Revised: September 16, 2011
Accepted: September 25, 2011
Published online: February 12, 2012
A major challenge in vaccine design is to identify antigen presentation and delivery systems capable of rapidly stimulating both the humoral and cellular components of the immune system to elicit a strong and sustained immunity against different viral isolates. Approaches to achieve this end involve live attenuated and inactivated virions, viral vectors, DNA, and protein subunits. This review reports the state of current antigen delivery, and focuses on two innovative systems recently established at our labs. These systems are the filamentous bacteriophage fd and an icosahedral scaffold formed by the acyltransferase component (E2 protein) of the pyruvate dehydrogenase complex of Bacillus stearothermophilus.
- Citation: Trovato M, Krebs SJ, Haigwood NL, De Berardinis P. Delivery strategies for novel vaccine formulations. World J Virol 2012; 1(1): 4-10
- URL: https://www.wjgnet.com/2220-3249/full/v1/i1/4.htm
- DOI: https://dx.doi.org/10.5501/wjv.v1.i1.4
Historically, live attenuated virus vaccines were used to control many viral diseases, such as measles, smallpox, mumps and rubella. These vaccines elicit both humoral and cell-mediated immune memory responses[1-5], however, they are associated with genetic instability and residual virulence[6-7]. For these reasons, current human clinical trials using live attenuated viral vaccines are rare, and research has been limited to non-human primate studies[8] to understand the mechanisms about the protection elicited. The possible causes of pathogenicity may include: (1) reversion to a virulent form of the attenuated virus[9]; (2) recombination of the vaccine strain with wild type, pathogenic virus[10]; (3) ability of viral genomes to persist in tissues or the proviral genomes to integrate into the host genome[11]; and (4) the dysregulation of the immune system by viral proteins[12]. As alternatives to live vectors, a variety of other antigen delivery systems are available for vaccine research.
One of the most pursued strategies is based upon the use of a huge range of viral and bacterial vectors, which are defined as non-pathogenic vehicles containing inserted genes from pathogens for their expression and subsequent induction of specific immune responses. Viral vectors such as poxvirus, adenovirus, vesicular stomatitis virus, adenovirus-associated virus (AAV), alphavirus, cytomegalovirus and lentivirus, to name some but not all, have been explored[13]. In addition, vectors have been developed from bacteria such as Shigella[14] and Salmonella[15], by exploiting their ability to be delivered via the oral route, which is the natural route of infection for a variety of bacteria. Because so many of the factors governing the induction of optimal immunity are incompletely understood (such as the activation of the innate immune system by various components of vectors and their effects upon adaptive immunity), it is hard to know exactly what would make the best vector for any given vaccine. Given the expensive and long development process required for clinical testing, the choice of one vector over another for each vaccine has been largely based upon the key characteristics of the vector and results of preclinical studies rather than comparison of vectors and administration protocols, since regimens, doses, and matrices of prime-boost combinations could lead to innumerable variables. Some of these vectors, in particular adenovectors and poxvectors, including both canarypox and modified vaccinia Ankara, have been employed in clinical trials for an anti-HIV-1 vaccine[16-20], all obtaining unsatisfactory results. More recently, "The STEP trial", which utilized an adenovirus vector encoding HIV proteins, was performed as a proof of concept for prevention or control of HIV infection[21,22]. The trial was stopped when it was clear that the vaccine was not demonstrating efficacy.
One major concern regarding the use of vectors is the immune response against the vector itself,[23,24] especially in persons who have been previously exposed to the virus or a related virus. Other concerns are the disputed risk of integration[25], the limited gene-insert capacity (in AAV and alphavirus vectors[26,27]), the safety issue in neurotropism (such as in the vectors based on herpes virus or vesicular stomatitis virus[28]), and in bacterial vaccine vectors given orally, the transfer of heterologous genes to other bacteria[29].
In addition to the use of vectors, antigens can be delivered by injecting plasmid DNA encoding immunogenic proteins under the control of eukaryotic promoters. DNA vaccination was first described in 1993[30] and accepted as one of the most promising ways of immunization. Overall, available evidence suggests that DNA vaccination is generally well tolerated[31-33], and two veterinary vaccines have been already licensed[34]. The fate of the DNA after injection has been a matter of concern due to the risk of integration of the nucleic acids into the host genome. However, all available data suggest that frequency of integration is below the frequency of spontaneous mutation[35,36]. Notwithstanding these premises, failures and triggering of disease exacerbation instead of protection have also been reported on the use of DNA vaccines. One of the main criticisms for DNA vaccination is their relatively low efficacy, mainly because a large amount of DNA is needed to be injected in order to achieve a strong response[37]. Improvement of the immune response to DNA vaccines has been attempted by aiming at enhanced plasmid uptake by exploring different routes of administration (gene-gun, electroporation), delivery of the antigen in combination with various immunostimulatory cytokines[38], and by using DNA vaccines in combination with adjuvants or cytokines in order to locally recruit different subsets of professional antigen-presenting cells (APCs)[39].
Virus-like particles (VLPs) represent another recent important biotechnological advancement. They are composed of one or several recombinantly expressed viral proteins, which spontaneously assemble into supramolecular structures resembling infectious viruses or, in some cases, subviral particles.
VLPs, like viruses, are comprised of one or more proteins arranged geometrically into dense, repetitive arrays. These structures are characteristics of microbial antigens, and the mammalian immune system has evolved to respond vigorously to this arrangement of antigens. Thus, due to their highly repetitive surface, VLPs are able to induce strong B-cell responses by efficiently crosslinking the membrane-associated immunoglobulin molecules that constitute the B-cell receptor. In addition to their ability to stimulate B-cell-mediated immune responses, VLPs have been shown to be highly effective in stimulating CD4 proliferative responses and cytotoxic T lymphocyte (CTL) responses[40-44]. Currently, VLP-based vaccines are in various stages of development, spanning preclinical evaluation to market. Vaccines for hepatitis B (Recombivax and Energix) and human papillomavirus (Gardasil and Cervarix) have been licensed commercially[41]. Furthermore, VLPs may also be used as a platform for inducing immune responses against antigens of choice. This can be achieved by constructing chimeric VLPs that display heterologous epitopes[45,46]. However, generating chimeric VLPs is largely empirical; it is almost impossible to predict whether individual peptides will be compatible with VLP assembly or whether the insertion will be immunogenic. Another important limitation of this approach is the size and the nature of the epitopes that can be inserted into the VLPs, in particular into their immunodominant regions.
Other promising strategies for vaccine delivery include the use of virosomes (liposomes carrying viral envelope proteins)[47-51], or subunit vaccines expressed in plant[52-56] or insect cells[57-61].
In summary, a variety of vaccine carriers and delivery modalities have been developed in recent years. However, the failure of clinical trials, based on available formulations, indicates that novel vaccine modalities are still very much needed to complement the existing array of options. In this context, we propose two innovative concepts for vaccine design based on the filamentous bacteriophage fd and the E2 protein from the pyruvate dehydrogenase (PDH) complex of Bacillus stearothermophilus (B. stearothermophilus). Both of the delivery systems combine the advantages of safety with the capability to elicit both humoral and cellular antigen-specific immune responses.
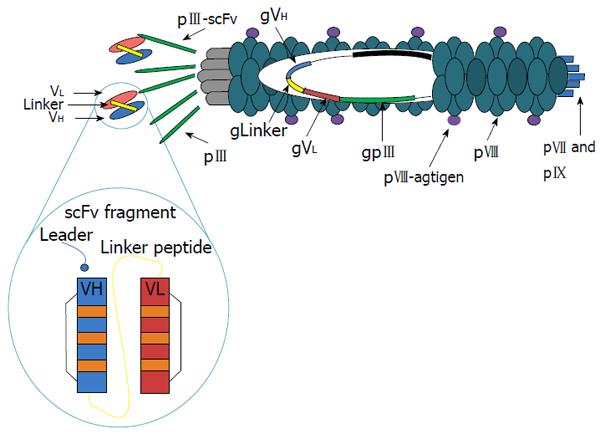
The system of antigen delivery by bacteriophage is based on modification of the phage display technology. Phage libraries with specific antisera have been screened to identify phage displaying peptides, which correspond to or mimic antigenic epitopes[62]. Immunization with phages displaying these peptides has proven capable of eliciting protective antibodies in animal models of various diseases[63,64]. The filamentous bacteriophage fd (others are M13, f1) is well understood at both the structural and genetic levels[65]. The capsid contains five types of coat protein, four minor (approximately five copies of each, encoded by phage genes III, VI, VII, and IX) and one major (approximately 2700 copies, encoded by phage gene VIII). The potential to display a large number of copies on the surface of these filamentous bacteriophages makes them an attractive vehicle for the expression of foreign peptides. There is evidence from immunoassays and nuclear magnetic resonance (NMR) spectroscopy that the exposed peptides have a stable three-dimensional structure closely resembling what they exhibit in the wild-type parent protein[66]. We have shown that the peptides expressed on the pVIII phage major coat protein can both produce specific antibodies and present T helper epitopes[67]. Moreover, we have observed that the filamentous bacteriophages are taken up and processed by major histocompatibility complex (MHC) class I and class II pathways[68]. In addition, because co-expression of linked helper T-cell and cytotoxic T-cell (CTL) epitopes on the surface of the same APC is a strict requirement for priming a CTL response, we designed hybrid bacteriophages simultaneously displaying helper and/or cytotoxic epitopes on the same virion. They were obtained by infecting bacterial cells, harboring a plasmid encoding a modified gene VIII, with an engineered bacteriophage carrying a second and different copy of a modified gene VIII. Using phage particles coexpressing both helper and CTL peptides, we demonstrated the ability of bacteriophage virions to evoke an antigen specific CTL response both in vitro and in vivo[69]. This surprising finding supports the further exploration of this benign bacteriophage as a powerful and versatile antigen-delivery system and makes the fd virions particularly attractive for vaccine design.
The efficacy of filamentous bacteriophage fd antigen delivery system can be further improved by exploiting the possibility of displaying additional peptides or target sequences on one of the minor coat proteins. In particular, targeting APCs can enhance subsequent immune responses and avoid tolerance induction, should this occur in the presence of a co-stimulatory signal. This has been shown, for example, for the DEC-205 receptor, a decalectin involved in the uptake and presentation by dendritic cells (DCs)[70]. For this purpose, we have engineered two unique restriction sites into the fdAMPLAY388 bacteriophage vector by site-directed mutagenesis, which allows the insertion of peptides/proteins at the N-terminus of the pIII gene. Thus, we cloned into the fdAMPLAY vector a single chain antibody fragments (scFvs) from NLDC-145 mouse monoclonal antibody, which is known to bind the mouse dendritic cell-surface molecule DEC-205 (Figure 1). Functional phage particles were purified and Western immunoblotting analysis confirmed the identity of the 90 kDa scFv fusion protein expressed by the recombinant fd. We have recently demonstrated that bacteriophage fd particles displaying the scFv from NLDC-145 mouse monoclonal antibody at the N-terminus of the pIII gene were able to induce activation and maturation of mouse DCs, both in vivo or in vitro, in absence of other stimuli. Furthermore, DCs targeting with fd particles induced a strong antigen specific immune response and, in experiments investigating inhibition of tumor growth, had a protective efficacy similar to adoptive transferred DCs[71]. These results further validate the potential employment of this safe, versatile, and inexpensive delivery system for vaccine formulation.
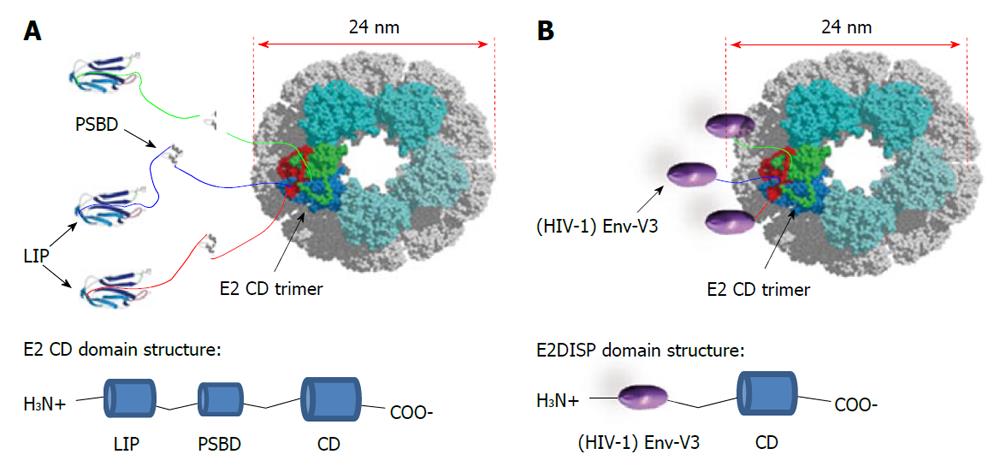
This novel antigen-presenting system is based on the PDH complex of B. stearothermophilus[72]. The PDH complex belongs to the family of 2-oxo acid dehydrogenase complexes. They consist of multiple copies of three different enzymes that catalyze the oxidative decarboxilation of 2-oxo acids. In the case of the PDH complex, two of its component enzymes, E1 and E3, assemble over the surface of a large structural scaffold formed by the core enzyme, E2, a dihydrolipoyl acyltransferase. The E2 chain of B. stearothermophilus is composed of three independently folded domains separated by flexible linker regions: a lipoyl domain (LIP) of 9.5 kDa, a peripheral subunit-binding domain (PSBD) of 5.3 kDa and a catalytic (acetyltransferase) core domain (CD) of 28 kDa. The core C-terminal catalytic domain of E2 self-assembles into trimers, which in turn aggregates to generate a 60-chain core with icosahedral symmetry[73]. The 60-mer has a molecular weight of > 1.5 MDa and a diameter of 24 nm as visualized under electron microscope[72]. Surprisingly, E2 can be renatured from denaturing conditions in vitro to assemble into the 60-mer virus-like particle, without the need of chaperonins[74]. This finding may be attributed to the heat-stable properties often found in proteins from this thermophilic bacterium. Efficient refolding of E2 into the 60-mer is also possible by replacing the natural peripheral domains with foreign peptides as N-terminal fusions to the CD. The display of exogenous peptides on the E2 scaffold is obtained by using engineered plasmids which allows the insertion of exogenous oligonucleotides at the 5’ of the gene coding for the acyltransferase domain of the E2 protein[72]. Thus, a suitably engineered E2 core (E2DISP) can display 60 copies of heterologous polypeptides on the surface of a high molecular mass scaffold[72]. This property can be exploited for vaccine design. Previous work from our laboratory has described that small epitopes (9-15 aa) displayed on the E2 scaffold elicit both cellular and humoral specific immune responses[75,76]. However, displaying larger protein antigens may be more useful, as they contain multiple T cell epitopes as well as antibody determinants.
In theory, there should be no limitation to the size of amino acid sequences displayed on the E2 scaffold. In fact, at the N-terminus of the acyltransferase CD each E2 chain in the B. stearothermophilus PDH complex naturally displays 187 amino acid residues in the form of the two folded protein domains (LIP and PSBD domains) and two flexible linkers (Figure 2A). Moreover, the E2 system is naturally designed to present up to 60 copies of the E1 (about 150 kDa) or E3 (about 100 kDa) enzymes noncovalently attached on its surface. Thus, up to 60 large polypeptides can be presented on the E2 scaffold as N-terminal fusions to the acetyltransferase CD. In practice, several proteins, such as the green fluorescent protein (GFP), have been expressed as an N-terminal fusions[72].
These properties of E2 VLPs compare favorably to other types of VLPs, such as the VLPs based on human papillomavirus that can only accept approximately 60 amino acids of foreign antigen[77] or on Hepatitis B surface antigen (HBsAg) that has a limit of approximately 36 amino acids[78]. E2DISP fusion proteins are produced in E. coli, and if soluble, the proteins can be purified directly and endotoxin can be removed using standard biochemical techniques. If insoluble, they can be denatured and refolded slowly by step-down dialysis to obtain soluble VLPs. Solubility can also be improved by refolding the fusion proteins in the presence of E2DISP monomers at different molar ratios, which reduces the number of displayed heterologous proteins per VLP. Thus, production of E2 particles is relatively simple and inexpensive, as compared with the mammalian or baculovirus cell culture. In this context we have successfully expressed and refolded a large array of HIV antigens and protein domains. In particular, we have constructed stable VLPs displaying the HIV-1 Gag (p17) protein as an N-terminal fusion to the E2 CD and found that mice immunized with the Gag (p17)-E2 60mer particles mounted a strong and sustained antibody response. Using transgenic mouse model systems, we also demonstrated that CD8+ T cells primed with E2 recombinant particles were able to exert lytic activity[79]. In addition, as schematically represented in Figure 2B, we displayed the HIV-1 Envelope (Env) third hypervariable region (V3), which is known to bind neutralizing antibodies[80] on the E2DISP system. We evaluated the immunogenicity of these purified 60mer VLPs in mice and rabbits, alone and in combination with an HIV Env glycoprotein (gp160) expression plasmid in an attempt to enhance immunogenicity. Using this system we demonstrated that simultaneous co-immunization with Env(V3)-E2 VLPs and gp160 encoded DNA was more effective than each individual component alone or the DNA-prime/protein-boost immunization regimen in eliciting both neutralizing antibodies in rabbits and CD8+ T cells responses in mice, even in the absence of adjuvants[81].
Vaccine is one of the most powerful and cost-effective tools of modern medicine. Currently, there is a renaissance in vaccine research due to the fact that healthcare authorities are increasingly recognizing the public health benefit and cost-effectiveness of vaccines. Since vaccines are administered a few times at most, they are also more cost-effective than many other drugs in treating the subsequent diseases. In recent years, several economists have argued convincingly that the cost savings are even more impressive because they go beyond the cost of medical care, and should include income lost due to illness and its sequelae[82]. The revival of interest in vaccines has of course also been underpinned by a rapidly expanding body of knowledge in the fields of immunology and by the impressive biotechnological advancement in recent years. Today, purification of microbial elements, genetic engineering, and improved knowledge of immune protection allow direct creation of attenuated mutants, expression of vaccine proteins in a variety of vectors, purification and even synthesis of microbial antigens, and induction of immune responses through manipulation of DNA, RNA, and proteins. A major aim is to integrate information gathered from classical pre-clinical trials with that derived from different fields of investigation. In this context, several approaches are being pursued to improve rationally designed delivery systems. We have been exploring the advantages of two innovative antigen display systems derived from non-pathogenic prokaryotic organisms, based on the filamentous bacteriophages fd and the acyltransferase component (the E2 protein) from the PDH complex of B. stearothermophilus. The fd and E2 antigen delivery systems combine the safety with the capability to trigger both humoral and cellular antigen-specific immune response, even in the absence of adjuvants. Both of these vehicles gain access to APCs, and intersect the human leukocyte antigen class I and II intracellular compartments, inducing specific cytotoxic T-lymphocyte responses. Moreover, the E2DISP delivery system offers a unique opportunity to display up to 60 copies of individual or multiple heterologous polypeptides, by preserving their native structure on the surface of a single VLP. These antigen presentation systems could be combined with other well-studied vaccines in an attempt to elicit full-spectrum immune responses.
Peer reviewer: Kenji Okuda, MD, PhD, Professor, Yokohama City University School of Medicine, Chouju Medical Institute, 441-8124 Az-ayamanaka, Noyorichou, Toyohasi, Aichi Prefecture, Japan
S- Editor Wang JL L- Editor Ma JY E- Editor Zheng XM
| 1. | Lauring AS, Jones JO, Andino R. Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol. 2010;28:573-579. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 2. | Jones T. Varivax (Merck & Co). Curr Opin Investig Drugs. 2002;3:54-57. |
| 3. | Kuwata T, Miura T, Hayami M. Using SHIVs to develop an anti-HIV-1 live-attenuated vaccine. Trends Microbiol. 2001;9:475-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 8] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 4. | Johnson RP, Desrosiers RC. Protective immunity induced by live attenuated simian immunodeficiency virus. Curr Opin Immunol. 1998;10:436-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 107] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Hu SL. Non-human primate models for AIDS vaccine research. Curr Drug Targets Infect Disord. 2005;5:193-201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 61] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 6. | Ruprecht RM. Live attenuated AIDS viruses as vaccines: promise or peril? Immunol Rev. 1999;170:135-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 7. | Ehrenfeld E, Modlin J, Chumakov K. Future of polio vaccines. Expert Rev Vaccines. 2009;8:899-905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 41] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 8. | Young KR, McBurney SP, Karkhanis LU, Ross TM. Virus-like particles: designing an effective AIDS vaccine. Methods. 2006;40:98-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 69] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 9. | Jenkins PC, Modlin JF. Decision analysis in planning for a polio outbreak in the United States. Pediatrics. 2006;118:611-618. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 14] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 10. | Monath TP, Levenbook I, Soike K, Zhang ZX, Ratterree M, Draper K, Barrett AD, Nichols R, Weltzin R, Arroyo J. Chimeric yellow fever virus 17D-Japanese encephalitis virus vaccine: dose-response effectiveness and extended safety testing in rhesus monkeys. J Virol. 2000;74:1742-1751. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 102] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 11. | Ramirez K, Barry EM, Ulmer J, Stout R, Szabo J, Manetz S, Levine MM, Pasetti MF. Preclinical safety and biodistribution of Sindbis virus measles DNA vaccines administered as a single dose or followed by live attenuated measles vaccine in a heterologous prime-boost regimen. Hum Gene Ther. 2008;19:522-531. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 12. | Cheng SM, Li JC, Lin SS, Lee DC, Liu L, Chen Z, Lau AS. HIV-1 transactivator protein induction of suppressor of cytokine signaling-2 contributes to dysregulation of IFN{gamma} signaling. Blood. 2009;113:5192-5201. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 13. | Liu MA. Immunologic basis of vaccine vectors. Immunity. 2010;33:504-515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 14. | Fennelly GJ, Khan SA, Abadi MA, Wild TF, Bloom BR. Mucosal DNA vaccine immunization against measles with a highly attenuated Shigella flexneri vector. J Immunol. 1999;162:1603-1610. [PubMed] |
| 15. | Fouts TR, Lewis GK, Hone DM. Construction and characterization of a Salmonella typhi-based human immunodeficiency virus type 1 vector vaccine. Vaccine. 1995;13:561-569. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 22] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 16. | Hanke T, McMichael AJ, Samuel RV, Powell LA, McLoughlin L, Crome SJ, Edlin A. Lack of toxicity and persistence in the mouse associated with administration of candidate DNA- and modified vaccinia virus Ankara (MVA)-based HIV vaccines for Kenya. Vaccine. 2002;21:108-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 41] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 17. | Peters BS, Jaoko W, Vardas E, Panayotakopoulos G, Fast P, Schmidt C, Gilmour J, Bogoshi M, Omosa-Manyonyi G, Dally L. Studies of a prophylactic HIV-1 vaccine candidate based on modified vaccinia virus Ankara (MVA) with and without DNA priming: effects of dosage and route on safety and immunogenicity. Vaccine. 2007;25:2120-2127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 85] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 18. | Vasan S, Schlesinger SJ, Chen Z, Hurley A, Lombardo A, Than S, Adesanya P, Bunce C, Boaz M, Boyle R. Phase 1 safety and immunogenicity evaluation of ADMVA, a multigenic, modified vaccinia Ankara-HIV-1 B'/C candidate vaccine. PLoS One. 2010;5:e8816. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 19. | Wee EG, Patel S, McMichael AJ, Hanke T. A DNA/MVA-based candidate human immunodeficiency virus vaccine for Kenya induces multi-specific T cell responses in rhesus macaques. J Gen Virol. 2002;83:75-80. [PubMed] |
| 20. | Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med. 2009;361:2209-2220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2345] [Cited by in RCA: 2394] [Article Influence: 149.6] [Reference Citation Analysis (0)] |
| 21. | Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet. 2008;372:1881-1893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1393] [Cited by in RCA: 1357] [Article Influence: 79.8] [Reference Citation Analysis (0)] |
| 22. | McElrath MJ, De Rosa SC, Moodie Z, Dubey S, Kierstead L, Janes H, Defawe OD, Carter DK, Hural J, Akondy R. HIV-1 vaccine-induced immunity in the test-of-concept Step Study: a case-cohort analysis. Lancet. 2008;372:1894-1905. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 586] [Cited by in RCA: 582] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 23. | Quirk EK, Mogg R, Brown DD, Lally MA, Mehrotra DV, DiNubile MJ, Robertson MN. HIV seroconversion without infection after receipt of adenovirus-vectored HIV type 1 vaccine. Clin Infect Dis. 2008;47:1593-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 24. | Casimiro DR, Chen L, Fu TM, Evans RK, Caulfield MJ, Davies ME, Tang A, Chen M, Huang L, Harris V. Comparative immunogenicity in rhesus monkeys of DNA plasmid, recombinant vaccinia virus, and replication-defective adenovirus vectors expressing a human immunodeficiency virus type 1 gag gene. J Virol. 2003;77:6305-6313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 348] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 25. | Mastrangelo MJ, Eisenlohr LC, Gomella L, Lattime EC. Poxvirus vectors: orphaned and underappreciated. J Clin Invest. 2000;105:1031-1034. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 26. | Romano G. Current development of adeno-associated viral vectors. Drug News Perspect. 2005;18:311-316. [PubMed] |
| 27. | Falkner FG, Holzer GW. Vaccinia viral/retroviral chimeric vectors. Curr Gene Ther. 2004;4:417-426. [PubMed] |
| 28. | Lachmann R. Herpes simplex virus-based vectors. Int J Exp Pathol. 2004;85:177-190. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 41] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 29. | Zhang XL, Jeza VT, Pan Q. Salmonella typhi: from a human pathogen to a vaccine vector. Cell Mol Immunol. 2008;5:91-97. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 51] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 30. | Ulmer JB, Donnelly JJ, Parker SE, Rhodes GH, Felgner PL, Dwarki VJ, Gromkowski SH, Deck RR, DeWitt CM, Friedman A. Heterologous protection against influenza by injection of DNA encoding a viral protein. Science. 1993;259:1745-1749. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1713] [Cited by in RCA: 1617] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 31. | Capua I, Terregino C, Cattoli G, Mutinelli F, Rodriguez JF. Development of a DIVA (Differentiating Infected from Vaccinated Animals) strategy using a vaccine containing a heterologous neuraminidase for the control of avian influenza. Avian Pathol. 2003;32:47-55. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 145] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 32. | Coelho-Castelo AA, Trombone AP, Rosada RS, Santos RR, Bonato VL, Sartori A, Silva CL. Tissue distribution of a plasmid DNA encoding Hsp65 gene is dependent on the dose administered through intramuscular delivery. Genet Vaccines Ther. 2006;4:1. [PubMed] |
| 33. | Gravier R, Dory D, Laurentie M, Bougeard S, Cariolet R, Jestin A. In vivo tissue distribution and kinetics of a pseudorabies virus plasmid DNA vaccine after intramuscular injection in swine. Vaccine. 2007;25:6930-6938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 34. | Brun A, Bárcena J, Blanco E, Borrego B, Dory D, Escribano JM, Le Gall-Reculé G, Ortego J, Dixon LK. Current strategies for subunit and genetic viral veterinary vaccine development. Virus Res. 2011;157:1-12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 35. | Faurez F, Dory D, Le Moigne V, Gravier R, Jestin A. Biosafety of DNA vaccines: New generation of DNA vectors and current knowledge on the fate of plasmids after injection. Vaccine. 2010;28:3888-3895. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 83] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 36. | Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, Harper LB, Pauley CJ, Niu Z, Denisova L. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11:711-721. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 203] [Cited by in RCA: 231] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 37. | Donnelly JJ, Wahren B, Liu MA. DNA vaccines: progress and challenges. J Immunol. 2005;175:633-639. [PubMed] |
| 38. | van Drunen Littel-van den Hurk S, Babiuk SL, Babiuk LA. Strategies for improved formulation and delivery of DNA vaccines to veterinary target species. Immunol Rev. 2004;199:113-125. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 40] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 39. | McKay PF, Barouch DH, Santra S, Sumida SM, Jackson SS, Gorgone DA, Lifton MA, Letvin NL. Recruitment of different subsets of antigen-presenting cells selectively modulates DNA vaccine-elicited CD4+ and CD8+ T lymphocyte responses. Eur J Immunol. 2004;34:1011-1020. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 68] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 40. | Buonaguro L, Tornesello ML, Buonaguro FM. Virus-like particles as particulate vaccines. Curr HIV Res. 2010;8:299-309. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 44] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 41. | Chackerian B. Virus-like particles: flexible platforms for vaccine development. Expert Rev Vaccines. 2007;6:381-390. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 232] [Cited by in RCA: 253] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 42. | Jennings GT, Bachmann MF. The coming of age of virus-like particle vaccines. Biol Chem. 2008;389:521-536. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 285] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 43. | Ramqvist T, Andreasson K, Dalianis T. Vaccination, immune and gene therapy based on virus-like particles against viral infections and cancer. Expert Opin Biol Ther. 2007;7:997-1007. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 44. | Roy P, Noad R. Virus-like particles as a vaccine delivery system: myths and facts. Adv Exp Med Biol. 2009;655:145-158. [PubMed] |
| 45. | Casal JI. Use of the baculovirus expression system for the generation of virus-like particles. Biotechnol Genet Eng Rev. 2001;18:73-87. [PubMed] |
| 46. | Pumpens P, Grens E. HBV core particles as a carrier for B cell/T cell epitopes. Intervirology. 2001;44:98-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 210] [Cited by in RCA: 216] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 47. | Perrin P, Sureau P, Thibodeau L. Structural and immunogenic characteristics of rabies immunosomes. Dev Biol Stand. 1985;60:483-491. [PubMed] |
| 48. | Cornet B, Vandenbranden M, Cogniaux J, Giurgea L, Dekegel D, Ruysschaert JM. Virosomes reconstituted from human immunodeficiency virus proteins and lipids. Biochem Biophys Res Commun. 1990;167:222-231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 49. | Grimaldi S, Giuliani A, Giuliani A, Ferroni L, Lisi A, Santoro N, Pozzi D. Engineered liposomes and virosomes for delivery of macromolecules. Res Virol. 1995;146:289-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 13] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 50. | Cusi MG. Applications of influenza virosomes as a delivery system. Hum Vaccin. 2006;2:1-7. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 43] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 51. | Bomsel M, Tudor D, Drillet AS, Alfsen A, Ganor Y, Roger MG, Mouz N, Amacker M, Chalifour A, Diomede L. Immunization with HIV-1 gp41 subunit virosomes induces mucosal antibodies protecting nonhuman primates against vaginal SHIV challenges. Immunity. 2011;34:269-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 237] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 52. | Gil F, Titarenko E, Terrada E, Arcalís E, Escribano JM. Successful oral prime-immunization with VP60 from rabbit haemorrhagic disease virus produced in transgenic plants using different fusion strategies. Plant Biotechnol J. 2006;4:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 53. | Gómez N, Wigdorovitz A, Castañón S, Gil F, Ordás R, Borca MV, Escribano JM. Oral immunogenicity of the plant derived spike protein from swine-transmissible gastroenteritis coronavirus. Arch Virol. 2000;145:1725-1732. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 43] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 54. | Arakawa T, Chong DK, Langridge WH. Efficacy of a food plant-based oral cholera toxin B subunit vaccine. Nat Biotechnol. 1998;16:292-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 200] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 55. | Komarova TV, Baschieri S, Donini M, Marusic C, Benvenuto E, Dorokhov YL. Transient expression systems for plant-derived biopharmaceuticals. Expert Rev Vaccines. 2010;9:859-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 119] [Cited by in RCA: 110] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 56. | Buriani G, Mancini C, Benvenuto E, Baschieri S. Plant heat shock protein 70 as carrier for immunization against a plant-expressed reporter antigen. Transgenic Res. 2011;20:331-344. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 57. | Matsuoka H, Kobayashi J, Barker GC, Miura K, Chinzei Y, Miyajima S, Ishii A, Sinden RE. Induction of anti-malarial transmission blocking immunity with a recombinant ookinete surface antigen of Plasmodium berghei produced in silkworm larvae using the baculovirus expression vector system. Vaccine. 1996;14:120-126. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 15] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 58. | Pang AL, Hashimoto CN, Tam LQ, Meng ZQ, Hui GS, Ho WK. In vivo expression and immunological studies of the 42-kilodalton carboxyl-terminal processing fragment of Plasmodium falciparum merozoite surface protein 1 in the baculovirus-silkworm system. Infect Immun. 2002;70:2772-2779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 59. | Pérez-Filgueira DM, Resino-Talaván P, Cubillos C, Angulo I, Barderas MG, Barcena J, Escribano JM. Development of a low-cost, insect larvae-derived recombinant subunit vaccine against RHDV. Virology. 2007;364:422-430. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 73] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 60. | Pérez-Martín E, Gómez-Sebastián S, Argilaguet JM, Sibila M, Fort M, Nofrarías M, Kurtz S, Escribano JM, Segalés J, Rodríguez F. Immunity conferred by an experimental vaccine based on the recombinant PCV2 Cap protein expressed in Trichoplusia ni-larvae. Vaccine. 2010;28:2340-2349. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 61. | Todolí F, Solano-Gallego L, de Juan R, Morell P, Núñez Mdel C, Lasa R, Gómez-Sebastián S, Escribano JM, Alberola J, Rodríguez-Cortés A. Humoral and in vivo cellular immunity against the raw insect-derived recombinant Leishmania infantum antigens KMPII, TRYP, LACK, and papLe22 in dogs from an endemic area. Am J Trop Med Hyg. 2010;83:1287-1294. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 9] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 62. | Folgori A, Tafi R, Meola A, Felici F, Galfré G, Cortese R, Monaci P, Nicosia A. A general strategy to identify mimotopes of pathological antigens using only random peptide libraries and human sera. EMBO J. 1994;13:2236-2243. [PubMed] |
| 63. | Irving MB, Pan O, Scott JK. Random-peptide libraries and antigen-fragment libraries for epitope mapping and the development of vaccines and diagnostics. Curr Opin Chem Biol. 2001;5:314-324. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 134] [Cited by in RCA: 138] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 64. | Chen X, Scala G, Quinto I, Liu W, Chun TW, Justement JS, Cohen OJ, vanCott TC, Iwanicki M, Lewis MG. Protection of rhesus macaques against disease progression from pathogenic SHIV-89.6PD by vaccination with phage-displayed HIV-1 epitopes. Nat Med. 2001;7:1225-1231. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 59] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 65. | Greenwood J, Willis AE, Perham RN. Multiple display of foreign peptides on a filamentous bacteriophage. Peptides from Plasmodium falciparum circumsporozoite protein as antigens. J Mol Biol. 1991;220:821-827. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 214] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 66. | Jelinek R, Terry TD, Gesell JJ, Malik P, Perham RN, Opella SJ. NMR structure of the principal neutralizing determinant of HIV-1 displayed in filamentous bacteriophage coat protein. J Mol Biol. 1997;266:649-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 67. | De Berardinis P, D'Apice L, Prisco A, Ombra MN, Barba P, Del Pozzo G, Petukhov S, Malik P, Perham RN, Guardiola J. Recognition of HIV-derived B and T cell epitopes displayed on filamentous phages. Vaccine. 1999;17:1434-1441. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 35] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 68. | Gaubin M, Fanutti C, Mishal Z, Durrbach A, De Berardinis P, Sartorius R, Del Pozzo G, Guardiola J, Perham RN, Piatier-Tonneau D. Processing of filamentous bacteriophage virions in antigen-presenting cells targets both HLA class I and class II peptide loading compartments. DNA Cell Biol. 2003;22:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 51] [Cited by in RCA: 56] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 69. | De Berardinis P, Sartorius R, Fanutti C, Perham RN, Del Pozzo G, Guardiola J. Phage display of peptide epitopes from HIV-1 elicits strong cytolytic responses. Nat Biotechnol. 2000;18:873-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 77] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 70. | Bonifaz LC, Bonnyay DP, Charalambous A, Darguste DI, Fujii S, Soares H, Brimnes MK, Moltedo B, Moran TM, Steinman RM. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J Exp Med. 2004;199:815-824. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 705] [Cited by in RCA: 718] [Article Influence: 34.2] [Reference Citation Analysis (0)] |
| 71. | Sartorius R, Bettua C, D'Apice L, Caivano A, Trovato M, Russo D, Zanoni I, Granucci F, Mascolo D, Barba P. Vaccination with filamentous bacteriophages targeting DEC-205 induces DC maturation and potent anti-tumor T-cell responses in the absence of adjuvants. Eur J Immunol. 2011;41:2573-2584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 41] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 72. | Domingo GJ, Orru' S, Perham RN. Multiple display of peptides and proteins on a macromolecular scaffold derived from a multienzyme complex. J Mol Biol. 2001;305:259-267. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 56] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 73. | Perham RN. Swinging arms and swinging domains in multifunctional enzymes: catalytic machines for multistep reactions. Annu Rev Biochem. 2000;69:961-1004. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 468] [Article Influence: 19.5] [Reference Citation Analysis (0)] |
| 74. | Lessard IA, Domingo GJ, Borges A, Perham RN. Expression of genes encoding the E2 and E3 components of the Bacillus stearothermophilus pyruvate dehydrogenase complex and the stoichiometry of subunit interaction in assembly in vitro. Eur J Biochem. 1998;258:491-501. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 39] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 75. | Domingo GJ, Caivano A, Sartorius R, Barba P, Bäckström M, Piatier-Tonneau D, Guardiola J, De Berardinis P, Perham RN. Induction of specific T-helper and cytolytic responses to epitopes displayed on a virus-like protein scaffold derived from the pyruvate dehydrogenase multienzyme complex. Vaccine. 2003;21:1502-1509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 24] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 76. | D'Apice L, Sartorius R, Caivano A, Mascolo D, Del Pozzo G, Di Mase DS, Ricca E, Li Pira G, Manca F, Malanga D. Comparative analysis of new innovative vaccine formulations based on the use of procaryotic display systems. Vaccine. 2007;25:1993-2000. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 77. | Dale CJ, Liu XS, De Rose R, Purcell DF, Anderson J, Xu Y, Leggatt GR, Frazer IH, Kent SJ. Chimeric human papilloma virus-simian/human immunodeficiency virus virus-like-particle vaccines: immunogenicity and protective efficacy in macaques. Virology. 2002;301:176-187. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 48] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 78. | Woo WP, Doan T, Herd KA, Netter HJ, Tindle RW. Hepatitis B surface antigen vector delivers protective cytotoxic T-lymphocyte responses to disease-relevant foreign epitopes. J Virol. 2006;80:3975-3984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 44] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 79. | Caivano A, Doria-Rose NA, Buelow B, Sartorius R, Trovato M, D'Apice L, Domingo GJ, Sutton WF, Haigwood NL, De Berardinis P. HIV-1 Gag p17 presented as virus-like particles on the E2 scaffold from Geobacillus stearothermophilus induces sustained humoral and cellular immune responses in the absence of IFNγ production by CD4+ T cells. Virology. 2010;407:296-305. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 80. | Zolla-Pazner S, Cardozo T. Structure-function relationships of HIV-1 envelope sequence-variable regions refocus vaccine design. Nat Rev Immunol. 2010;10:527-535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 126] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 81. | Jaworski JP, Krebs S, Trovato M, Kovarik DN, Brower Z, Sutton WF, Waagmeester G, Sartorius R, D’Apice L, Caivano A. Co-immunization with Multimeric Scaffolds and DNA Rapidly Induces Potent Autologous HIV-1 Neutralizing Antibodies and CD8+ T Cell. PlosOne. 2012;In Press. |
| 82. | Bloom DE, Canning D, Weston M. The value of vaccination. World Economics. 2005;6:15-39. |