Copyright
©The Author(s) 2015.
World J Virology. Aug 12, 2015; 4(3): 219-244
Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.219
Published online Aug 12, 2015. doi: 10.5501/wjv.v4.i3.219
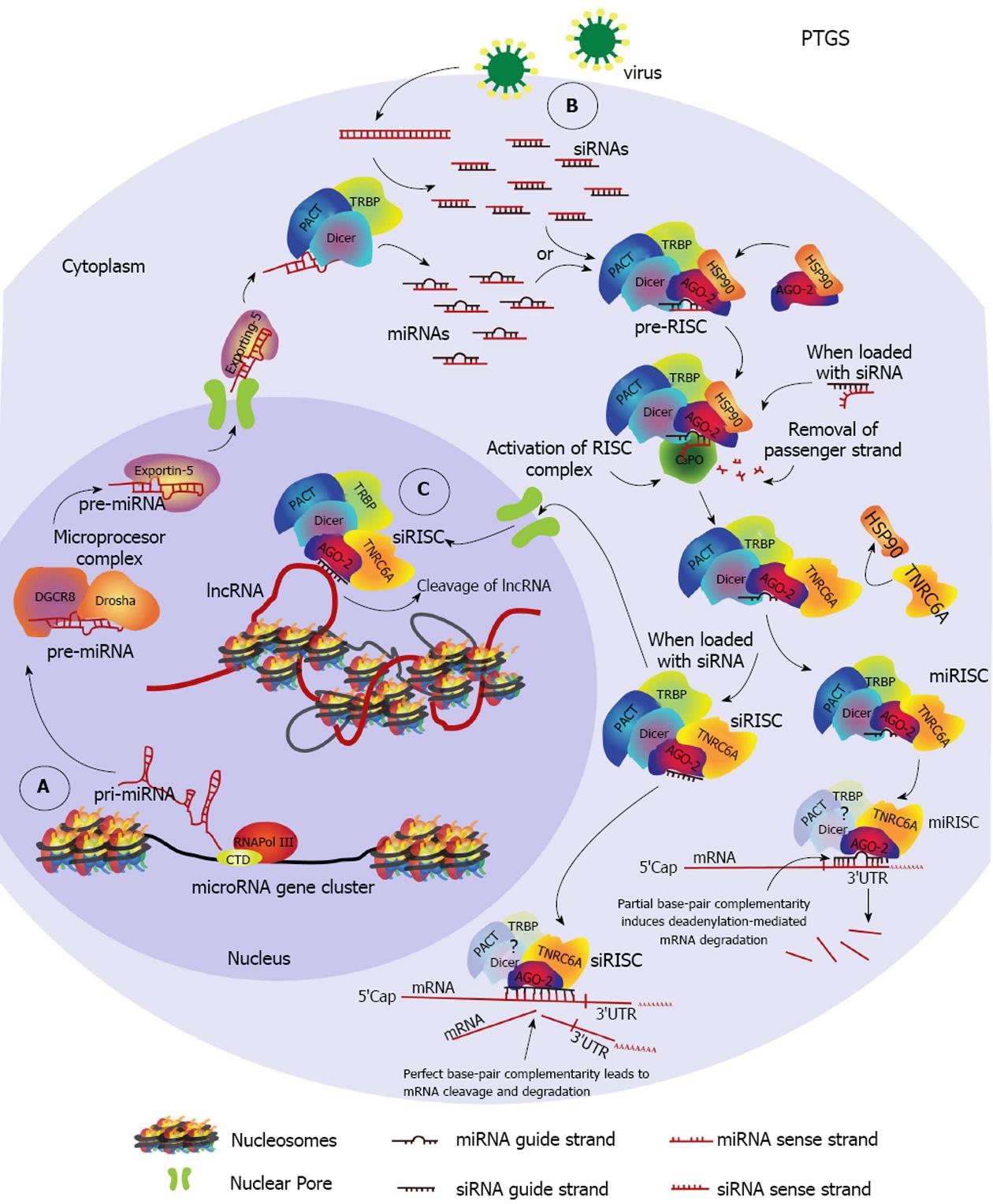
Figure 1 Cytoplasmic and nuclear post-transcriptional gene silencing pathways.
A: A primary-microRNA (pri-miRNA) is transcribed by the RNA Polymerase III (RNA Pol III) from a miRNA gene cluster. The pri-miRNA is then processed by the microprocessor complex into the precursor-miRNA (pre-miRNA) which is exported to the cytoplasm by exportin-5. In the cytoplasm, dicer in complex with Tar-RNA-binding protein (TRBP) and protein kinase R activator of transcription (PACT), process the pre-miRNA into miRNA duplexes. MiRNA duplexes are ioaded into argonaute (AGO) proteins 1-4 with help from heat shock protein 90 (HSP90), forming the miRNA pre-RNA-induced silencing complex (pre-miRISC). The pathway is shown for AGO-2. The pre-RISC complex is activated after removal of the passenger strand from the duplex by C3PO, becoming the miRISC. TNRC6A becomes part of the complex. MiRISC finds a target region within the 3’UTR of an mRNA and induces deadenylation-dependent mRNA degradation; B: During viral infections double-strand RNA (dsRNA) intermediates of viral replication are processed by DICER/TRBP/PACT and are loaded into AGO-2 to form the siRISC complex after removal of the passenger strand. Complete complementarity between the guide strand siRNA and the target region induces cleavage of the targeted mRNA. MiRNAs can also induce mRNA cleavage if this condition is satisfied; C: A nuclear post-transcriptional gene silencing pathway can occur when an activated siRISC is imported into the nucleus and identifies a target within a nuclear RNA such as a Long-non-coding-RNA (lncRNA) resulting in cleavage of the RNA molecule.
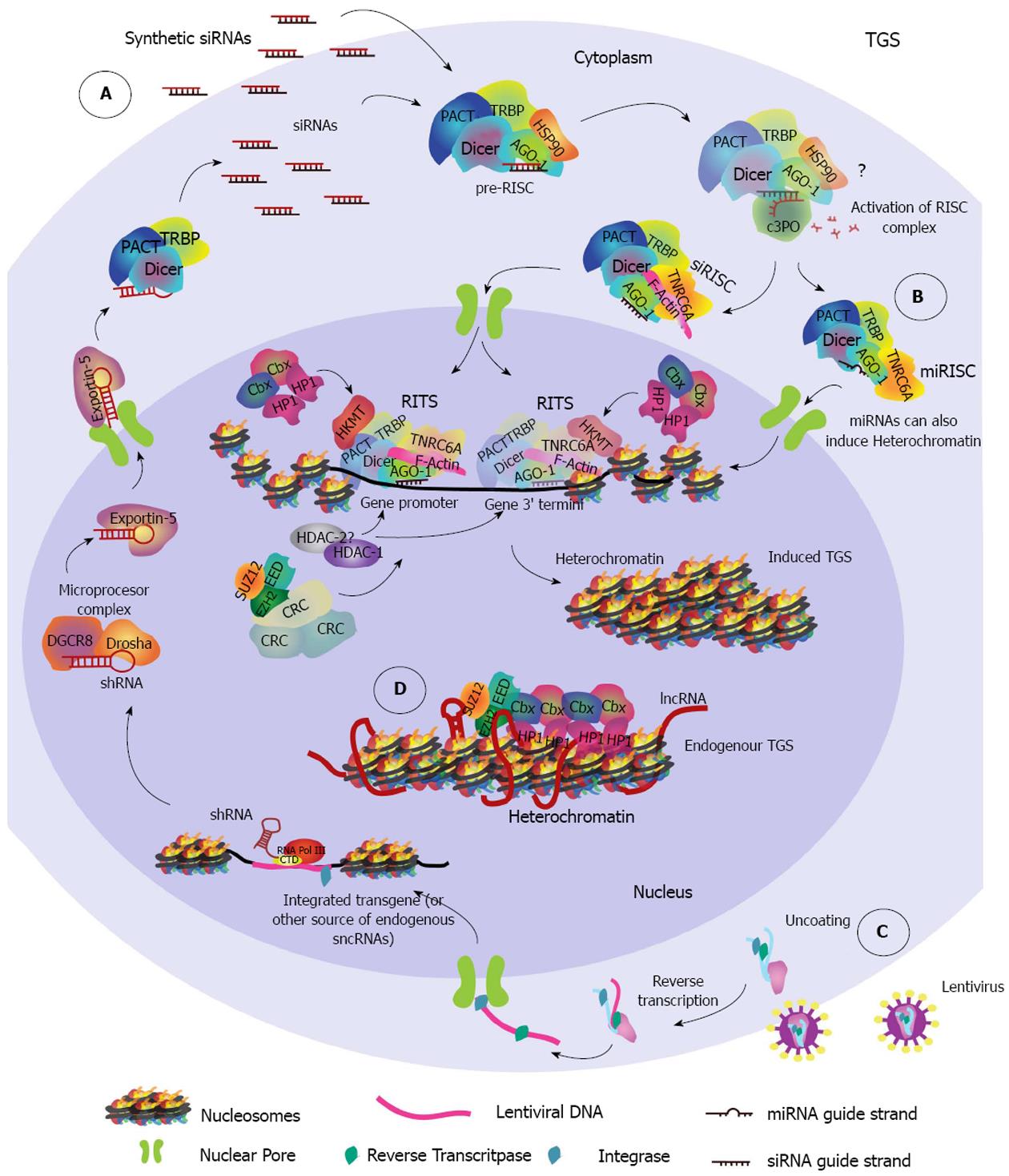
Figure 2 Endogenous and induced transcriptional gene silencing pathways.
A: Synthetic siRNAs that have been designed to target the promoter region or the 3’ end termini of a gene are loaded into AGO-1, forming the pre-RISC complex. It is currently unknown if removal of passenger strand is required, nonetheless if it occurs it probably takes place in the cytoplasm following the same steps as used in PTGS. F-actin participates in the nuclear import of the RISC complex which, once in the nucleus becomes the RITS complex as histone-lysine-methyltransferases (HKMTs) and other epigenetic related proteins such as Histone-deacetylases (HDAC), DNA methyltransferases (DNMTs), Histone Protein 1 (HP1) and others assemble with it. It is unknown whether RISC related proteins remain in the RITS complex. The RITS complex may vary in its composition depending on the chromatin microenvironment and the small non-coding RNA (sncRNA) target region, therefore only some proteins are shown as an example. Establishment of repressive epigenetic marks (not highlighted for simplicity) and further recruitment of chromatin remodeling complexes (CRCs) results in heterochromatin formation and induces transcriptional gene silencing (TGS). Two independent target regions are pictured together to show the different regions of a gene that can be targeted to induce TGS; B: miRISC complexes whose guide strand targets a promoter region may be exported into the nucleus, form a RITS complex and induce TGS in the same way as was described for siRISC complexes; C: Lentiviruses can be used to drive transgene integration of a DNA cassette designed to express a shRNA that induces TGS. ShRNAs are transcribed by RNA Pol III and are processed through the microRNA pathway. In the cytoplasm they are converted to siRNA duplexes that are loaded into RISC complexes and follow the same import pathway to induce TGS as explained in A; D: Endogenous transcriptional gene silencing is induced by a long-non-coding RNA (lncRNA) whose secondary structure is recognized by members of the Polycomb Group repressive complex 1 (PRC1), such as enhancer of zeste 2 (EZH2), embryonic ectoderm development (EED) and suppressor of zeste 12 (SUZ12). The interaction recruits HP1 and other proteins of PRC1 complexes like the (Chromobox) Cbx family that contain a chromodomain able to induce heterochromatin formation.
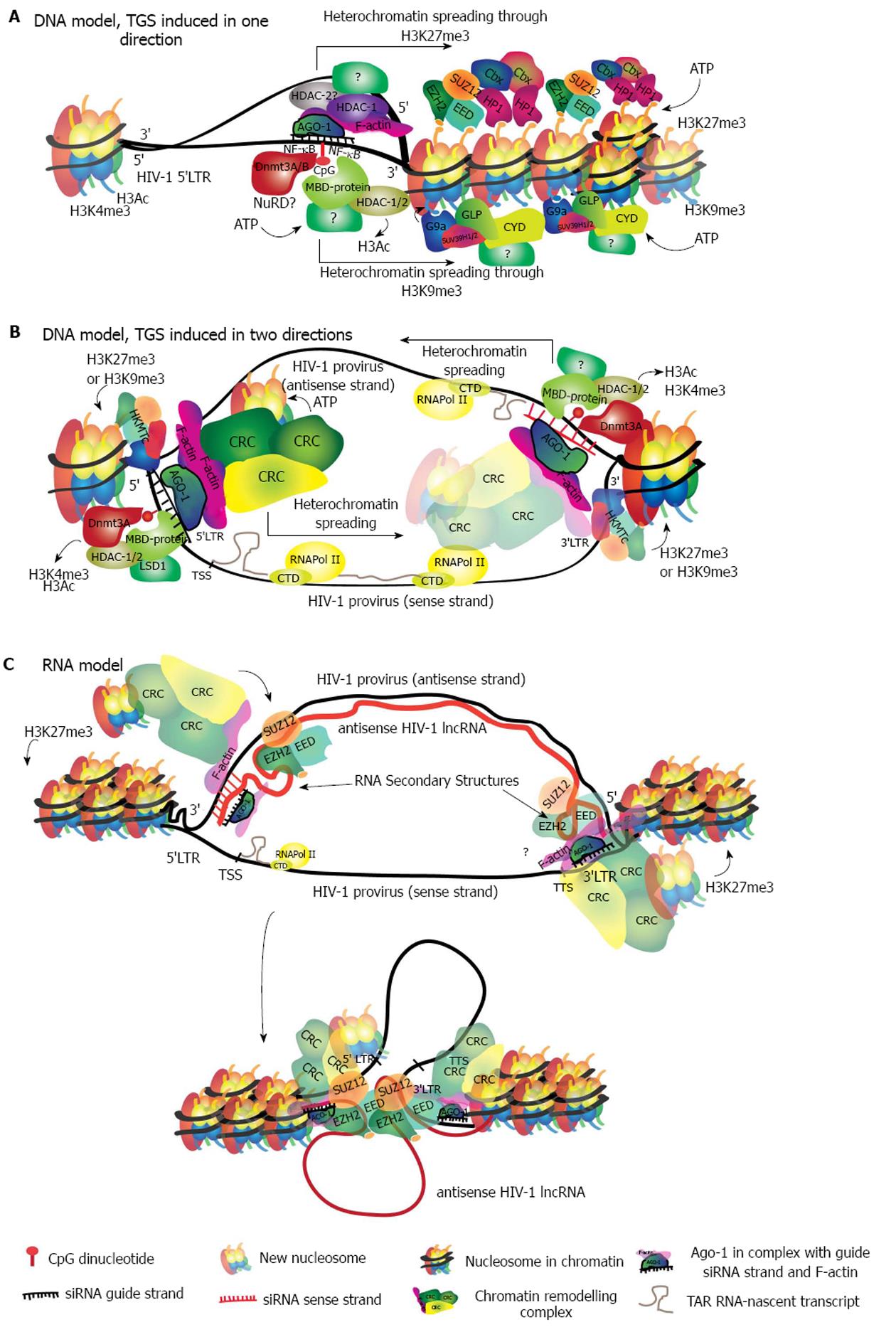
Figure 3 Models describing possible molecular mechanisms of siRNA-induced transcriptional gene silencing in human immunodeficiency virus 1.
A: DNA model in which the siRNA guide-strand finds its target in the 5’LTR promoter of the HIV-1 genome binding directly to the DNA. This binding triggers the recruitment of HDACs and HKMTs, which further recruit CRCs to induce chromatin compaction. Two mutually exclusive pathways are shown simultaneously, for simplicity. While both pathways may be initiated with the DNA methylation of CpG dinucleotides, they differ in the proteins that are recruited to the locus. The pathway characterized by H3K27me3 is shown above and involves initial recruitment of PCR2 (EZH2-SUZ12-EED) followed by the specific CRCs readers of H3K27me3. The H3K9me3 recruits G9a or SUV39H1/2 followed by specific CRCs as well. In this model heterochromatin is likely to spread in only one direction; B: In this DNA model, both strands of the siRNA duplex find a target on opposite DNA strands given that both, the 5’LTR and 3’LTR from the HIV-1 genome, have the same sequence. Regardless of the epigenetic pathway that is induced, heterochromatin will spread in the 5’ to 3’ direction from each end of the HIV-1 genome; C: In the RNA binding model, antisense transcription generates a HIV-1 specific lncRNA that covers all the HIV-1 genome. The siRNA guide-strand will bind the 3’UTR of this transcript, which corresponds to the 5’LTR sequence. The binding recruits PCR2, which establishes H3K27me3 and may also interact with the secondary structures of the lncRNA. Higher order interactions may bring together the 3’end of the HIV-1 genome, recruiting CRCs and inducing heterochromatin. A binding site for the same siRNA strand remains in the DNA sense strand at the 3’LTR, which could potentially contribute to heterochromatin formation. HIV-1: Human immunodeficiency virus 1; DNMT3 A/B: DNA methyl-transferase A/B; HDAC: Histone deacetylase; MBD-protein: Methy-CpG-binding protein; Cbx: Chromobox family; HKMT: Histone lysine (K) methyl-transferase; CRC: Chromatin remodelling complex.
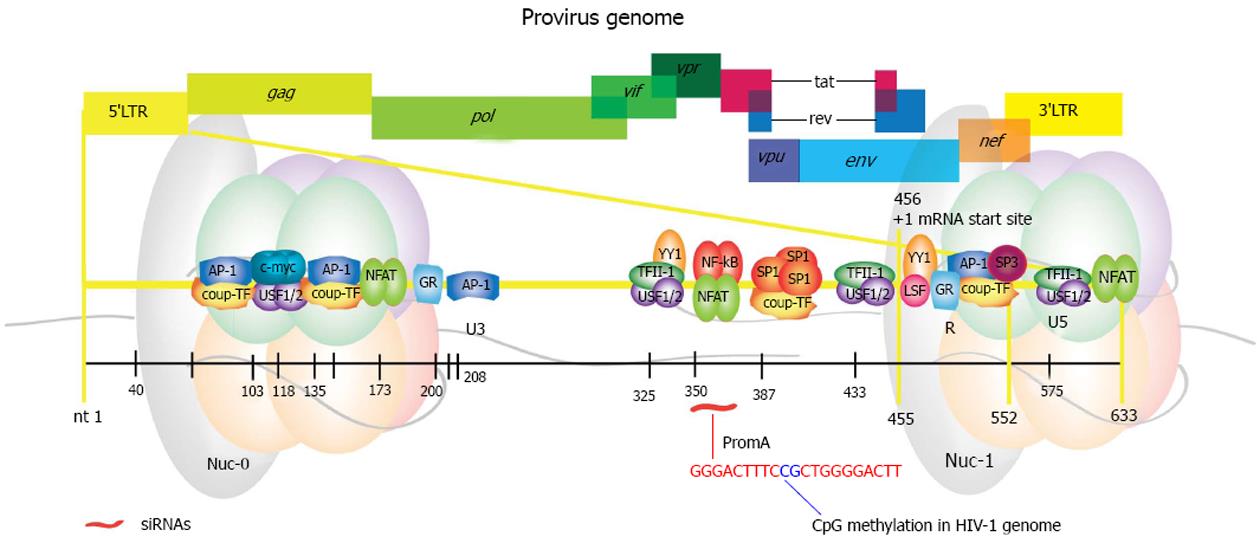
Figure 4 Map of the human immunodeficiency virus 1 genome showing in magnification the 5’LTR region with the location of transcription factor binding sites.
The specific coordinates within the HIV-1 genome for each of the shown DNA regulatory elements is listed on Table 1. 5’LTR: 5’ long terminal repeat; gag: Group specific antigen; pol: Polymerase; vif: Viral infectivity factor; vpr: Viral protein R; vpu: Viral protein unique; tat: Trans-activator of transcription; rev: RNA export element; env: Envelope; nef: Negative factor; 3’LTR: 3’ long terminal repeat; Ap-1: Activator protein 1; COUP-TF: Chicken ovalbumin upstream transcription factor; c-myc: V-myc avian myelocytomatosis viral oncogene homolog; USF 1/2: Upstream stimulatory factor 1 or 2; NFAT: Nuclear factor activated T cells; GR: Glucocorticoid receptor responsive element; YY1: Ying-yang 1; TFII-I: Transcription factor II-I; NF-κB: Nuclear factor κ beta; SP: Specificity protein; LSF: Late SV40 factor; U3: Untranslated region 3; R: R region; U5: Untranslated region 5; Nuc-0: Nuclesome 0; Nuc-1: Nucleosome 1.
- Citation: Méndez C, Ahlenstiel CL, Kelleher AD. Post-transcriptional gene silencing, transcriptional gene silencing and human immunodeficiency virus. World J Virology 2015; 4(3): 219-244
- URL: https://www.wjgnet.com/2220-3249/full/v4/i3/219.htm
- DOI: https://dx.doi.org/10.5501/wjv.v4.i3.219