INTRODUCTION
It is widely accepted that many diseases, in particular disorders of the Central Nervous System are multifactorial of origin. Consequently, reasonable pharmacotherapies should aim at addressing those multiple factors causing and sustaining the diseases, simultaneously. However, only recently strategies of “polypharmacology” (one drug adressing several targets) have appeared in research and in the literature. Apparently, the drug industry has relied for a long time on a small number of targets that had already been validated, subsequently generating an abundance of “follow-on” drugs. Statistical analyses of large component size, degree distribution and clustering coefficient quantitatively confirm this bias. This bias has been reinforced by the conviction, that the “major” targets known for most mental disorders were sufficient for the development of efficient pharmacotherapies. However, with the advent of modern sequencing technologies facilitating the sequencing of whole genomes within reasonable times and declining costs, intensive efforts to search for more targets, preferably on the gene level, have gained new steam.
GENOME-WIDE ASSOCIATION STUDIES
The idea behind genome-wide association studies (GWAS) was to identify all genetic changes eventually causing a disease. Until very recently, however, browsings of whole genomes for sites of associations with a mental disorder have not met with the expectations, because they did not stand up to rigorous statistical analysis or were not sufficiently specific for only one mental illness[1]. Moreover, it has to be mentioned, that these tremendous efforts have been burdened by the fact, that an estimated 99% of all single nucleotide polymorphisms (SNPs) are benign and without adverse effects. Nevertheless, it has to be acknowledged that these are unbiased approaches with the potential to discover new pharmacological targets.
Many so-called “candidates” have surfaced in schizophrenia (SCZ) research (such as COMT, NRG, dysbindin), but for instance in a larger European ancestry sample, where 14 candidate genes had been detected[2], associations were equivocal. In another more recent study on psychosis, the authors came to the conclusion, that “No individual SNP showed compelling evidence for association with psychosis”[3]. Furthermore, in a recent study on anorexia nervosa (AN) encompassing 5551 AN cases and 21080 controls, there were “No findings that reached genome-wide significance”, concluding that the sample, the largest yet reported for this disorder, was underpowered for their detection[4]. Apparently, these statistical hurdles were overcome by the efforts of the Psychiatric Genetics Consortium, resulting in the identification of 22 loci significant for association to SCZ, and considered to be statistically sufficiently robust[5]. It was estimated that more than 8000 SNPs independently contribute to SCZ, which confirms that SCZ is a highly polygenic disorder[6]. A subsequent analysis by the same consortium encompassing very large samples (approximately 37000 cases and 113000 controls) revealed 128 independent associations spanning 108 defined loci (including at least 600 genes) that are genome-wide significant for SCZ[7].
In an attempt to assign a physiological meaning to some genes belonging to those loci, genes linked to voltage-gated calcium channel subunits (CACNA1C, CACNB2, and CACNA1I), involved in glutamatergic neurotransmission and synaptic plasticity (GRM3, GRIN2A, SRR, GRIA1) and the dopamine D2 receptor gene (DRD2), “the target of all effective anti-psychotic drugs” have been discussed. Apart from doubts raised recently that the symptoms of psychosis or SCZ are caused by the over activity of dopamine[8], hypothesis-free investigations should rather serve to broaden our view of the disorder by searching for additional genes of interest not belonging to the main stream of thinking. Dopamine and glutamate neurotransmission appear to be involved in too many essential brain functions as to be suitable for drug therapies specific for SCZ. Another very popular category of genes identified in the GWAS loci were genes with important functions in the immune system, nourishing the belief brought up in quite some publications before, that SCZ is an autoimmune disorder[9]. However, the data published in this respect are weak and controversial, so that repetition of this hypothesis does not render it more likely. Moreover, a recently published investigation on SCZ and multiple sclerosis (MS) samples[10], focusing exactly on risk alleles of the HLA group of genes, reports on associations of these genes with both SCZ and MS, but with an opposite directionality of effect of associated HLA alleles (that is, MS risk alleles were associated with decreased SCZ risk). Our studies may also be in this line of results, revealing that many more genes of the immune system were downregulated in post-mortem brains of SCZ patients, than upregulated. However, more importantly, as pointed out in our paper, many of those gene products may have functions in the Central Nervous System distinct from their immunological functions[11]. This notion had already been raised before by other groups who discovered the involvement of MHC genes in signal transduction of glutamate receptors and synaptic plasticity[12,13], which attributed a feature to MHC distinct from its immune function in the aetiology of SCZ[14].
Since all these recent studies required enormous logistic coordination and substantial financial support, they deserve to be scrutinized for their cost-benefit ratio in more detail.
As already alluded to above, associations have been found with genomic regions (“loci”), not with single genes. Genomic loci typically encompass more than one gene, so that it remains elusive, which gene is affected. Moreover, a SNP may or may not influence expression of genes within - or outside - the locus. And SNPs may or may not interfere with non-protein-coding genes (or unknown genes), or with annotated protein-coding genes[15] .
Very unlikely the identified SNP is the causative SNP, but only tags the stretch of DNA where the causal variant is located. Furthermore, all the SCZ -associated SNPs show up in non-coding regions of DNA (e.g., intergenic, or intronic) or are synonymous exonic polymorphisms[16]. And above all, it has been underscored in a recent publication, that within approximately 7300 GWAS associations to common diseases or traits, only 20 could be clearly attributed to a causal variant[17].
Typically (and expectedly) the odds ratios associated with each SCZ risk SNP are around 1.10, indicating a very small effect on disease risk.
Some of the SNPs are not specific for the disorder, but also show associations e.g. with bipolar disorder or, to a lesser extent, with major depression, with attention-deficit hyperactivity disorder, and with autism (genetic pleiotropy)[18,19]. Hence, the clinical overlap between these disorders may arise to some extent from a shared genetic predisposition.
It is well known that SCZ is a broad spectrum psychiatric disorder which poses substantial problems of reliabilities of inclusion/exclusion criteria of patients according to diagnostic manuals ICD-10 or DSM IV, manuals that have been repeatedly critisized and are in the course of being thoroughly revisited[20-22]. In this light, the extensive increase of n-numbers in GWAS is bound to increase the probability of irreproducibility of results. Therefore, it remains at least controversial, to what extent further increases in sample sizes will improve our understanding of the disorder[23,24].
It is still surprising that even from apparently robust “candidate gene” approaches of the pre-GWAS era[25,26], almost all GWAS studies, including Ripke et al[5] were unable to replicate any of those genes. For these reasons, it is still a matter of debate to what extent genome-wide statistical significance can be reliably used to decide which gene is a risk gene or not[27].
Only a minor part of the heritability of SCZ can be explained by the existing results[28]. As concerns the major part, one assumption implies, that rather than simply arising from cumulative effects of multiple independent genes, gene-gene interactions (epistasis) may have higher impacts on the genetic risk to develop the disorder[29].
There are preliminary clinical[29,30], and experimental data[31,32] supporting a role for epistasis in SCZ. Unfortunately, systematic investigations on epistatic events in SCZ are not available, and GWAS studies do not help that matter[33].
Conversely, the majority of genetic heritability cannot be explained by GWAS studies, neither individually nor collectively[34,35]. And for most of the associated variants no functional links have been provided[36].
Therefore, GWAS results are of limited value for closer insights into molecular mechanisms of the identified loci/genes, and far away from applications in translational medicine or pharmacotherapeutic interventions. A very important, additional point to be made here is that all those studies depend on statistical significant results, which require prohibitively high numbers of samples due to multiple testing and other statistical problems, which in turn increase the “noise” level churning up the spiral more and more. On the other hand, gene modifications that do not result in any statistical significances because of their subtle effects, may synergize metabolically on the gene product level with each other or with additional genes not modified to exert significant contributions to the development of the disorder, meaning that not only genes reaching statistical significance are important. Very likely, there is much more passing undetected in the “noise” (see below, summary). Along these lines, it has to be assumed, that variation anywhere in the genome affects every character. In keeping with this, the notion was put forward, that there are “no special genes for psychosis”. Instead, the “normal gene” model proposes that any gene or allele that influences the development of the human brain can tentatively act as vulnerability gene or allele, as well[37].
The disappointing failure to identify even one etiological candidate gene during many years of genetic studies on psychosis may be explained by the possibility that genetic vulnerability to psychosis is due to random mutations. Therefore, given the hypothesis that gene alleles that are endowed with general functions in brain development could also act as vulnerability gene alleles, it appears plausible that there is no need to postulate the existence of specific “psychosis genes or polymorphisms”.
FROM GENETICS TO EPIGENETICS
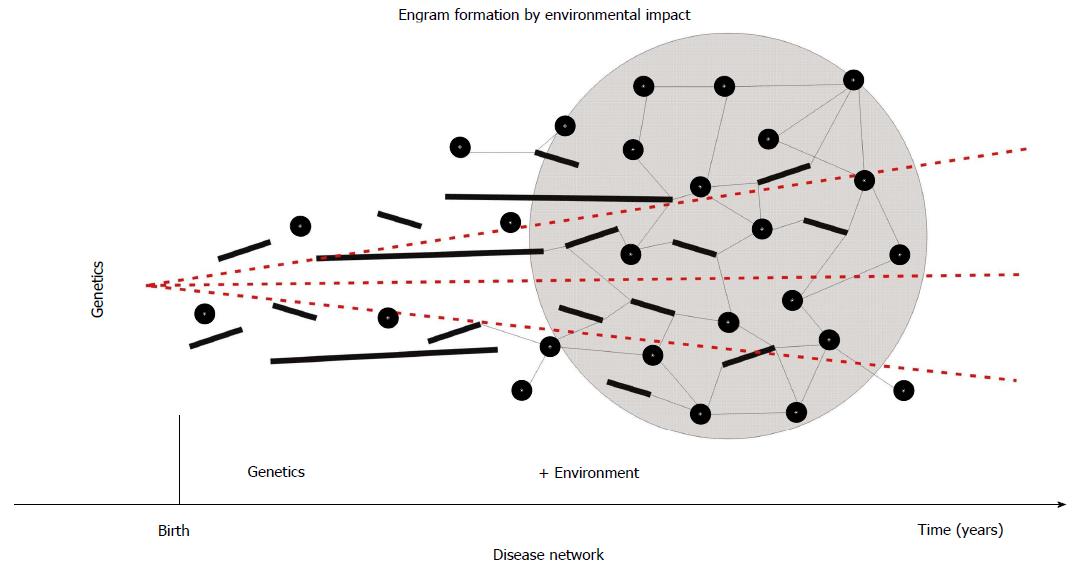
Moreover, epigenetic modifications of various susceptibility genes with minor effects may well reinforce the development of psychosis[38]. Possibly, the combined effects of those genes along with their interaction with environmental factors results in a number of distinct phenotypes. In other words, a wide array of genes is working through various intermediate developmental and physiological pathways on different molecular levels. Changes of gene expression impact on protein expression, which interferes with cell metabolism (e.g., in neurons)[39]. However, often directionality is reversed feeding back from “higher” levels down to ongoing activities at lower levels - thus the interactions are bidirectional[37]. This would account, at least in part, for the substantial heterogeneity repeatedly observed in the psychotic phenotype. As a matter of fact, there is strong evidence for environmental interference with regulation of neurodevelopment by means of epigenetic modifications. What is more, it has to be recalled that almost 80% of human brain growth falls in the postnatal period of life - this includes axonal growth, arborisation of dendrites, synapse formation, and myelination of axons. Therefore, this life span provides ample room for adaptation to environmental conditions and to organize brain development on the epigenetic level[40]. Apparently, the complex construction of brain, mind, and consciousness to enable the organism to respond adequately in a world of social interactions can better be optimized after the individual is born. Needless to mention, that the development of human intelligence and many more typical human characteristics (writing, language, abstraction, anticipation, etc.) are subject to environmental impact. Therefore, it comes of no surprise, that also the majority of psychiatric disorders arise primarily through problems of social functioning, social navigation, or social understanding. It appears safe to say that psychosis is not just a gene-driven by-product of brain evolution. Eventually, each society with its inherent adverse conditions of life gives rise to its specific psychosomatic or brain disorders. In this way, psychosis is an example of “socially sensitive” diseases and reflects structural deficits inherent of human societies. But also “simple” traumatic events, such as lack of oxygen supply during delivery may leave behind a traumatic “imprint” or “engram” that may be accelerated and reinforced by multiple, additional adverse events in childhood and adolescence (short or long-lasting, such as the influence of the family) and eventually result in the formation of a molecular “disease module” or a neuronal “disease circuit” (Figure 1).
Figure 1 Theoretical formation of a disease network in human brain.
Short-term (e.g., traumatic = circles), medium-term (short bars), and long-term (e.g., family = long bars) adverse impact emboss molecular engrams that eventually synergize to form a disease module or network (as shown by connections between nodes). Because the development of the human brain occurs predominantly postnatally, these environmental (epigenetic) influences appear to have more importance than the underlying genetic vulnerability (dashed lines).
This reasoning does not deny the presence of a genetic vulnerability being modified or reshaped postnatally by environmental influences and contributing its burden to the progression of the disease. However, even if a defect (polymorphism, copy number variant, etc.) is detected in a gene of known function, the phenotypic impact of that defect can only be seen in the context of the functions of the gene product’s interaction partners, i.e., by its network context[41]. There is no question that the most immediate reactions on environmental stimuli are located on the cellular level. These reactions may change or modify cellular components or subcellular, molecular interconnectivities. They may not only impact on predisposed genetic abnormalities but influence transcription on the epigenetic level (DNA methylations and posttranslational modifications of histone proteins)[39,40] and post-transcriptional events, such as editing of mRNAs or mRNA degradation by small interfering RNAs or micro (mi)RNAs[42]. The latter way of posttranscriptional regulation of gene expression has become very popular to experimentally silence single genes as an alternative to produce gene knock-out animals. In the context of this review, however, it appears to be even more interesting, because those short RNAs (21-25 nucleotides in length) typically are not specific for one mRNA, hence display multi-target functions[43]. Expression levels of several hundreds of mRNAs can be modified by one miRNA, which results in “fine-tuning” of target gene expression. There are several reports delineating the occurrence of altered miRNA expression profiles in psychiatric disorders[44,45]. For example, miRNAs 1202 and 135 turned out to be involved in major depression disorder (MDD), supporting their role in influencing higher brain functions[46,47]. Owing to this relatively new field of research, the molecular mechanisms leading to altered miRNA expression in those disorders are largely unknown. Until recently, experimental and/or computational methodologies capable of detecting accurately and with high resolution miRNA gene transcription start sites (TSSs) were not available, although efforts in this direction have been made several years ago[48,49]. The latter, however, lacked reliable accuracy in experimental techniques, or presented in silico algorithms providing low resolution/high false positive rate predictions and heuristics. The first algorithm that surpassed the barrier of 54% sensitivity and 64.5% precision in miRNA TSS identification of the earlier studies by achieving 93.6% sensitivity and 100% precision is MicroTSS[50]. MicroTSS can accurately identify miRNA TSSs in single nucleotide resolution[51]. The interconnection between miRGen v3.0 and other DIANA resources enables users to identify in silico as well as experimentally verified miRNA targets on lncRNAs with LncBase[52]. This is very promising progress in getting more insight into regulatory mechanisms of miRNA gene expression and their influence on target mRNAs. A great challenge will be to identify methylation patterns and posttranslational modifications of histones of these genes in health and disease. As a result, effects on protein expression and disturbances of molecular networks in brain disorders may be better understood. Along these lines, it would be important to know, to what extent the products of genes targeted by miRNAs belong to disease networks (see below). Pharmacological interventions on miRNA gene expression would then be reasonable strategies to tackle the problem of the multifactorial origin of chronic psychiatric disorders. Consequently, a key hypothesis is that various pathobiological processes interacting within complex networks are continuously embossing a disease phenotype[39]. Generally speaking, it can be concluded that reductionistic biology (concentrated on single molecules) will not provide insights into the workings of those interconnected networks nor result in improvements of therapeutical solutions.
MOLECULAR NETWORKS IN HEALTH AND DISEASE
There are quite some efforts to abandon reductionistic biology and pave the way for a broader understanding of the maintenance of health and the initiation and progress of disease. Models of oscillating molecular networks could play key functions in identifying weak, but crucial variations in molecular interactions characterizing disease processes. For example, genome-scale metabolic (GSM) networks[53] have been used to investigate metabolic interactions at the cellular level. Additionally, as an attempt to improve the work with GSMs by computational modelling, “Flux Balance Analysis” (FBA) has been elaborated. This constraint-based modelling approach, or constraint-based reconstruction and analysis method, characterizes and predicts aspects of an organism’s metabolism[54]. As a matter of fact, efforts to integrate mRNA expression data into metabolic networks could be significantly improved using FBA as an analytical platform[55]. For example, when growth of mutant E. coli was simulated with FBA, 86% of the mutant phenotypes (i.e., growth or no growth) were accurately predicted[56]. Current GSMs stand out by their large sizes and by a rich source of annotations, but can be applied only to model protein relationships and reactions. More detailed modelling would need more sophisticated resources, which are hard to recover within a drug industry setting. FBA uses rates of uptake of extracellular metabolites and their production as input. The most important challenge appears to reside in the unclear relationship between gene expression and reaction flux[57]. As an advantage, this strategy can be used without biochemical data of enzyme kinetics or concentrations of intracellular metabolites. However, modelling a system using GSM networks is restricted to conditions of a pseudo-steady state, assuming, for instance, that cell proliferation is constant. There is quite some disagreement that the pseudo-steady state assumption is valid. Exact reaction kinetics, which would better reflect in vivo activity[58], cannot be reconciled with this approach. Nevertheless, these mathematical tools to study network behaviour paved the way to describe intracellular molecular interactions. They could be further improved, if they were replenished by modules of protein-protein interactions (PPIs) and by the influence of modules of signal transduction pathways.Overall, studies in this area would greatly benefit from the development of a good metabolite resource[59].
Some time ago, the total number of protein interactions within the so-called human “interactome” has been estimated to fall in the range of 130000-650000 interactions[60,61]. The wide range of variance is due to the fact that only subsets of these interactions have been experimentally identified. Networks have not only been used to gain insight into disease mechanisms[62,63], but also to study comorbidities[64], and to analyze the actions of drugs and effects on their targets[65,66].
Some topological properties and mathematical tools to analyse those networks (Figure 2): Degree distribution in a network shows the number of connections of each single node, hence, identifies nodes with many connections as opposed to nodes with only few connections. In random networks, this distribution would look like a Gaussian curve of distribution, where few nodes have low numbers, and few have high numbers of connections whereas the majority of nodes have similar (mean) numbers of connections. Biological networks are different in that typically most of their nodes have very low connections (degrees), and with increasing connections there is a rapid dropoff of number of nodes with only very few nodes displaying high numbers of connections, obeying the mathematics of a power law distribution. For instance, the degrees of Scale-Free networks, as observed often in social networks, like Facebook, follow a power law, where a small number of nodes (people) are highly connected (hubs, see below). Hence, the low end of a power law distribution may indicate increasing importance within a network. However, the question arises here, if the importance of a node in a network is only dependent on its number of connections, which leads us to measures of centrality. A node with a high degree may occupy a central position within a network, or a peripheral position. Conversely, a node with a low degree (very few connections), may also be located in a central position. Hence, the question arises if it is possible to compute the centrality of a given node or how close any node is to any other node in a network. Closeness centrality is the most easiest way to determine it. It measures the average of the shortest pathlength of one node to every other node in the network. The result of this may be that nodes with low degrees may display higher closeness centrality, or display more importance, than nodes with higher degrees. Consequently, network structure and function may be affected more easily by attacks on these types of nodes compared to others with less closeness centrality. It may also give some hints as to the efficiency of transmission of information from those nodes to any other nodes of the network. The most widely used centrality measure is betweenness centrality[67], characterizing a node’s influence. It is the percentage of shortest paths from every pair of nodes in the network. It gives us an idea what amount of information has to pass through each individual node. It has been proposed that edges located “between” highly connected subgraph clusters (so-called “community structures”) are edges with high betweenness; consequently, a network could be disrupted by disabling these edges[68].
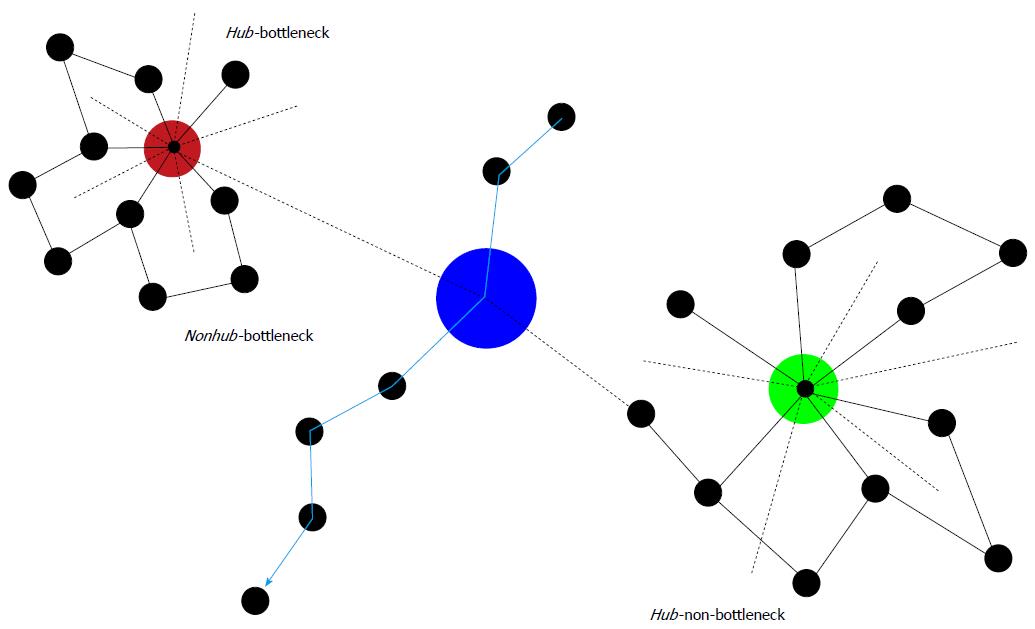
Figure 2 Some characteristics of protein-protein interaction networks.
Hubs are important nodes (proteins) due to their numerous connections (dashed lines = connections in 3rd dimension). Directly inserted into a biochemical pathway, they can also represent a bottleneck (Hub-bottleneck). Otherwise, they are Hub-non-bottlenecks. Nonhub-bottlenecks are nodes inserted in biochemical pathways, but lacking numerous connections. Evidently, disturbances of any of these three important types of nodes result in serious consequences. Therefore, although associations of chronic mental illness with these types of nodes may exist, more unwanted side effects than benefits would ensue from targeting these types of nodes by pharmacotherapy.
Eigenvector centrality is more sophisticated in adding weights to nodes (e.g., ranking web pages) enabling identification of heavily used nodes vs nodes being used infrequently. Another important topological feature of networks is their clustering coefficient, which is an indication of neighbourhood connectivities. In metabolic and other biological networks these connectivities are not random but some are favoured over others. Clustering algorithms are used to group sets of proteins in PPIs showing greater similarities among proteins of the same cluster than in different clusters which identifies functional protein modules or densely connected subgraphs. Several methods have been developed to search for such complexes. The Markov Cluster algorithm simulates a flow on the graph by calculating successive powers of the associated adjacency matrix[69]. Restricted Neighborhood Search Clustering is a cost-based local search algorithm calculated according to the numbers of intra-cluster and inter-cluster edges, that explores the solution space to minimize a cost function[70]. Molecular Complex detection is based on node weighting to isolate densely connected regions by local neighborhood density and outward traversal from a locally dense seed protein[71]. Spirin et al[72] developed a means to detect highly connected subgraphs (cliques) in combination with Monte Carlo optimization. The authors described two types of clusters: Protein complexes and dynamic functional modules. Furthermore, a highly connected subgraphs algorithm was used for discovery of protein complexes by Przulj et al[73], and spectral clustering for generating modules, and possible functional relationships among the members of the cluster for predicting new protein-protein connections has been proposed by Sen et al[74]. Along these lines, networks displaying small world characteristics should be mentioned. Compared to random networks, small world networks show intense local connectivities combined with more or less frequent large connectivities, displaying both low average shortest path lengths of random graphs and high clustering coefficients. The wiring of neuronal networks as well as of PPI networks shows features typical of small world graphs[75].
Hubs: A typical feature of hubs is their multi-connectedness or degree. Hub proteins appear to be encoded by essential genes[76]. These genes are older than genes encoding non-hub proteins and they are more stable over time[77]. Reportedly, products of essential genes are located in hub-like positions or, if expressed in multiple tissues, in functional centers of metabolic networks[78]. Due to this importance of hubs, the hypothesis arose that hub proteins in human molecular networks ought to be encoded by disease genes. However, it turned out later, that in human cells essential genes, but not disease genes, are encoding hubs.
Bottlenecks: Interestingly, protein clusters in interaction networks constructed by the method of edge betweenness show a strong tendency to display related functions[79]. These high-betweenness proteins were called bottlenecks[80] in contrast to hubs as proteins with high degree. Betweenness reflects the important role nodes would play in information transmission in the network. Proteins encoded by essential genes very often are positioned as bottlenecks (both nonhub-bottlenecks and hub-bottlenecks), whereas, surprisingly, proteins in positions of hub-nonbottlenecks are expressed by non-essential genes. The majority of proteins expressed by these genes are structural proteins, whereas proteins located in hub-bottlenecks rather belong to signal transduction pathways. A large part of proteins of nonhub-bottlenecks do not belong to complex members but to regulatory proteins or to proteins of the signal transduction machinery. Their coexpression with their neighbors in the networks is less well correlated than with proteins of nonhub-nonbottlenecks,, which is in agreement with the finding that betweenness is a good predictor of average correlation with neighbors. Apparently, inhibition or blockage of both hubs and bottleneck nodes may severely affect network integrity and function (Figure 2).
One critical feature to be considered in PPIs are post-translational modifications (PTM). More than 200 different types of PTMs have been identified, such as phosphorylations, glycosylations, methylations, acetylations, amidations, as detailed in http://www.uniprot.org/docs/ptmlist curated by UniProt[81] and other databases, like dbPTM, PTMCuration, PTMcode. For additional systematic searches of individual protein interactions, more databases, such as STRING[82], MINT and IntAct[83,84], for protein interactions within pathways, Wikipathways[85], Reactome[86] and Ingenuity (Ingenuity Pathway Analysis: http://www.ingenuity.com) are available. The volume of data held by the IntAct database alone, which includes just under 10% of the estimated human interactome[60], and currently encompassing a range of 50000 binary human interactions, may grow to 750000 binary interaction evidences in the next 5 years. More recently the issue was investigated if proteins exhibiting a particular type of PTM from a collected series of protein sets (displaying 12 types of PTMs) showed characteristic PPI network properties, such as scope of impact (interaction degree), diversity of responses (clustering coefficient), or position in a signalling pathway (closeness centrality)[87]. Interestingly, the 12 PTM-types could be grouped into 2 major groups with (1) sumoylation, nitrosylation, methylation, acetylation, phosphorylation, ubiquitination, and (2) disulfide bond, carboxylation, hydroxylation, proteolytic cleavage, glycosylation, and amidation. Not surprisingly, it turned out that there is a considerable overlap of PTMs, occurring in the same protein, especially with methylations, acetylations, and phosphorylations, indicating their joint associations with histones and their epigenetic functions. Results show that all PTM-types show a tendency of higher degrees, lower clustering coefficients, and higher closeness centralities than protein sets devoid of the respective PTMs. Furthermore, high degree proteins carrying the PTMs acetylation, phosphorylation, ubiquitination showed larger overlaps with human disease proteins than proteins with low degree.
Additionally, it is a challenge to involve the dynamic aspect into network studies by integrating complex data sets across time, space and different organizational levels, providing a systems-level understanding. Therefore, in contrast or in extension to the above mentioned pseudo-steady state approaches, a recent review[88] welcomes various strategies to include the dynamics of biological networks and assumes that these approaches will be the de-facto network modelling in the future. As a matter of fact, the MINT-IntAct consortium is just beginning to implement this in their database, generating dynamic interaction data, in which dynamic changes in protein complex composition in response to stimuli are to be presented as animations driven by radio-buttons. All those mathematical tools and more sophisticated algorithms developed in the future will pave the way to analyze cellular (neuronal) and molecular (gene or protein) interaction networks in great detail, and identify modules or single nodes pivotal for their normal functioning. Moreover, simulations will provide insights into temporary profiles of those networks and detect sites of malfunction that may accumulate over time and set the stage of modules of disease, which reiterates to the model depicted in Figure 1.
DISEASE NETWORKS
Increased attention has been payed by the bioinformatics society to establish disease networks. These can be grouped into the following types of molecular networks: PPIs networks, whose nodes are proteins linked to each other via physical (binding) interactions; metabolic networks, whose nodes are metabolites that are linked if they participate in the same biochemical reactions; regulatory networks, or protein signalling networks whose directed links represent regulatory relationships, such as links between a transcription factor and a gene, or by other signalling molecules on downstream events or on PTM, such as those between a kinase and its substrates; and RNA networks, encompassing RNA-DNA interactions, such as small non-coding miRNAs and siRNAs in regulating gene expression.
PPI networks in particular have become very popular in this context[41]. PPIs entail binding characteristics between proteins and can also include PTMs and protein-protein dimerizations (see below).
Closer insights into the influences of molecular interconnectedness on disease progression could reveal gene products linked to disease and disease pathways, which, in turn, could offer more suitable targets for drug development. Additionally, these new targets could serve as better biomarkers that more accurately monitor the functional integrity of the network perturbed by the diseases. In this way, they could directly impact on clinical practice to achieve better classifications of disease and enabling earlier diagnosis and prognosis, which eventually aims at personalized therapies and treatment[20]. Therefore, analyses of disease networks are believed to permit a better understanding of the pathophysiology of chronic psychiatric diseases with the potential to design combinatorial pharmacotherapies[59].
Moreover, if infections have to be included in the consideration of disease progression, efforts have been undertaken to develop host-pathogen PPI networks[89]. These networks can lead to a better understanding of host-pathogen interactions and to identification of pivotal points for pharmacotherapeutic treatments. In all these approaches, the available databases have to be standardized to seamlessly enable sharing of data, such as being developed in BioPAX[90]. Along those lines, attempts were made to discover functional subnetworks tentatively related to the progression of colorectal cancer by combining analysis of mRNA expression with PPI data[91]. Here, a new computational algorithm was used to search for subnetworks embedded in a PPI network, that entailed genes differentially expressed in the disease. In another study, trying to elaborate predictions on metastasis in breast cancer, it has been shown that markers differentially expressed in subnetworks were more precise than single gene markers[92]. Hence, analysis of PPI networks is very useful to identify candidate biomarkers, to get more insights into disease mechanisms, and to obtain a better understanding of their biology. PPI network analysis also revealed significantly elevated protein interactions specific for the disorder. Two hundred and ninety of such interactions were identified, which corresponded to a 10-fold increase compared to random expectation (P < 10-6)[93]. In other studies, similar results have been reported, i.e., there are significantly increased, direct interactions of gene products associated with disorders of similar phenotypes[94,95]. From those observations, it can be concluded that once some disease components have been identified in the network, more disease-related components should be located in their neighbourhood. In other words, it seems likely that there are interactomes linked to diseases that are embedded in PPI-networks in well-circumscribed neighbourhoods. These interactomes frequently are named disease modules.
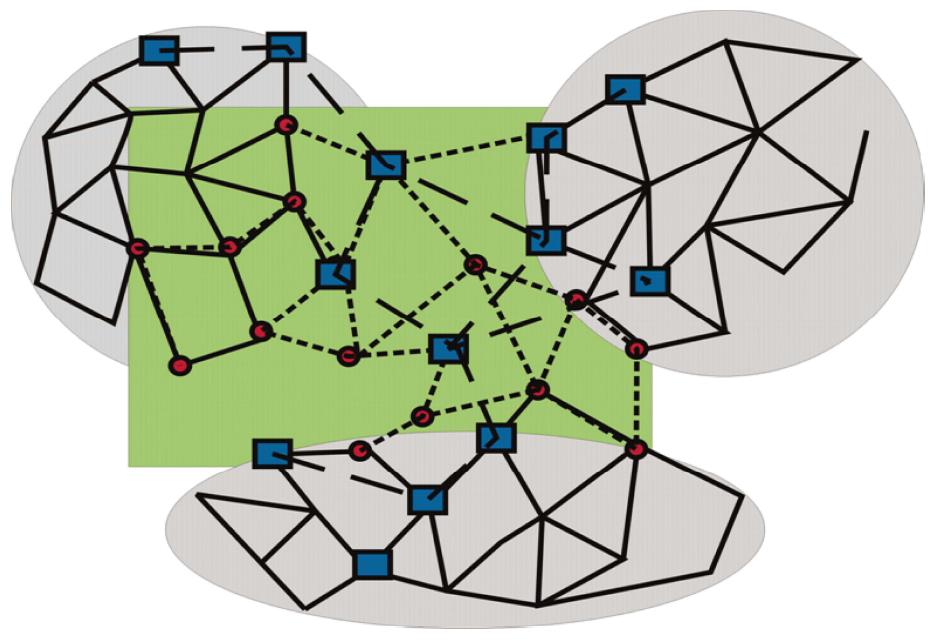
Along these lines, three distinct network modules should be considered (Figure 3): (1) topological modules; (2) functional modules; and (3) disease modules. (1) Topological modules stand out due to locally densely connected neighbourhoods of the interactome, i.e., intra-modular nodes preferentially interact within the module rather than with nodes outside of the module. In this respect, topological modules represent a pure network property; (2) Functional modules are distinguished for their significant segregation of nodes of related function (shown as circles in Figure 3, and connected by short dashed lines), and thus require to define some nodal characteristics. Their belonging to the same network neighbourhood is grounded on the assumption that intensities of nodal interactions are determined by their joint cellular functions; and (3) A disease module is a group of nodes showing changes (of expression, due to mutations, or epigenetic modifications) connected to a specific disease phenotype (drawn as squares in Figure 3, and connected by long, dashed lines).
Figure 3 Topological, functional, and disease modules.
Locally densely connected topological modules (grey circles) contrast with functional modules (circles connected by short, dashed lines), showing more (upper left), less or no overlap (center) with topological modules. The latter are preferentially associated with signal transduction pathways. Disease modules (nodes shown as dark squares, and connected by long, dashed lines) overlap with topological and functional modules, but may be less intensively connected and occupy more peripheral sites of networks.
The tacit assumption in network medicine is that the topological, functional, and disease modules partially overlap: Cellular components that form a topological module have closely related functions, thus being part of a functional module; and a disease is a result of disturbances in some functional module, which means that a functional module is also part of a disease module. However, several characteristics of disease modules are important to bear in mind. As pointed out, a disease module likely overlaps with the topological and/or functional modules, but, because a disease module is defined in relation to a particular disease, each disease has its own unique module. Optionally, a gene, protein, or metabolite can be implicated in several disease modules. There is general agreement in network medicine of mutual, partial overlaps of the topological, functional, and disease modules, which means that on the cellular level, topological modules typically are also closely related in their functions, hence being part of a functional module; and a disease results from disturbances in some functional module, which means that a functional module is also part of a disease module. However, it has to be pointed out, that albeit overlapping features of disease modules with the other modules, the definition of disease modules is specified by each particular disease, endowing these modules with some unique characteristics. Finally, when looking at single genes, proteins or metabolites in a particular disease module, it should be mentioned that each of these components can be a part of other disease modules.
A few years ago, PPI networks (disease networks) were constructed using data from genes differentially expressed in some psychiatric disorders[96]. The study revealed several disease markers (nodes or vortices) characteristic for SCZ (SBNO2), for bipolar disorder (SEC24C), and for MDD (SRRT). Furthermore, similar networks were constructed for Parkinson’s disease (PD), using proteins differentially expressed only in substantia nigra and frontal cerebral cortex[97]. Construction of those networks was based on the following assumptions[96]: (1) There is a positive correlation between expression levels of most proteins and mRNAs in brain; (2) Proteins with similar expression patterns more likely interact with each other; and (3) The abundance of proteins correlates with their participation in biological processes.
In this way, thirty seven unreported disease marker genes were identified. Eight of them belonged to the core functional modules and four were strongly associated with some neurotransmitters, including dopamine. The results of this study may pave the way for addressing new targets in search for more efficient pharmacotherapy of PD. A more general study on the animal model of PD induced by the chemical MPTP used proteomics meta-data from the literature where neuronal alterations due to the metabolite of MPTP MPP+ had been reported[98]. The topological analysis of the protein networks generated on physical or functional interactions revealed a close interaction between nodes as identified by an average shortest path length smaller than in random networks. Moreover, specific alterations in the mitochondrial proteome underlined in what way this model can recapitulate some pathogenic events of PD.
As mentioned above, the construction of those networks was based on the assumption that there is a positive correlation between expression levels of most proteins and mRNAs in brain. This may not be true in any case. Moreover, there are more steps between transcription of a gene and its product, such as post-transcriptional and-translational regulations, all of which complicate the correspondence between expression of a gene and its protein product.
DRUG-TARGET NETWORKS AND NETWORK PHARMACOLOGY
Traditionally, the focus in drug development is to interfere with the activity of one target molecule tentatively crucially involved in the onset or the progression of a disease. Typically, they evoke additional biological responses in patients, some of them leading to adverse or even toxic effects. Others may be benign. These benign effects may result from additional drug effects on additional targets beyond specificity (“dirty drug”). Research to identify those additional targets may open up new therapeutic options for the drug, which is well known in pharmacology as “drug repositioning”[99]. These basic principles constitute the basis of drug-target networks (DTN). Drug discovery can benefit from concepts of DTN in two ways: As polypharmacology and computational drug repositioning. Polypharmacology takes into account the above mentioned features of many drugs to be “promiscuous” in their specificities addressing more than one target molecule[100]. Specific changes of gene expression profiles in a cell are results of direct or indirect responses to a drug or a disease. Disease-induced disturbances of expression profiles could be conceived as perturbations of the dynamic, equilibrium state of a PPI. Drugs ideally may (re-) organize these molecular networks (drug-induced profile) and reverse disease-induced profiles into a state towards the dynamic, equilibrium state (Figure 4). Often, drug discovery is accompanied by investigating biochemical pathways. Because, however, in complex diseases multiple pathways may not be functioning in dynamic equilibrium, it may be advisable to use DTN to search for targets not necessarily being connected at the pathway level, but interacting more specifically on the disease level. Drugs expressing multiple target specificities with little adverse effects or displaying better tolerance have frequently been discovered in natural sources. In contrast to many currently approved drugs, natural products can be viewed as multi-component complex systems with therapeutic potential for a variety of diseases. They display many biological activities and good drug-like properties, show vast chemical diversity and can interact with multiple cellular target proteins[101]. Moreover, biologically active natural products are able to influence disease-related pathways, could provide selective ligands for disease-related targets[102], and could eventually shift the biological network from disease status to the healthy status.
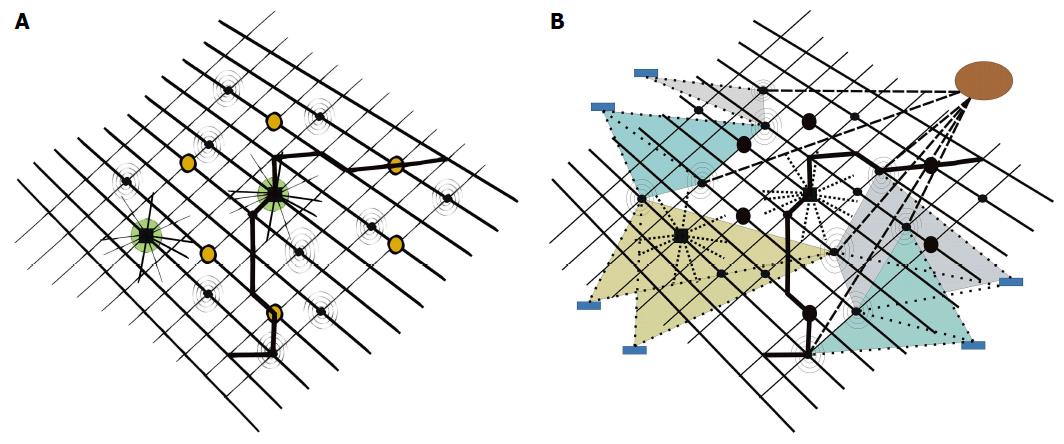
Figure 4 Molecular disease network.
A: With two “hubs” (big, dark circles, showing high connectivity profiles) and six “bottlenecks” (dark circles), inserted in pathways (one pathway highlighted in bold), plus additional molecular nodes, tentatively disturbing network oscillations (spiral nodes, belonging to disease network); B: Subtle disturbances of network harmony by molecular nodes of minor importance (spiral nodes) may substantially interfere on a long-term scale in their summation with network oscillations and result in disease. Drugs with multi-target properties (circle upper right, strong, dashed lines), or multiple drugs with specificities for only one or a few targets (rectangles in periphery, weak, dashed lines) may address nodes of disease networks and reset disease networks to networks characteristic of healthy states.
One very well-known drug in this respect is acetyl-salicylic acid. Salicylates have been known for centuries as anti-inflammatory substances and were already used as extracts from the willow tree (Salix Alba) in the form of tea in times when these active substances had not been identified. While the anti-inflammatory properties of salicylates are due to their inhibition of the NF-κB pathway[103], the serendipitous modification of salicylic acids by an acetyl group endows aspirin with a variety of additional characteristics. The substance inactivates cyclooxygenases through acetylation of serine residues[104]. Some 33 cellular proteins have been identified as acetylation targets of aspirin, one being the tumor suppressor protein, p53 at K382, inducing expression of its target genes[105]. Furthermore, enzymes of the glycolytic pathway (such as glyceraldehyde-3-phosphate dehydrogenase, enolase, aldolase, pyruvate kinase M2, and lactate dehydrogenase A and B chains), cytoskeletal proteins, histones, ribosomal and mitochondrial proteins are targets of aspirin modification. Aspirin also acetylates enzymes involved in ribonucleotide biosynthesis, such as glucose-6-phosphate dehydrogenase and transketolase[106]. Additionally, induction of apoptosis by activation of p38 kinases[107], and catabolism of polyamines[108] have been related to anti-cancer mechanisms of aspirin[109,110]. Regular intake of aspirin has been shown to reduce the risk of cancer of the esophagus by 73%, of the colon by 63%, of the stomach by 62%, of the breast and prostata by 39%, and of the lung by 36%[111]. The inhibition of G6PDH with increasing concentrations of aspirin is believed to be a crucial event in its anti-cancer effect, because this enzyme regulates the synthesis of nucleotides and nucleic acids. Along these lines, the activation of the ERK pathway mediated by the high levels of Ras mutations observed in many cancers[112], has been shown to be inhibited by aspirin, as well. Antiproliferative effects have also been reported from actions of salicylic acid, that is able to reduce mitochondrial calcium uptake[113]. Finally, aspirin has been shown to activate CREB, facilitating its binding to a cAMP-response element in the promoter of the neurotrophic factor CNTF and increasing its gene expression[114]. Aspirin (salicylates) is only one example of multi target effects exerted by many natural substances. Very likely, many other substances occurring in plants may be superior to synthetic drugs with high specificity for one target. For that reason, it may be more beneficial to learn from nature in what ways pharmacotherapies could become more efficient. Polypharmacology, indicating multi-target strategies in pharmacotherapies, encompasses those attempts to identify the multi-target nature of natural substances, but also to develop synthetic drugs with multiple target properties[115]. Especially in chronic brain disorders, these drugs may offer higher efficiency and less unwanted adverse effects[116]. In these terms, successful drug discovery in the pharmaceutical industry is still in its infancy, slowly trying to abandon the point of view that successful drugs should interact only with single, individual targets and approaching concepts of systems biology models of human metabolic networks[117]. Investigations on DTN on one hand, and disease networks on the other hand can aid to find overlapping targets and achieve better understanding of the mechanisms of action of multi-target (natural) compounds. The goal, hence, is to understand drug targets in the context of cellular and disease networks using systems pharmacological approaches[118].
Generally, hundreds of different biologically active substances are found in herbal extracts[119] and in contrast to most of synthetic drugs designed to bind single targets, most of the ingredients of herbal formulae display only weak to moderate effects, but address multiple cellular targets during treatment of complex diseases[120]. Normally, the underlying mechanisms are not clear. As a newly emerging field, network pharmacology[121] could help to understand the mechanisms of multiple action drugs across multiple scales from the molecular and cellular level to the tissue and organism level by analyzing the features of biological networks[122]. Network pharmacology is supposed to integrate polypharmacology and network biology and considered as upcoming paradigm in drug development[123].
It is exciting to study network pharmacology in product-target networks using natural products. Recent results on network properties suggested a marked enrichment of polypharmacology with respect to nodes (compounds) with large degree and high betweenness centrality. These nodes turned out to be highly influential in the whole network. Despite a slow change of direction in drug research and development, it has to be acknowledged that until recently approximately every second drug approved by the FDA was a natural product or a derivative thereof[124]. That means, that also in the industrial context increased efforts have been made to explore in more detail the mechanisms of action of herbal formulae employing network pharmacological approaches, such as DTN[125], PPI networks[126], metabolic networks[127], or disease networks (see above)[128]. In order to speed up virtual screening of natural products on a large scale, the Universal Natural Products Database has been constructed. Until now, it entails 197201 natural products (http://pkuxxj.pku.edu.cn/UNPD). Due to the complexity of studies using network pharmacology approaches, most of them are based on static networks[129]. However, network pharmacology also provides a systems-level approach to understanding the development and pathogenesis of disease, taking into account the dynamics of biological networks[130].
The mechanisms of action of natural products have been studied by well established, modern technologies, such as gene expression microarrays[131], technologies in proteomics[132], and metabolomics[133]. The concept of ‘‘network targets’’ emerging from these investigations extends the widely used concept of a drug tailored for a single biological component to the concept of a drug or group of drugs exerting multiple effects on a biological network[134]. The problem presently remaining is to develop mathematical algorithms able to configure herbal formulae holistically as a gestalt, whose emergent and tentatively synergistic properties no longer rely on analyzing each substance separately. Exemplarily, components of the Liu-Wei-Di-Huang pill have been investigated completely in silico to predict their effects and mechanisms[135].
After analyzing the composition of chemical groups in LWDH, their chemical characteristics and distribution in chemical space were studied. Then their pharmacological properties were explored. Based on these results, predictions were made as to what biological molecules could be targeted by LWDH components. It was suggested that a biological molecule would be a “good” candidate target if there were several components specific for that biological molecule in the natural product. The results revealed that PPI networks constructed from the predicted candidate targets of LWDH displayed high intranet connectivity. Therefore, a compound-target network was constructed from the PPI network, where compounds were connected to their targets[136]. Then a disease network could be constructed by adding edges between candidate target proteins and a disease if the target protein’s gene was in a gene list related to the disease. The results show that some target proteins belonging to hormone signalling, such as ESR1, NCOA1 and AR, are not only highly connected to the components of LWDH (high degree), attributing hub-like properties to them, but also highly connected to disease. However, a disease might be influenced by many biological processes that are targeted by different groups of ingredients, and hormone signalling is only one example. Like in many studies before, this study reveals, as well, that often “key players”, here hubs and bottlenecks in molecular networks, become a focus of major interest.
CONCLUSION
There are numerous mathematical tools available to analyze networks both on the static and dynamic levels, and to distinguish structural components required for maintenance of their connectivities or responsible for their malfunctioning. Apparently, one major challenge for future studies is to improve algorithms taking into account changes of PTM over time and incorporate many more molecular nodes into these observations than just some “important” ones. The notion to be delineated here, is that efforts to identify “key” players in molecular networks may be misleading. Although they are often involved in maintenance and progression of a chronic mental disorder, they are also crucial for maintenance of many other metabolic functions independent of the disease. Therefore, interference by drugs with these crosspoints may easily destroy network integrity and, hence, be accompanied with numerous unwanted adverse effects. We rather want to conclude from the features of molecular networks outlined above, that it is more desirable to search for targets with more subtle effects on network functions, but specifically perturbed in a mental illness, and hence belonging to a disease network, as well (Figure 3). Along these lines, Lamb[137] have shown that mere node connectivity (degree) might not be the only influential parameter to characterize biological networks. And Goñi et al[138] reported that in case of neurodegenerative diseases, less extensively connected proteins are much more appropriate therapeutic targets than highly connected ones, as the critical role of highly connected nodes (hubs) in the network modules prevents them from substantial fluctuation. Exceptions probably are genes of miRNAs displaying “hub” features as well, that nevertheless may be good pharmacological targets. Moreover, it was shown that the above mentioned betweenness centrality can also be used as an important parameter to search for lowly connected nodes[80]. Chronic degenerative disorders of the brain extend over considerable periods of time, which strongly argues for disturbances of multiple nodes with weak influences, each. Or conversely, if abnormally functioning hubs or bottlenecks were the only players, the disorders would not be long-lasting and chronic. Consequently, in order to correct “damages” in disease networks, future pharmacological strategies to treat mental disorders may be aimed at targeting “peripheral” molecules with only subtle effects using polypharmacological approaches. The great challenge, hence, is to identify those “peripheral” targets and develop wide spectrum drugs aimed at those targets. In summary, it has to be kept in mind, that the human brain both in health and disease is a biological system distinguished by its extremely high complexity especially on the molecular level. Mathematical approaches to investigate changes on this level still require many improvements, and even greater challenges are confronted when it comes to address dynamic changes of the system. Because, however, this is at the core of biological systems, there is no way around. Reductionistic attempts to understand molecular mechanisms of mental illness are not able to address these issues adequately.
ACKNOWLEDGMENTS
The valuable comments and helpful discussions on the manuscript by F. Tretter, Bayerische Akademie für Suchtfragen in Forschung und Praxis, München, are greatly acknowledged. Moreover the support of the Faculty of Medicine, University of Chile, Santiago, Chile, is highly appreciated.