Copyright
©2013 Baishideng.
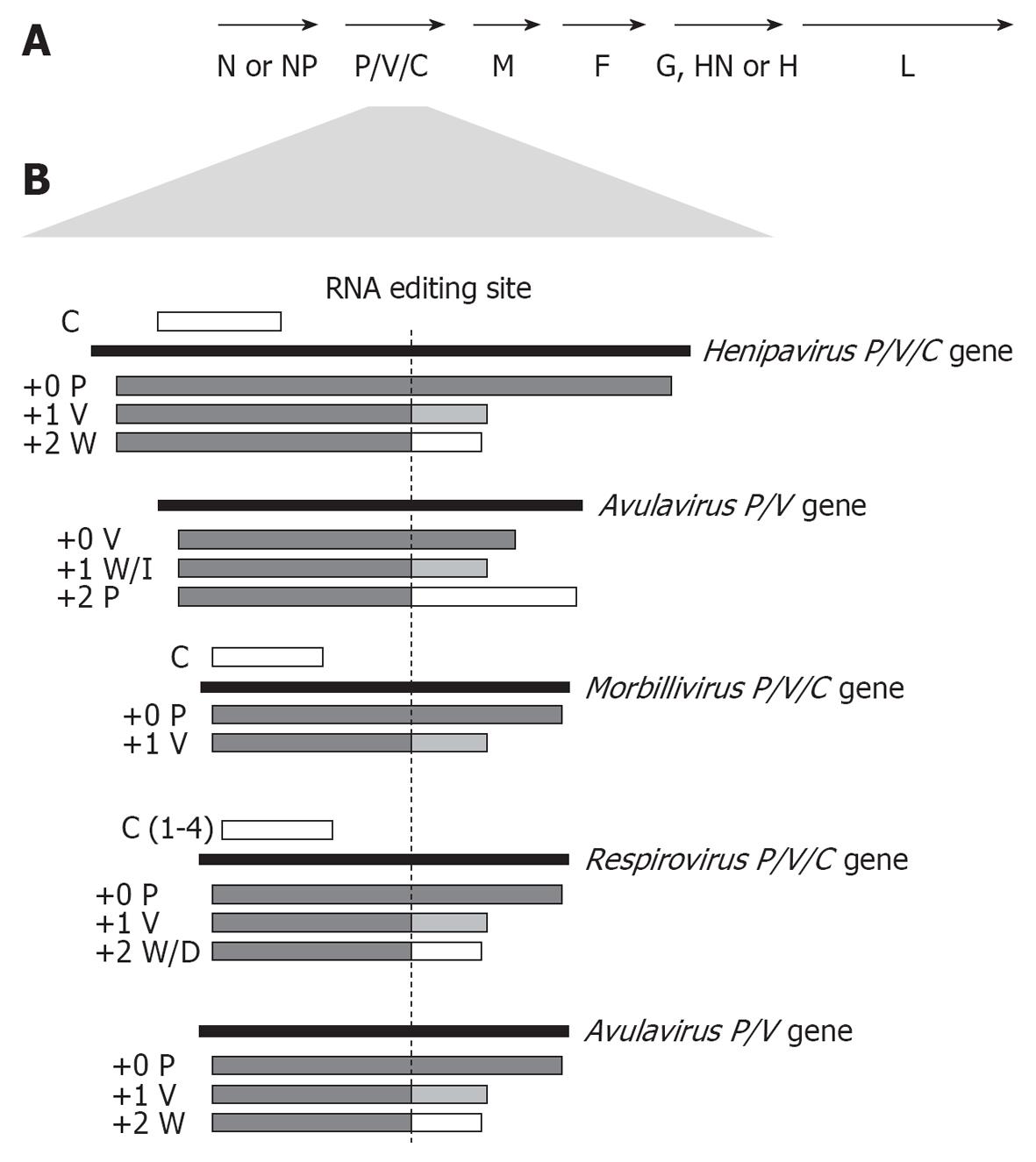
Figure 1 Coding strategies of paramyxovirus P genes.
A: Genome organisation of the Paramyxovirinae subfamily; B: Paramyxoviruses express multiple proteins from the P gene through RNA editing to insert additional non-coded G nucleotides into P gene transcripts at the editing site (indicated), causing a frameshift in the downstream open reading frame (ORF) to generate distinct C-termini. Editing strategies of the 5 best-studied genera are shown, with proteins produced from unedited (+0), or edited (+1 or +2 frameshift) mRNA indicated below the P gene. Several members of the henipavirus, respirovirus and morbillivirus genera, but not the rubulaviruses or avulaviruses, produce one or more C proteins by translation from internal start codon(s) in alternate ORF(s) (indicated as a white bar above the P gene).
Figure 2 Conserved residues in the paramyxovirus V C-terminal domain.
Paramyxovirus V protein C-terminal sequences are aligned with identical and similar residues highlighted. Asterisks indicate absolutely conserved histidine and cysteine residues involved in zinc-binding (see text for details). Residue numbers are indicated in the sequence titles. MuV: Mumps virus; PIV5: Parainfluenza virus 5; hPIV: Human PIV; MPRV: Mapuera virus; PoRV: Porcine rubulavirus; SeV: Sendai virus; MeV: Measles virus; CDV: Canine distemper virus; RPV: Rinderpest virus; PDV: Phocine distemper virus; NDV: Newcastle disease virus; APMV2: Avian paramyxovirus 2; HeV: Hendra virus; NiV: Nipah virus; ASPV: Atlantic Salmon Paramyxovirus; FDLV: Fer-de-Lance virus; SalV: Salem virus; MoV: Mossman virus; MenV: Menangle virus; PPV-1: Pigeon paramyxovirus 1; BeV: Beilong virus; J-V: J-virus; TioV: Tioman virus; NarPV: Nariva virus.
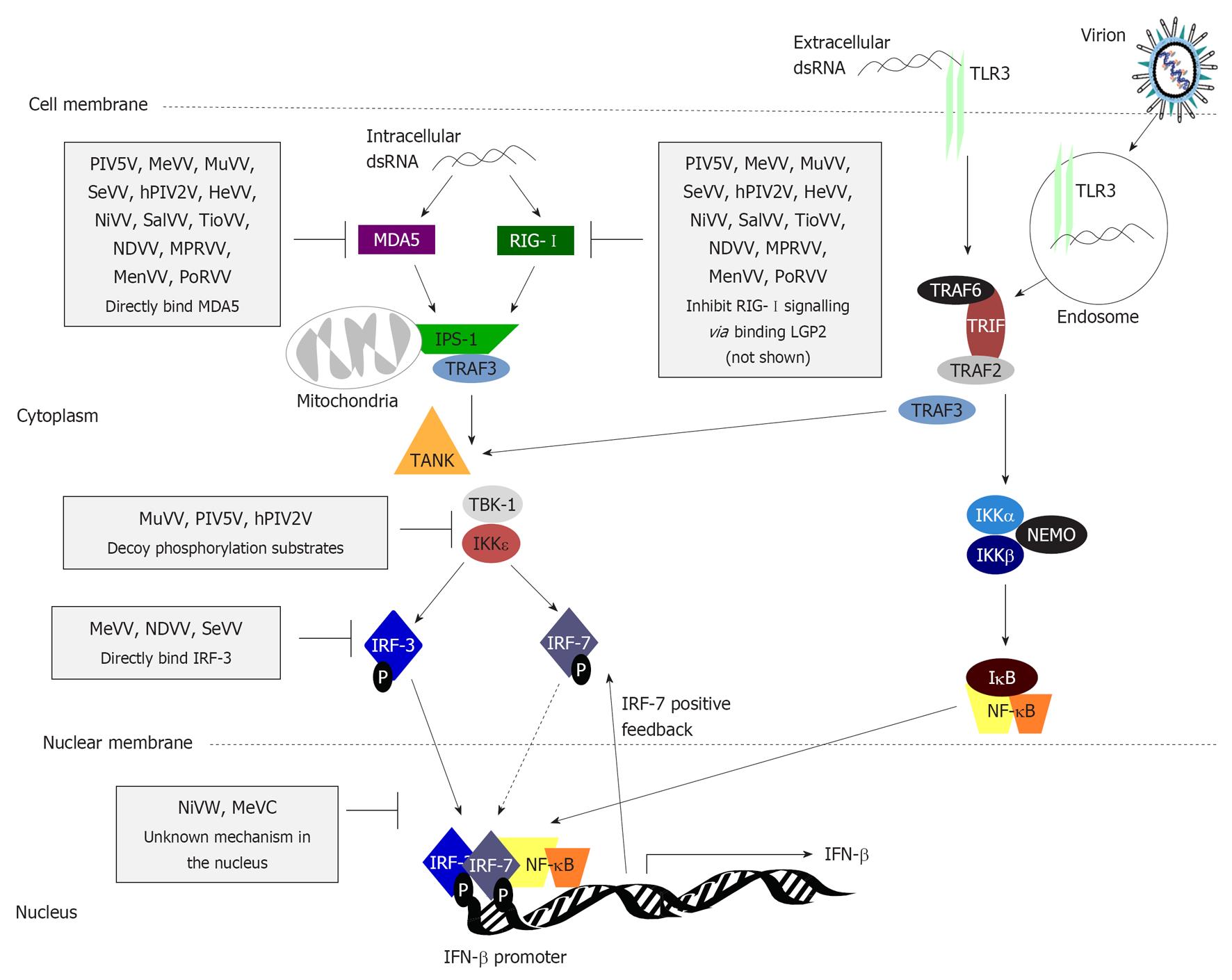
Figure 3 Type I interferon induction is inhibited by paramyxovirus interferon-antagonist proteins at multiple stages.
Pathogen-associated molecular patterns (PAMPs) generated during virus infection, such as dsRNA, are recognised by PRRs including endosomal/surface expressed Toll-like receptor 3 (TLR3) and cytoplasmic retinoic acid-inducible gene-I (RIG-I)/melanoma differentiation associated protein 5 (MDA5). TLR3 signals through the adaptor molecule Toll/interleukin-1 receptor domain-containing adaptor inducing interferon (IFN)β (TRIF), which recruits tumor necrosis factor receptor-associated factor (TRAF)2 to activate the inhibitor of nuclear factor κB (NF-κB) kinase (IKK)α/β kinases to phosphorylate inhibitory inhibitor of NF-κB (IκB), triggering its degradation and activation/nuclear translocation of NF-κB. TLR3 signalling via TRAF3 results in phosphorylation/activation of IFN regulatory factor (IRF)-3, causing its homodimerisation, or heterodimerisation with IRF-7 in professional IFN producing/IFN-primed cells, and translocation into the nucleus where, with NF-κB and activating transcription factor 2 (ATF2)/c-jun (not shown), it activates early type I IFN transcription. RIG-I and MDA5 also induce phosphorylation of IRF-3 following recognition of cytoplasmic PAMPs in infected cells via interaction with the mitochondrial membrane protein IFNβ promoter stimulator 1 (IPS-1), which recruits and activates TANK and the TANK-binding kinases (TBKs). TBK-1 and IKKε via the E3 ubiquitin ligase TRAF3. Many paramyxoviruses target this pathway; steps commonly targeted are indicated (black bars) with specific examples of the paramyxovirus proteins responsible (see text for details). DC: Dendritic cell; PIV5: Parainfluenza virus 5; MeV: Measles virus; MuV: Mumps virus; SeV: Sendai virus; hPIV: Human PIV; HeV: Hendra virus; NiV: Nipah virus; SalV: Salem virus; TioV: Tioman virus; NDV: Newcastle disease virus; MPRV: Mapuera virus; MenV: Menangle virus; PoRV: Porcine rubulavirus; LGP2: Laboratory of genetics and physiology 2.
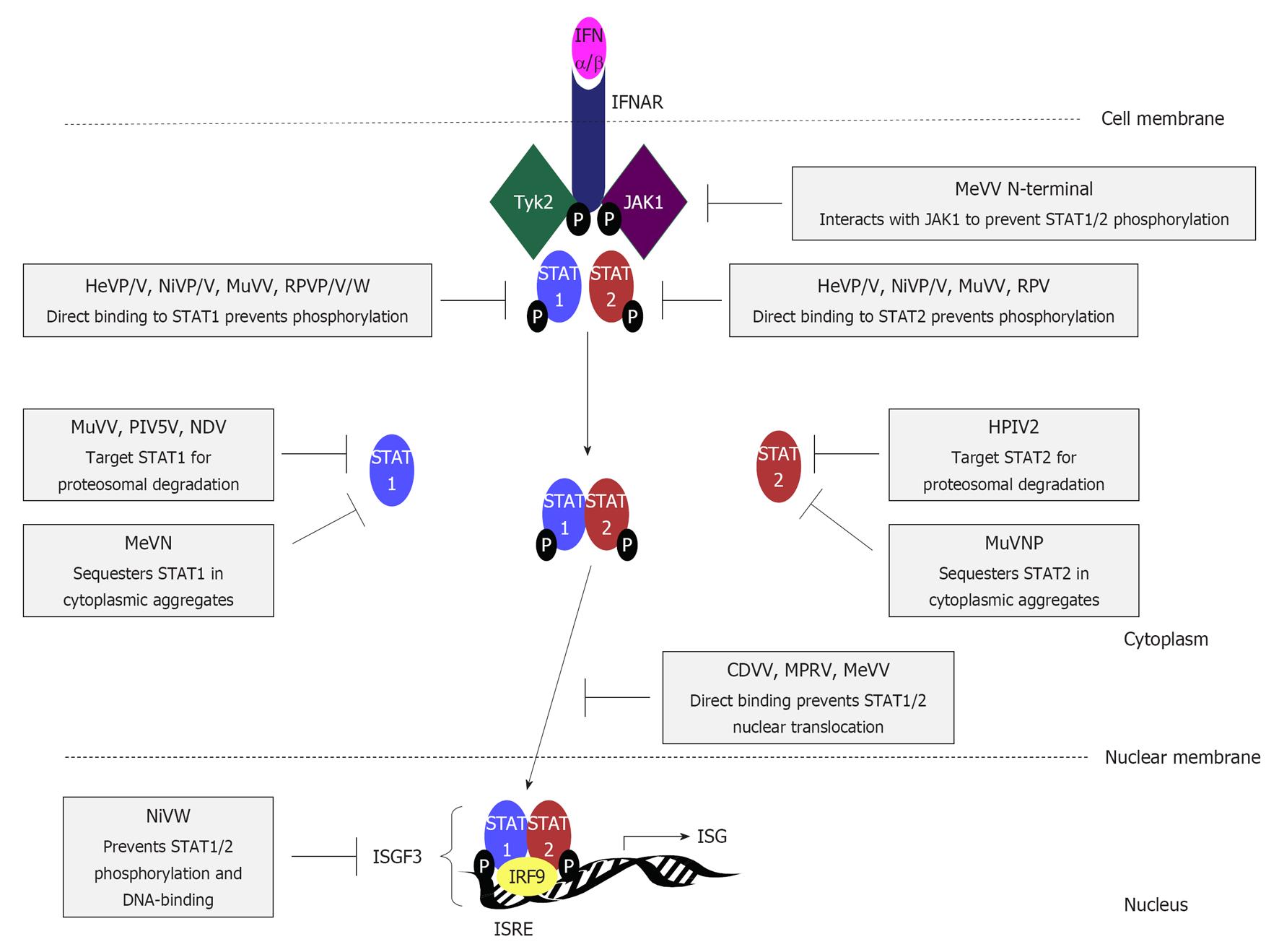
Figure 4 Interferon signalling pathways are targeted by paramyxovirus interferon-antagonist proteins through diverse mechanisms.
Interferon (IFN)β binds to type I IFN receptor subunits IFNα/β receptor (IFNAR)1 and IFNAR2, causing dimerization, activation and phosphorylation of the receptor-associated kinases Janus kinase 1 (JAK1) and tyrosine kinase 2 (Tyk2), to create docking sites for the SH2 domains of signal transducers and activators of transcription (STAT)1 and 2. STAT1 and 2 are phosphorylated by Tyk2 and JAK1 respectively, and form a heterodimer that translocates into the nucleus, forming the heterotrimeric transcription factor complex IFN-stimulated gene factor 3 (ISGF3) with IFN regulatory factor (IRF)-9. ISGF3 binding to IFN stimulatory response element (ISRE) sequences in the promoters of hundreds of IFN-stimulated genes (ISGs) activates the transcription of antiviral and immune-modulatory proteins to establish an antiviral state in infected and neighbouring cells, and contribute to shaping the adaptive immune response. STAT1 and/or STAT2 are targeted by almost all paramyxoviruses through the activity of several IFN antagonists by mechanisms that are reported to differ significantly; mechanisms and specific viral proteins responsible are indicated (see text for details). HeV: Hendra virus; NiV: Nipah virus; MuV: Mumps virus; RPV: Rinderpest virus; MeV: Measles virus; PIV5: Parainfluenza virus 5; NDV: Newcastle disease virus; hPIV: Human PIV; CDV: Canine distemper virus; MPRV: Mapuera virus.
- Citation: Audsley MD, Moseley GW. Paramyxovirus evasion of innate immunity: Diverse strategies for common targets. World J Virol 2013; 2(2): 57-70
- URL: https://www.wjgnet.com/2220-3249/full/v2/i2/57.htm
- DOI: https://dx.doi.org/10.5501/wjv.v2.i2.57