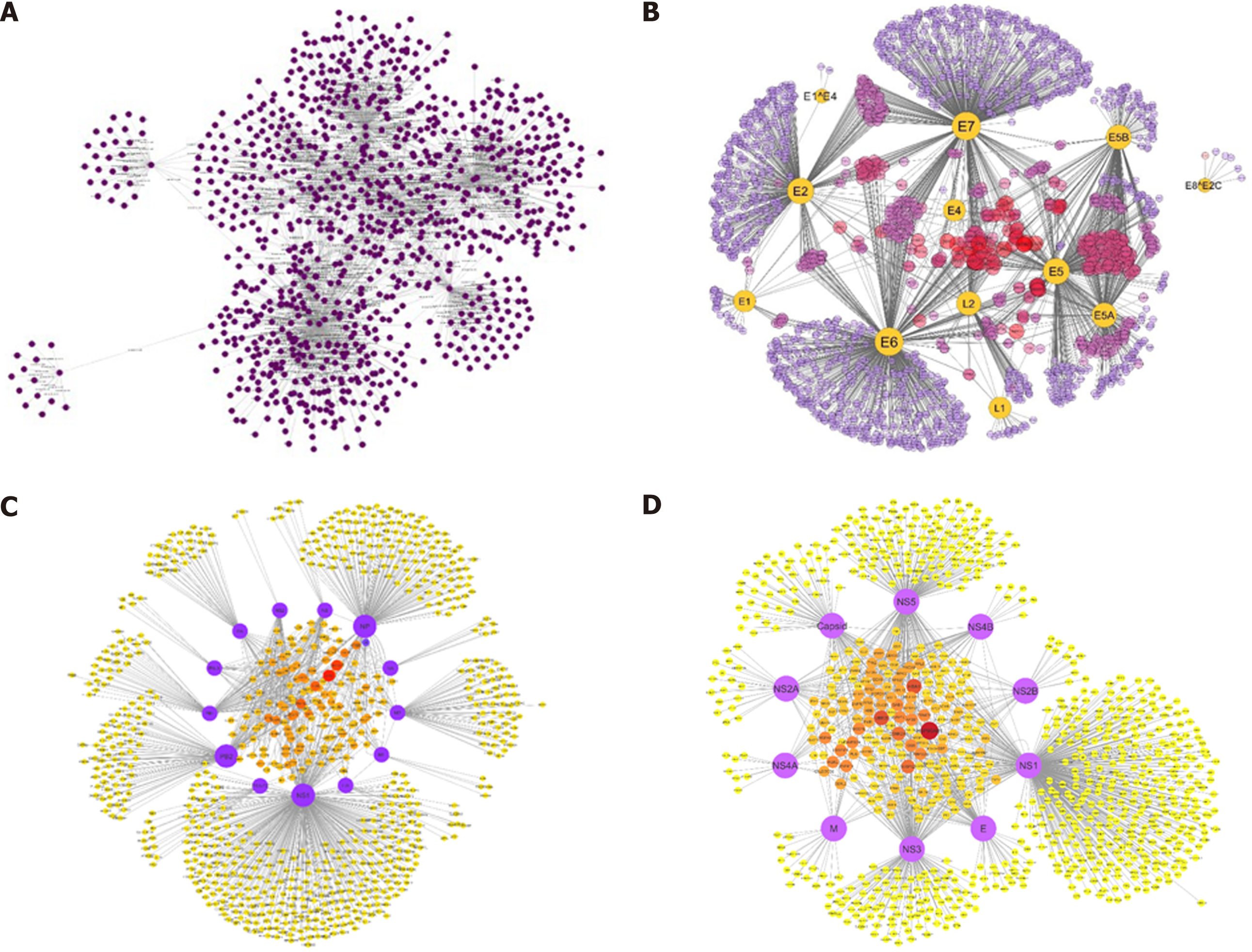
The quickly developing knowledge of human interactome map and the availability of different host-pathogen networks have paved us the way for a better understanding of diseases. Viral genomes code for a very small number of proteins, which makes it easy to understand the mechanisms of the infections by viruses[64,65]. The network-based study on the infection of host with viral pathogenesis is progressing over time. In one of our previous studies, we constructed a comprehensive protein interaction network of HCV with its host Homo sapiens[66] and found out many crucial insights into finding potential targets against HCV and some other disease pathways, such as cancer pathways (Figure 4). In fact, certain viruses such as papilloma and herpesvirus have been reported to be causing up to 20% of the cancers[67]. Additionally, virus-host relationship was also studied by us for human papillomavirus[68], influenza A virus (IAV)[69], and dengue virus with Homo sapiens. Interestingly, in a study performed by Navratil et al[70], they compared a set of virus targets with a list of 1729 human genetic disease-related proteins, and found that 13% of human virus targets are also linked with at least one human genetic disease. In short, there are so many types of viruses causing a wide variety of infections worldwide. From Ebola virus outbreak in Africa to Middle East respiratory syndrome coronavirus outbreak, viruses have killed thousands of people with no specific effective treatment. Every viral infection involves PPIs between the virus and its host including the viral entry to the host cell and hijacking the host transcription machinery. Identification of PPIs between the viruses and their hosts lets us understand the infection mechanisms of the viruses and find a way to combat the infections using antiviral drugs or vaccines[71].
When we talk about human interactome, more than 645000 PPIs are reported to be disease-associated while only 2% of these proteins are targeted by drugs[72]. The reason for most of the proteins considered to be undruggable is because of the absence of detectable pockets for binding ligands[73]. Researchers have been significantly investigating PPI inhibitors and stabilizers and have succeeded in developing new technologies that have enabled the systematic discovery of drugs focused on PPIs[74,75]. Zhang et al[76] and Robertson and Spring[77] have extensively explained the use of peptidomimetics to find the ‘hot spots’ on the protein surfaces for drug design. Targeting PPIs for designing therapeutics was once considered a difficult and impossible task. However, during the past two decades, the concept has changed and PPI drug targets have gained considerable interest from the scientific community. Some researchers have been conducting drug target studies in both wet and dry labs, hoping to find potential hot spot regions in PPIs’ binding interfaces for designing therapeutic drugs. The discovery of small molecule PPI modulators by the emergence of new technologies has made the PPIs significant drug targets[72,78]. Until now, three databases have been dedicated to modulators of PPIs: (1) 2P2I database[79]; (2) TIMBAL[80]; and (3) iPPI-DB[81], and more than 40 PPIs have been targeted successfully[82]. To our knowledge, some of the druggable hotspots for well-studied PPI targets identified by various studies are: MDM2/p53, IL-2/IL-2Ra, HPV-11 E2/HPV-11 E1, TNF-α/TNFR1, and several others[83].
Currently, much focus has been diverted towards the recent COVID-19 pandemic, and many studies have been carried out to combat the deadly virus experimentally and computationally. Gordon et al[84] performed affinity purification-MS and identified 332 physical interactions between human proteins and severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) proteins. The study helped researchers to dig deep down into the host molecular machineries and identify potential hotspots for developing therapeutic compounds to treat COVID-19. PPI identification will also help in predicting the behavior of the virus and the biological processes targeted by the virus. Khorsand et al[85] developed a three-layered network model to predict SARS-CoV-2-human PPIs and reported the most central human proteins in the network by investigating host proteins that are targeted by the viral proteins.
In summary, network biology has become the focus of attention in the recent era by scientists for understanding diseases and the biological processes targeted by the disease. Interaction networks are playing a significant role in understanding virus-host relationship and drug discovery.
The study on PPIs is not just a new field, but a new era in study of virus-host relationships, and we can say that PPIs are at the core of any viral infection. Scientists can use PPIs to gain innumerable novel insights into the functional constitution of a proteome by analyzing all kinds of network parameters. Network biology can help scientists find many potential drug targets that might be involved in certain viral pathways. Many studies have used network biology to construct protein interaction networks of lethal pathogens such as HCV, IAV, dengue virus, and human papilloma virus with their host Homo sapiens to dig deep down into the molecular constitution of the disease pathways, and have successfully found multiple potential drug targets against the viruses. In short, the future of PPI-induced network biology is quite clear and scientists can perform plenty of useful studies against any disease or pathway. Computational prediction of PPIs has become a mandatory tool for finding out the functionalities of unknown proteins.
Provenance and peer review: Invited article; Externally peer reviewed.
Specialty type: Virology
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Ahmad S S-Editor: Gao CC L-Editor: Wang TQ P-Editor: Zhang YL
| 1. | Gonzalez MW, Kann MG. Chapter 4: Protein interactions and disease. PLoS Comput Biol. 2012;8:e1002819. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 144] [Cited by in RCA: 177] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 2. | Berggård T, Linse S, James P. Methods for the detection and analysis of protein-protein interactions. Proteomics. 2007;7:2833-2842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 411] [Cited by in RCA: 444] [Article Influence: 24.7] [Reference Citation Analysis (0)] |
| 3. | Zahiri J, Bozorgmehr JH, Masoudi-Nejad A. Computational Prediction of Protein-Protein Interaction Networks: Algo-rithms and Resources. Curr Genomics. 2013;14:397-414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 86] [Cited by in RCA: 76] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 4. | De Las Rivas J, Fontanillo C. Protein-protein interactions essentials: key concepts to building and analyzing interactome networks. PLoS Comput Biol. 2010;6:e1000807. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 374] [Cited by in RCA: 386] [Article Influence: 25.7] [Reference Citation Analysis (0)] |
| 5. | von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1668] [Cited by in RCA: 1410] [Article Influence: 61.3] [Reference Citation Analysis (0)] |
| 6. | Wang PI, Marcotte EM. It's the machine that matters: Predicting gene function and phenotype from protein networks. J Proteomics. 2010;73:2277-2289. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 7. | Gillis J, Pavlidis P. "Guilt by association" is the exception rather than the rule in gene networks. PLoS Comput Biol. 2012;8:e1002444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 150] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 8. | Pedamallu CS, Posfai J. Open source tool for prediction of genome wide protein-protein interaction network based on ortholog information. Source Code Biol Med. 2010;5:8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 25] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 9. | Skrabanek L, Saini HK, Bader GD, Enright AJ. Computational prediction of protein-protein interactions. Mol Biotechnol. 2008;38:1-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 171] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 10. | Brückner A, Polge C, Lentze N, Auerbach D, Schlattner U. Yeast two-hybrid, a powerful tool for systems biology. Int J Mol Sci. 2009;10:2763-2788. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 315] [Cited by in RCA: 370] [Article Influence: 23.1] [Reference Citation Analysis (0)] |
| 11. | Blikstad C, Ivarsson Y. High-throughput methods for identification of protein-protein interactions involving short linear motifs. Cell Commun Signal. 2015;13:38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 40] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Walhout AJ, Boulton SJ, Vidal M. Yeast two-hybrid systems and protein interaction mapping projects for yeast and worm. Yeast. 2000;17:88-94. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 13. | Legrain P, Selig L. Genome-wide protein interaction maps using two-hybrid systems. FEBS Lett. 2000;480:32-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 90] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 14. | Uetz P, Hughes RE. Systematic and large-scale two-hybrid screens. Curr Opin Microbiol. 2000;3:303-308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 83] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. PNAS. 2001;98:4569. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2632] [Cited by in RCA: 2348] [Article Influence: 97.8] [Reference Citation Analysis (0)] |
| 16. | Ito T, Tashiro K, Muta S, Ozawa R, Chiba T, Nishizawa M, Yamamoto K, Kuhara S, Sakaki Y. Toward a protein–protein interaction map of the budding yeast: A comprehensive system to examine two-hybrid interactions in all possible combinations between the yeast proteins. PNAS. 2000;97:1143. [RCA] [DOI] [Full Text] [Cited by in Crossref: 569] [Cited by in RCA: 499] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 17. | Uetz P, Giot L, Cagney G, Mansfield TA, Judson RS, Knight JR, Lockshon D, Narayan V, Srinivasan M, Pochart P, Qureshi-Emili A, Li Y, Godwin B, Conover D, Kalbfleisch T, Vijayadamodar G, Yang M, Johnston M, Fields S, Rothberg JM. A comprehensive analysis of protein-protein interactions in Saccharomyces cerevisiae. Nature. 2000;403:623-627. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3468] [Cited by in RCA: 3136] [Article Influence: 125.4] [Reference Citation Analysis (0)] |
| 18. | Rao VS, Srinivas K, Sujini GN, Kumar GN. Protein-protein interaction detection: methods and analysis. Int J Proteomics. 2014;2014:147648. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 304] [Cited by in RCA: 436] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 19. | Farooq QUA, Haq NU, Aziz A, Aimen S, ul Haq MI. Mass Spectrometry for Proteomics and Recent Developments in ESI, MALDI and other Ionization Methodologies. Curr Proteom. 2019;16:267-276. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 2] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 20. | Causier B. Studying the interactome with the yeast two-hybrid system and mass spectrometry. Mass Spectrom Rev. 2004;23:350-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 21. | Di Tullio A, Reale S, De Angelis F. Molecular recognition by mass spectrometry. J Mass Spectrom. 2005;40:845-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 49] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 22. | Abu-Farha M, Elisma F, Figeys D. Identification of Protein–Protein Interactions by Mass Spectrometry Coupled Techniques. In: Werther M, Seitz H, editors. Protein – Protein Interaction. Berlin, Heidelberg: Springer Berlin Heidelberg; 2008: 67-80. |
| 23. | Smits AH, Vermeulen M. Characterizing Protein-Protein Interactions Using Mass Spectrometry: Challenges and Opportunities. Trends Biotechnol. 2016;34:825-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 124] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 24. | Young MM, Tang N, Hempel JC, Oshiro CM, Taylor EW, Kuntz ID, Gibson BW, Dollinger G. High throughput protein fold identification by using experimental constraints derived from intramolecular cross-links and mass spectrometry. PNAS. 2000;97:5802. [RCA] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 346] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 25. | Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sørensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2682] [Cited by in RCA: 2515] [Article Influence: 109.3] [Reference Citation Analysis (0)] |
| 26. | Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3480] [Cited by in RCA: 3274] [Article Influence: 142.3] [Reference Citation Analysis (0)] |
| 27. | Sinz A. Cross-Linking/Mass Spectrometry for Studying Protein Structures and Protein-Protein Interactions: Where Are We Now and Where Should We Go from Here? Angew Chem Int Ed Engl. 2018;57:6390-6396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 126] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 28. | Yugandhar K, Gupta S, Yu H. Inferring Protein-Protein Interaction Networks From Mass Spectrometry-Based Proteomic Approaches: A Mini-Review. Comput Struct Biotechnol J. 2019;17:805-811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 38] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 29. | Gavin AC, Aloy P, Grandi P, Krause R, Boesche M, Marzioch M, Rau C, Jensen LJ, Bastuck S, Dümpelfeld B, Edelmann A, Heurtier MA, Hoffman V, Hoefert C, Klein K, Hudak M, Michon AM, Schelder M, Schirle M, Remor M, Rudi T, Hooper S, Bauer A, Bouwmeester T, Casari G, Drewes G, Neubauer G, Rick JM, Kuster B, Bork P, Russell RB, Superti-Furga G. Proteome survey reveals modularity of the yeast cell machinery. Nature. 2006;440:631-636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1990] [Cited by in RCA: 1861] [Article Influence: 97.9] [Reference Citation Analysis (0)] |
| 30. | Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FCP, Weissman JS, Krogan NJ. Toward a Comprehensive Atlas of the Physical Interactome of Saccharomyces cerevisiae. Molecul Cellul Proteom. 2007;6:439. [RCA] [DOI] [Full Text] [Cited by in Crossref: 600] [Cited by in RCA: 575] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 31. | Xu X, Song Y, Li Y, Chang J, Zhang H, An L. The tandem affinity purification method: an efficient system for protein complex purification and protein interaction identification. Protein Expr Purif. 2010;72:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 63] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 32. | Rohila JS, Chen M, Cerny R, Fromm ME. Improved tandem affinity purification tag and methods for isolation of protein heterocomplexes from plants. Plant J. 2004;38:172-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 164] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 33. | Legrain P, Wojcik J, M Gauthier J. Protein-protein interaction maps: A lead towards cellular functions. Trend Genet. 2001;17:346-52. [RCA] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 134] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 34. | MacBeath G, Schreiber SL. Printing proteins as microarrays for high-throughput function determination. Science. 2000;289:1760-1763. [PubMed] |
| 35. | Michnick SW, Ear PH, Landry C, Malleshaiah MK, Messier V. Protein-Fragment Complementation Assays for Large-Scale Analysis, Functional Dissection and Dynamic Studies of Protein–Protein Interactions in Living Cells. In: Luttrell LM, Ferguson SSG. Signal Transduction Protocols. Totowa, NJ: Humana Press, 2011: 395-425. |
| 36. | Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H, Andrews B, Tyers M, Boone C. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364-2368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1635] [Cited by in RCA: 1598] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 37. | Pei D, Xu J, Zhuang Q, Tse HF, Esteban MA. Induced Pluripotent Stem Cell Technology in Regenerative Medicine and Biology. In: Kasper C, van Griensven M, Pörtner R. Bioreactor Systems for Tissue Engineering II: Strategies for the Expansion and Directed Differentiation of Stem Cells. Berlin, Heidelberg: Springer Berlin Heidelberg, 2010: 127-141. |
| 38. | Pazos F, Valencia A. In silico two-hybrid system for the selection of physically interacting protein pairs. Proteins. 2002;47:219-227. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 157] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 39. | Lee SA, Chan CH, Tsai CH, Lai JM, Wang FS, Kao CY, Huang CY. Ortholog-based protein-protein interaction prediction and its application to inter-species interactions. BMC Bioinformatics. 2008;9 Suppl 12:S11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 76] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 40. | Enright AJ, Iliopoulos I, Kyrpides NC, Ouzounis CA. Protein interaction maps for complete genomes based on gene fusion events. Nature. 1999;402:86-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 732] [Cited by in RCA: 700] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 41. | Wojcik J, Schächter V. Protein-protein interaction map inference using interacting domain profile pairs. Bioinformatics. 2001;17 Suppl 1:S296-S305. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 147] [Cited by in RCA: 154] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 42. | Grigoriev A. A relationship between gene expression and protein interactions on the proteome scale: analysis of the bacteriophage T7 and the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 2001;29:3513-3519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 165] [Cited by in RCA: 152] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 43. | Zhang QC, Petrey D, Deng L, Qiang L, Shi Y, Thu CA, Bisikirska B, Lefebvre C, Accili D, Hunter T, Maniatis T, Califano A, Honig B. Structure-based prediction of protein-protein interactions on a genome-wide scale. Nature. 2012;490:556-560. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 521] [Cited by in RCA: 525] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 44. | Sato T, Yamanishi Y, Kanehisa M, Toh H. The inference of protein-protein interactions by co-evolutionary analysis is improved by excluding the information about the phylogenetic relationships. Bioinformatics. 2005;21:3482-3489. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 94] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 45. | Orchard S, Kerrien S, Abbani S, Aranda B, Bhate J, Bidwell S, Bridge A, Briganti L, Brinkman FS, Cesareni G, Chatr-aryamontri A, Chautard E, Chen C, Dumousseau M, Goll J, Hancock RE, Hannick LI, Jurisica I, Khadake J, Lynn DJ, Mahadevan U, Perfetto L, Raghunath A, Ricard-Blum S, Roechert B, Salwinski L, Stümpflen V, Tyers M, Uetz P, Xenarios I, Hermjakob H. Protein interaction data curation: the International Molecular Exchange (IMEx) consortium. Nat Methods. 2012;9:345-350. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 407] [Cited by in RCA: 404] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 46. | Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O'Donnell L, Reguly T, Nixon J, Ramage L, Winter A, Sellam A, Chang C, Hirschman J, Theesfeld C, Rust J, Livstone MS, Dolinski K, Tyers M. The BioGRID interaction database: 2015 update. Nucleic Acids Res. 2015;43:D470-D478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 715] [Cited by in RCA: 676] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 47. | Oughtred R, Stark C, Breitkreutz BJ, Rust J, Boucher L, Chang C, Kolas N, O'Donnell L, Leung G, McAdam R, Zhang F, Dolma S, Willems A, Coulombe-Huntington J, Chatr-Aryamontri A, Dolinski K, Tyers M. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019;47:D529-D541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1068] [Cited by in RCA: 961] [Article Influence: 160.2] [Reference Citation Analysis (0)] |
| 48. | Szklarczyk D, Franceschini A, Kuhn M, Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork P, Jensen LJ, von Mering C. The STRING database in 2011: functional interaction networks of proteins, globally integrated and scored. Nucleic Acids Res. 2011;39:D561-D568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2562] [Cited by in RCA: 2632] [Article Influence: 188.0] [Reference Citation Analysis (0)] |
| 49. | Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015;43:D447-D452. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6477] [Cited by in RCA: 7598] [Article Influence: 690.7] [Reference Citation Analysis (0)] |
| 50. | Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47:D607-D613. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10161] [Cited by in RCA: 11661] [Article Influence: 1943.5] [Reference Citation Analysis (1)] |
| 51. | Ammari MG, Gresham CR, McCarthy FM, Nanduri B. HPIDB 2.0: a curated database for host-pathogen interactions. Database (Oxford). 2016;2016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 200] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 52. | Orchard S, Ammari M, Aranda B, Breuza L, Briganti L, Broackes-Carter F, Campbell NH, Chavali G, Chen C, del-Toro N, Duesbury M, Dumousseau M, Galeota E, Hinz U, Iannuccelli M, Jagannathan S, Jimenez R, Khadake J, Lagreid A, Licata L, Lovering RC, Meldal B, Melidoni AN, Milagros M, Peluso D, Perfetto L, Porras P, Raghunath A, Ricard-Blum S, Roechert B, Stutz A, Tognolli M, van Roey K, Cesareni G, Hermjakob H. The MIntAct project--IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358-D363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1504] [Cited by in RCA: 1363] [Article Influence: 123.9] [Reference Citation Analysis (0)] |
| 53. | IMEx Consortium Curators; Del-Toro N, Duesbury M, Koch M, Perfetto L, Shrivastava A, Ochoa D, Wagih O, Piñero J, Kotlyar M, Pastrello C, Beltrao P, Furlong LI, Jurisica I, Hermjakob H, Orchard S, Porras P. Capturing variation impact on molecular interactions in the IMEx Consortium mutations data set. Nat Commun. 2019;10:10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 88] [Cited by in RCA: 103] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 54. | Goodacre N, Devkota P, Bae E, Wuchty S, Uetz P. Protein-protein interactions of human viruses. Semin Cell Dev Biol. 2020;99:31-39. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 55. | Licata L, Briganti L, Peluso D, Perfetto L, Iannuccelli M, Galeota E, Sacco F, Palma A, Nardozza AP, Santonico E, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2012 update. Nucleic Acids Res. 2012;40:D857-D861. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 746] [Cited by in RCA: 768] [Article Influence: 54.9] [Reference Citation Analysis (0)] |
| 56. | Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449-D451. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1525] [Cited by in RCA: 1456] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 57. | Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, Balakrishnan L, Marimuthu A, Banerjee S, Somanathan DS, Sebastian A, Rani S, Ray S, Harrys Kishore CJ, Kanth S, Ahmed M, Kashyap MK, Mohmood R, Ramachandra YL, Krishna V, Rahiman BA, Mohan S, Ranganathan P, Ramabadran S, Chaerkady R, Pandey A. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767-D772. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2600] [Cited by in RCA: 2527] [Article Influence: 157.9] [Reference Citation Analysis (0)] |
| 58. | Kwofie SK, Schaefer U, Sundararajan VS, Bajic VB, Christoffels A. HCVpro: hepatitis C virus protein interaction database. Infect Genet Evol. 2011;11:1971-1977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 65] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 59. | Guirimand T, Delmotte S, Navratil V. VirHostNet 2.0: surfing on the web of virus/host molecular interactions data. Nucleic Acids Res. 2015;43:D583-D587. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 169] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 60. | Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498-2504. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24663] [Cited by in RCA: 33278] [Article Influence: 1584.7] [Reference Citation Analysis (0)] |
| 61. | Enright AJ, Ouzounis CA. BioLayout--an automatic graph layout algorithm for similarity visualization. Bioinformatics. 2001;17:853-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 85] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 62. | Hu Z, Mellor J, Wu J, Yamada T, Holloway D, Delisi C. VisANT: data-integrating visual framework for biological networks and modules. Nucleic Acids Res. 2005;33:W352-W357. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 120] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 63. | Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, Timm J, Mintzlaff S, Abraham C, Bock N, Kietzmann S, Goedde A, Toksöz E, Droege A, Krobitsch S, Korn B, Birchmeier W, Lehrach H, Wanker EE. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1772] [Cited by in RCA: 1706] [Article Influence: 85.3] [Reference Citation Analysis (0)] |
| 64. | Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644-652. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 658] [Cited by in RCA: 542] [Article Influence: 31.9] [Reference Citation Analysis (0)] |
| 65. | de Chassey B, Navratil V, Tafforeau L, Hiet MS, Aublin-Gex A, Agaugué S, Meiffren G, Pradezynski F, Faria BF, Chantier T, Le Breton M, Pellet J, Davoust N, Mangeot PE, Chaboud A, Penin F, Jacob Y, Vidalain PO, Vidal M, André P, Rabourdin-Combe C, Lotteau V. Hepatitis C virus infection protein network. Mol Syst Biol. 2008;4:230. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 295] [Cited by in RCA: 303] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 66. | Farooq QUA, Khan FF. Construction and analysis of a comprehensive protein interaction network of HCV with its host Homo sapiens. BMC Infect Dis. 2019;19:367. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 67. | Morales-Sánchez A, Fuentes-Pananá EM. Human viruses and cancer. Viruses. 2014;6:4047-4079. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 109] [Cited by in RCA: 142] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 68. | Farooq QUA, Shaukat Z, Zhou T, Aiman S, Gong W, Li C. Inferring Virus-Host relationship between HPV and its host Homo sapiens using protein interaction network. Sci Rep. 2020;10:8719. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 69. | Farooq QUA, Shaukat Z, Aiman S, Zhou T, Li C. A systems biology-driven approach to construct a comprehensive protein interaction network of influenza A virus with its host. BMC Infect Dis. 2020;20:480. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 4] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 70. | Navratil V, de Chassey B, Combe CR, Lotteau V. When the human viral infectome and diseasome networks collide: towards a systems biology platform for the aetiology of human diseases. BMC Syst Biol. 2011;5:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 71. | Pujol A, Mosca R, Farrés J, Aloy P. Unveiling the role of network and systems biology in drug discovery. Trends Pharmacol Sci. 2010;31:115-123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 208] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 72. | Mabonga L, Kappo AP. Protein-protein interaction modulators: advances, successes and remaining challenges. Biophys Rev. 2019;11:559-581. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 136] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 73. | Cukuroglu E, Engin HB, Gursoy A, Keskin O. Hot spots in protein-protein interfaces: towards drug discovery. Prog Biophys Mol Biol. 2014;116:165-173. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 122] [Article Influence: 11.1] [Reference Citation Analysis (0)] |
| 74. | Bosch J. PPI inhibitor and stabilizer development in human diseases. Drug Discov Today Technol. 2017;24:3-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 22] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 75. | Mabonga L, Kappo AP. Peptidomimetics: A Synthetic Tool for Inhibiting Protein–Protein Interactions in Cancer. Int J Pept Res Ther. 2020;26:225-241. [RCA] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 55] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 76. | Zhang G, Andersen J, Gerona-Navarro G. Peptidomimetics Targeting Protein-Protein Interactions for Therapeutic Development. Protein Pept Lett. 2018;25:1076-1089. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 18] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 77. | Robertson NS, Spring DR. Using Peptidomimetics and Constrained Peptides as Valuable Tools for Inhibiting Protein⁻Protein Interactions. Molecules. 2018;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 75] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 78. | Feng Y, Wang Q, Wang T. Drug Target Protein-Protein Interaction Networks: A Systematic Perspective. Biomed Res Int. 2017;2017:1289259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 65] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 79. | Basse MJ, Betzi S, Bourgeas R, Bouzidi S, Chetrit B, Hamon V, Morelli X, Roche P. 2P2Idb: a structural database dedicated to orthosteric modulation of protein-protein interactions. Nucleic Acids Res. 2013;41:D824-D827. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 110] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 80. | Higueruelo AP, Schreyer A, Bickerton GR, Pitt WR, Groom CR, Blundell TL. Atomic interactions and profile of small molecules disrupting protein-protein interfaces: the TIMBAL database. Chem Biol Drug Des. 2009;74:457-467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 131] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 81. | Labbé CM, Laconde G, Kuenemann MA, Villoutreix BO, Sperandio O. iPPI-DB: a manually curated and interactive database of small non-peptide inhibitors of protein-protein interactions. Drug Discov Today. 2013;18:958-968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 81] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 82. | Bakail M, Ochsenbein F. Targeting protein–protein interactions, a wide open field for drug design. C R Chim. 2016;19:19-27. [RCA] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 74] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 83. | Kozakov D, Hall DR, Chuang GY, Cencic R, Brenke R, Grove LE, Beglov D, Pelletier J, Whitty A, Vajda S. Structural conservation of druggable hot spots in protein-protein interfaces. Proc Natl Acad Sci U S A. 2011;108:13528-13533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 185] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 84. | Gordon DE, Jang GM, Bouhaddou M, Xu J, Obernier K, White KM, O'Meara MJ, Rezelj VV, Guo JZ, Swaney DL, Tummino TA, Hüttenhain R, Kaake RM, Richards AL, Tutuncuoglu B, Foussard H, Batra J, Haas K, Modak M, Kim M, Haas P, Polacco BJ, Braberg H, Fabius JM, Eckhardt M, Soucheray M, Bennett MJ, Cakir M, McGregor MJ, Li Q, Meyer B, Roesch F, Vallet T, Mac Kain A, Miorin L, Moreno E, Naing ZZC, Zhou Y, Peng S, Shi Y, Zhang Z, Shen W, Kirby IT, Melnyk JE, Chorba JS, Lou K, Dai SA, Barrio-Hernandez I, Memon D, Hernandez-Armenta C, Lyu J, Mathy CJP, Perica T, Pilla KB, Ganesan SJ, Saltzberg DJ, Rakesh R, Liu X, Rosenthal SB, Calviello L, Venkataramanan S, Liboy-Lugo J, Lin Y, Huang XP, Liu Y, Wankowicz SA, Bohn M, Safari M, Ugur FS, Koh C, Savar NS, Tran QD, Shengjuler D, Fletcher SJ, O'Neal MC, Cai Y, Chang JCJ, Broadhurst DJ, Klippsten S, Sharp PP, Wenzell NA, Kuzuoglu-Ozturk D, Wang HY, Trenker R, Young JM, Cavero DA, Hiatt J, Roth TL, Rathore U, Subramanian A, Noack J, Hubert M, Stroud RM, Frankel AD, Rosenberg OS, Verba KA, Agard DA, Ott M, Emerman M, Jura N, von Zastrow M, Verdin E, Ashworth A, Schwartz O, d'Enfert C, Mukherjee S, Jacobson M, Malik HS, Fujimori DG, Ideker T, Craik CS, Floor SN, Fraser JS, Gross JD, Sali A, Roth BL, Ruggero D, Taunton J, Kortemme T, Beltrao P, Vignuzzi M, García-Sastre A, Shokat KM, Shoichet BK, Krogan NJ. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature. 2020;583:459-468. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3145] [Cited by in RCA: 3176] [Article Influence: 635.2] [Reference Citation Analysis (0)] |
| 85. | Khorsand B, Savadi A, Naghibzadeh M. SARS-CoV-2-human protein-protein interaction network. Inform Med Unlocked. 2020;20:100413. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 32] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 86. | Oti M, Brunner HG. The modular nature of genetic diseases. Clin Genet. 2007;71:1-11. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 308] [Cited by in RCA: 281] [Article Influence: 14.8] [Reference Citation Analysis (0)] |