Copyright
©The Author(s) 2021.
World J Virol. Sep 25, 2021; 10(5): 256-263
Published online Sep 25, 2021. doi: 10.5501/wjv.v10.i5.256
Published online Sep 25, 2021. doi: 10.5501/wjv.v10.i5.256
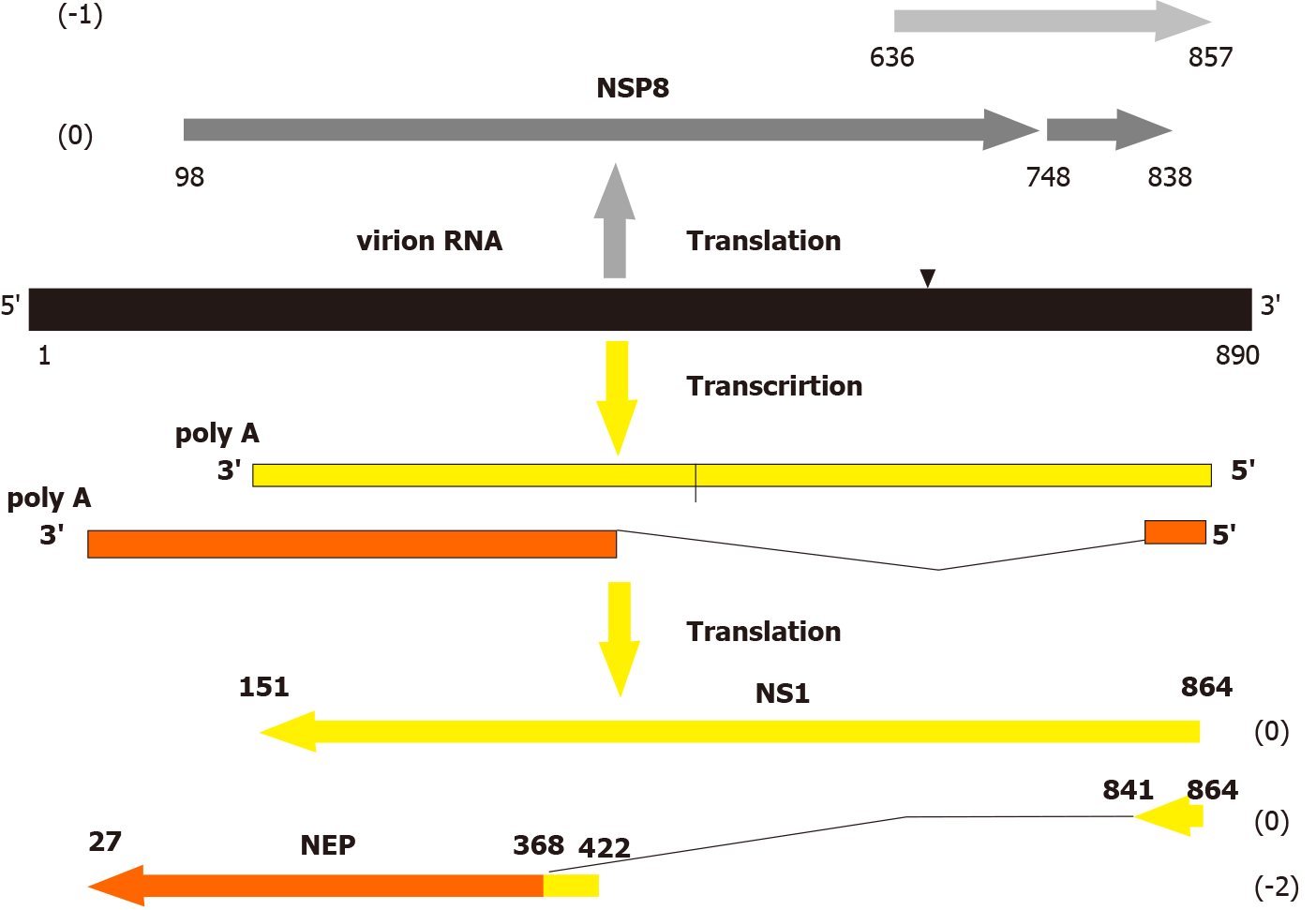
Figure 1 The scheme of expression of the genome negative sense segment of influenza A virus.
The negative sense (NS) segment of influenza A/Aichi/2/68 (H3N2) virus is displayed. The horizontal arrows show the open reading frames (ORFs) of the negative strand protein 8, non-structural anti-interferon protein (NS1), and nuclear export protein (NEP). Numbers in brackets indicate the ORF translation phase. Numbers under the lines indicate nucleotide positions from the 5’ end of the virion genome RNA. The broken line shows the splicing segment of the NEP gene mRNA. Triangle in the virion RNA molecule shows a site position of possible translation frameshifting[10]. NS: Negative sense; NSP8: Negative strand protein 8; NS1: non-structural anti-interferon protein.
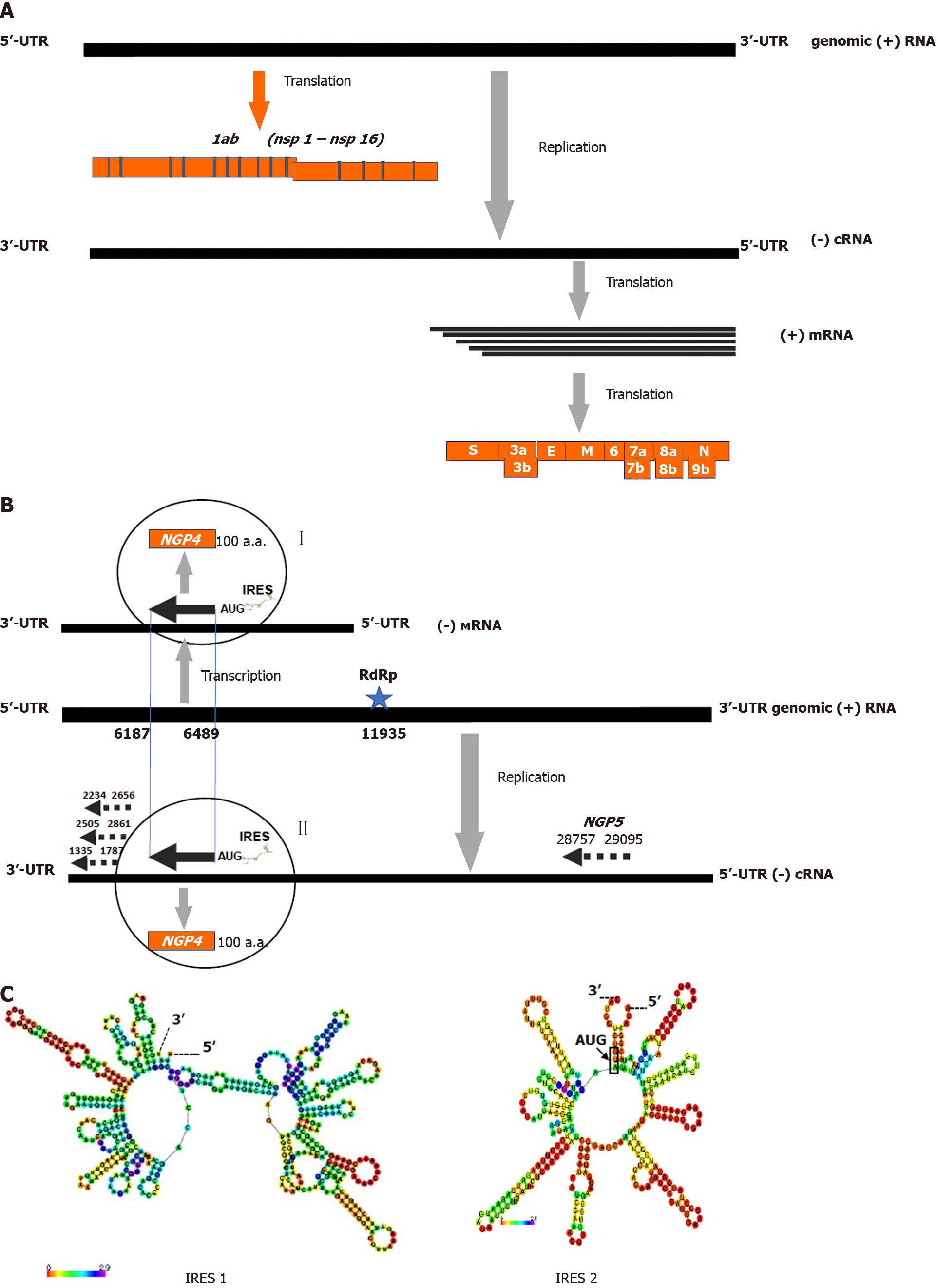
Figure 2 Positive sense genome strategy and translation cassette unit at the 3’ end of the negative sense complimentary RNA of coronavirus severe acute respiratory syndrome coronavirus 2 genome.
A: Replication scheme of the RNA genome of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) coronavirus (ac.n. MT890462.1). UTR means untranslated RNA region; B: A 3’ end area of the subgenomic (-)cRNA complimentary to the virus genome 5’ end (+)vRNA of SARS-CoV-2 (ac.n. MT635445.1) is displayed. Five ORF containing cassette for NGP1-NGP5 beginning either with classical AUG (NGP4) or noncanonical CUG (NGP1-3, NGP5) codons are shown by arrows. Nucleotides counting from the 5’ end of (+)vRNA are shown for each ORFs. Phases of the translation frame (fr) are estimated regarding the frame of NGP4 (fr.0) as follows: NGP1and 2 (fr. +1), NGP3 (fr.0). Poly A tract (11935-11940 nt) functioning as a viral RNA dependent RNA polymerase binding site is shown by star; C: IRES-like structures enriched with 16 and 10 canonical “hair-pins” RNA elements in the regions 8100-8599 nt (IRES 1) and 6488-6792 nt (IRES 2), respectively, were predicted by the IRESpred program[22]. The IRES-like structures 1 and 2 have significant free energy value as low as -99,4 and -73,8 kkal/mol, respectively. The data were partially presented in[15]. These partial elements were used here with the Publisher’s permission.
- Citation: Zhirnov O. Ambisense polarity of genome RNA of orthomyxoviruses and coronaviruses. World J Virol 2021; 10(5): 256-263
- URL: https://www.wjgnet.com/2220-3249/full/v10/i5/256.htm
- DOI: https://dx.doi.org/10.5501/wjv.v10.i5.256