Published online Feb 19, 2025. doi: 10.5498/wjp.v15.i2.100738
Revised: November 30, 2024
Accepted: December 20, 2024
Published online: February 19, 2025
Processing time: 142 Days and 16.6 Hours
The NaV1.1 sodium channel alpha subunit, encoded by SCN1A, is crucial for initiating and propagating action potentials in neurons. SCN1A gene has long been an established target in the etiology and therapy of epilepsy. However, very few studies have investigated the relevance of genetic variations in epilepsy and anti-epileptic drug resistance.
To investigate associations between polymorphisms, rs121917953 T/A and rs121918623 C/T, and drug resistance in epilepsy patients in the north Indian population.
A total of 100 age- and sex-matched epilepsy patients (50 drug responsive and 50 drug resistant subjects) were recruited and SCN1A rs121918623 C/T* and rs121917953 T/A* polymorphisms were analyzed by the allele specific-PCR technique. χ2 and Fisher’s exact test were used to estimate differences between the distribution of SCN1A rs121918623 and rs121917953 gene polymorphisms among various groups. The association between distinct rs121917953 genotypes and drug resistance was analyzed using logistic regression analysis.
For the SCN1A rs121917953 T/A* (D188V) polymorphism, a significantly higher proportion of individuals with AT genotype were observed in the drug-resistant group as compared to the drug-responsive group. Additionally, a higher risk association was exhibited by AT genotype for drug resistance with an odds ratio of 3.51 and P value = 0.017. For the SCN1A rs121918623 C/T* (T875M) polymorphism, no significant difference in genotype distribution was observed between the drug-resistant and drug-sensitive groups.
Our findings indicate that the SCN1A polymorphism D188V is associated with a higher risk of drug resistance for the AT variant as compared to the homozygous TT wild-type. Further research is needed at the functional level and in larger cohorts to determine the potential of these genes as a therapeutic target in epilepsy subjects.
Core Tip: Epilepsy is the most common paroxysmal neurological disorder and a significant global health burden. A subset of patients are resistant to anti-epileptic drugs, leading to increased mortality and decreased quality of life. Mutations in genes encoding neuronal voltage-gated sodium channels have been linked to inherited forms of epilepsy. This study investigates the correlation between SCN1A gene variants rs121918623 and SCN1A rs121917953 and drug resistance in epilepsy suggesting specific genotypes as potential biomarkers in drug-resistant idiopathic epilepsy.
- Citation: Dabla PK, Gupta S, Singh S, Viswas A, Yadav M, Sonkar SC, Koner BC. Sodium channel mutation SCN1A T875M, D188V and associated dysfunction with drug resistant epilepsy. World J Psychiatry 2025; 15(2): 100738
- URL: https://www.wjgnet.com/2220-3206/full/v15/i2/100738.htm
- DOI: https://dx.doi.org/10.5498/wjp.v15.i2.100738
Epilepsy is the most common paroxysmal and diverse neurological disorder, affecting approximately 42 million people globally. It has diverse symptoms, causes, prognosis, and treatments[1]. The overall prevalence of epilepsy is around 5-10 per 1000 people, with developing countries having a higher rate. While several anti-epileptic drugs (AEDs) are available to treat these seizures, a significant percentage of epilepsy patients continue to experience treatment resistance[2]. Individuals with drug-resistant epilepsy experience uncontrolled seizures and increased mortality rates. The majority of epilepsy phenotypes are the product of gene-environment interactions[3,4].
Mutations in genes encoding neuronal voltage-gated sodium channels have been linked to inherited forms of epilepsy[5]. The majority of these mutations occur in the SCN1A gene and are all dominantly inherited[6,7]. The alpha subunit of the NaV1.1 sodium channel, encoded by SCN1A, is crucial for initiating and propagating action potentials in neurons. The SCN1A mutations that cause epilepsy are in different functional domains of the channel, but they all may lead to decreased inactivation resulting in hyperactivity of neurons leading to seizures[8]. rs121917953 T/A* transversion occurring in exon 4 of the SCN1A gene, results in Asp188Val (D188V) substitution. This substitution lies just outside the S3 segment of domain I, in another highly conserved region of SCN1A. During seizures, neurons undergo prolonged membrane depolarization, during which they continuously fire action potentials at high frequency. The cumulative inactivation of sodium currents during this activity lowers the neuronal excitability and attenuates firing. It is suggested that the neurons with D188V mutation in sodium channels are more resistant to high-frequency firing, making them more capable of sustaining and propagating seizures[9]. rs121918623 C/T* SCN1A transversion, resulting in the amino acid substitution Thr875Met, is found in highly conserved S4 transmembrane regions of the channel, which is known to play a role in channel gating. Several recent studies have assessed the functional effect of this mutation in SCN1A[10]. Taken together, these findings indicate that the D188V and T875M mutations in SCN1A genes play a crucial role in the pathogenesis of epilepsy.
Apart from their role in epilepsy pathogenesis, the voltage-gated sodium channels also play a role as AED targets[8,11–13]. Kahlig et al[14] investigated the functional consequences of SCN1A mutations, including T825M, on sodium channel activity and drug sensitivity using cellular models. The association between SCN1A mutation and drug resistance has been commonly reported in many other SCN1A genetic variants. However, there are only a few studies in the literature that establish the link between SCN1A rs121918623 and rs121917953 mutations with drug resistance[15–18]. Furthermore, no targeted studies are available to establish findings in the Indian population. The present study explores the role of sodium channel alpha receptor subunit mutations, SCN1A rs121918623 and rs121917953, in drug resistance in epilepsy patients.
The study was conducted at the Department of Biochemistry, G.B. Pant Institute of Postgraduate Medical Education & Research (GIPMER), a neurological disease referral center in New Delhi, India. We included 100 age- and sex-matched patients, divided into two groups: Drug-resistant epilepsy (group I) and drug-responsive epilepsy (group II). All participants provided informed consent. Patients were recruited from the Department of Neurology, GIPMER. Diagnosis and classification followed the International League Against Epilepsy guidelines. We collected detailed histories of ethnicity, seizure duration, frequency and medication adherence. Patients with poor anti-epileptic medication compliance, severe adverse medication reactions or inaccurate seizure records were excluded. The study complied with the Declaration of Helsinki, revised in October 2013.
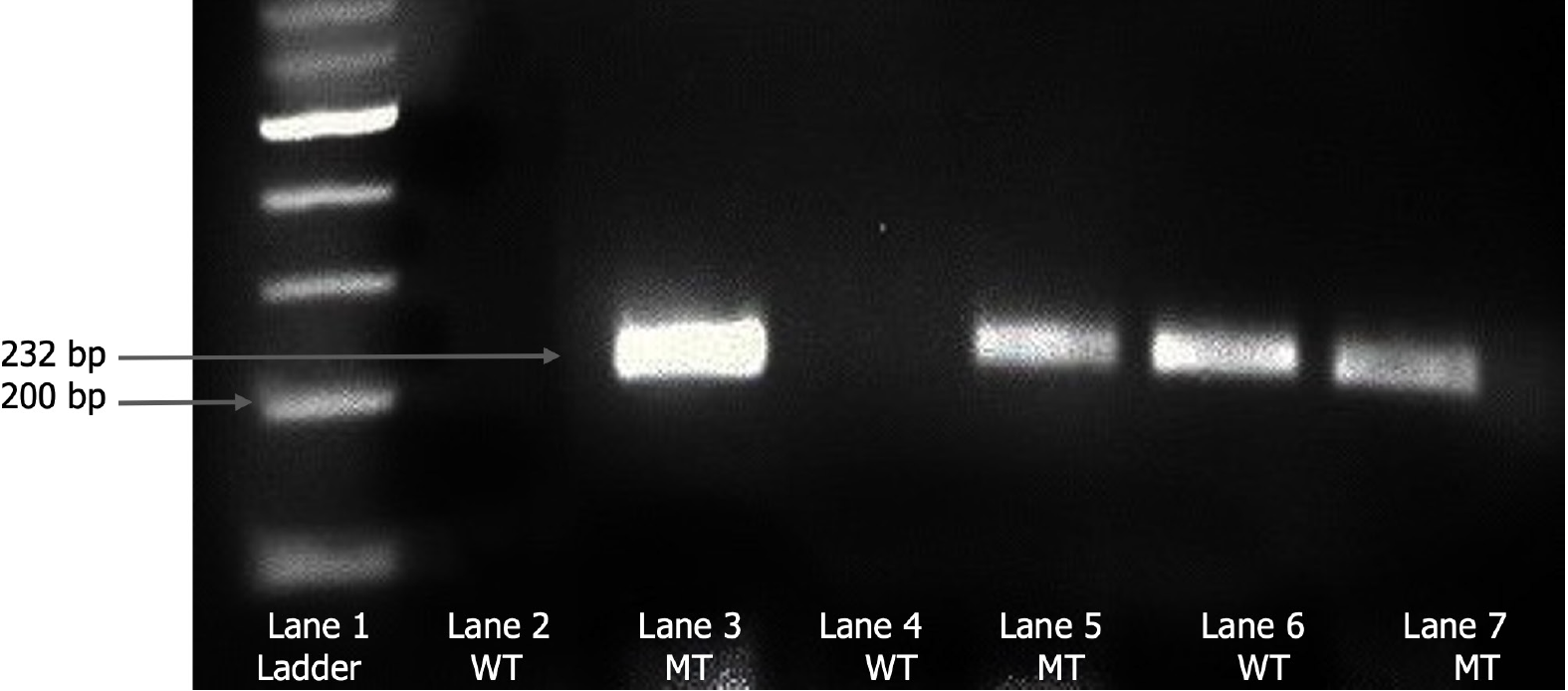
We collected 5 mL of total peripheral whole blood from each participant in ethylenediaminetetraacetic acid-containing vacutainers. Plasma was separated by centrifugation at 3500 rpm for 8 minutes and stored at -80°C for further analysis. DNA was isolated usingReliaPrep™ Blood gDNA Miniprep (column binding method). For the analysis of rs121918623 C/T* and rs121917953 T/A* polymorphism, allele specific-PCR was performed for the amplification of SCN1A gene's relevant region. Both rs121918623 C/T* and rs121917953 T/A* polymorphic sites in fragments were amplified using specific forward and reverse primers in a final volume of 25 μL comprising 50-100 ng genomic DNA. For rs121918623 C/T* polymorphism, PCR amplifications were performed under the following conditions: Denaturation step at 94°C for 5 minutes, then 35 cycles at 94°C for 45 seconds, 60°C for 1 minute, 72°C for 1 minute, and a final incubation at 72°C for 10 minutes. For rs121917953 T/A* polymorphism, the PCR amplification conditions were as follows: Denaturation step at 94°C for 5 minutes, then 35 cycles at 94°C for 45 seconds, 54°C for 1 minute, 72°C for 1 minute, and a final incubation at 72°C for 10 minutes. Primers were designed such that amplification progressed only if the nucleotide at the 3’-end of the primer was complementary to the base at the wild-type or variant-type DNA sample. The products were resolved using electrophoresis in a 2% agarose gel. Visualization and documentation of the results was performed using the Chemidoc gel documentation system (Bio-Rad ChemidocTM XRS+). SCN1A rs121918623 wild-type ‘C’ allele and mutant ‘T’ allele were identified as bands at 222 bp with respective primers. SCN1A rs121918623 heterozygotes showed a band with both forward primers. For SCN1A rs121917953, wild-type ‘T’ and mutant ‘A’ allele were identified as bands at 232 bp with respective primers. SCN1A rs121917953 heterozygotes showed bands with both forward primers (Figure 1).
Statistical analysis was performed using the Statistical package for Social Sciences version 26. Descriptive statistics are shown as frequencies and percentages for categorical variables. The comparison between various groups for SCN1A rs121918623 and rs121917953 gene polymorphism was carried out using χ2 and Fisher’s exact test. To examine the relationship between rs121917953 variants and drug resistance, binary logistic regression analysis was performed. A P value of < 0.05 was considered significant.
The distribution of SCN1A genotypic variants in drug-sensitive and drug-resistant epilepsy patients is shown in Table 1. Our results on the SCN1A rs121917953 T/A* polymorphism revealed that the percentage of homozygous wild-type genotype (TT) was higher in the drug-responsive group (34.6%) as compared to the drug-resistant group (13.9%). Whereas, the percentage of heterozygous mutant genotype (AT) was higher in the drug-resistant group (83.7%) as compared to the drug-responsive group (63.4%). Furthermore, on comparing the distribution of SCN1A rs121917953 T/A* genotypes, a significant difference was observed across both the study groups (P value of 0.001). Whereas, on comparing the distribution of SCN1A rs121918623 C/T* genotypic variants, no significant difference was observed across both the study groups, with a P value of 1.00 (Table 2).
| SNP | Primers | Sequence |
| rs121918623 | Forward primers | WT- 5’ATGATCTTTATTAGCATATTTAACG3’ |
| MT-5’ATGATCTTTATTAGCATATTTAATG3’ | ||
| Reverse primer | R-5’CAGCTTTTCCTAGGGAGTCCAAAAA3' | |
| rs121917953 | Forward primers | WT-5’ACAGTGAAATCGAGCCAGTTCCATGGAT3’ |
| MT-5’ACAGTGAAATCGAGCCAGATCCATGG3’ | ||
| Reverse primer | R-5’TCTGCTTAGTTTTCTTTTTTAGTATTT3’ |
Risk analysis was performed to examine the relationship between distinct SCN1A rs121917953 T/A* genotypes and drug resistance. The AT genotype exhibited significantly higher risk association with drug resistance with an odds ratio of 3.51 (95%CI = 1.256-3.826 and a P value = 0.017; Table 3).
| Genotype | Drug-responsive, n = 52 | Drug-resistant, n = 43 | OR (95%CI) | P value |
| TT | 18 (34.6) | 6 (13.9) | Reference | Reference |
| AT | 33 (63.4) | 36 (83.7) | 3.51 (1.256-3.826) | 0.017 |
| AA | 1 (1.9) | 1 (2.3) | 3.33 (0.180-61.686) | 0.419 |
In this tertiary care hospital-based study, we attempted to investigate the role of SCN1A rs121917953 T/A* and SCN1A rs121918623 C/T* genetic polymorphisms in the sodium voltage-gated channel alpha receptor subunits in epilepsy. To our knowledge, this is the first study to investigate the association between SCN1A rs121917953 T/A* and SCN1A rs121918623 C/T* polymorphisms with anti-epileptic drug resistance among epilepsy patients.
In our study, we found that the percentage of SCN1A rs121917953 T/A* homozygous wild-type genotype (TT) was higher in the drug-responsive group as compared to the drug-resistant group (P value = 0.001). SCN1A rs121917953 T/A* polymorphism exhibited a significantly higher risk association with drug resistance for mutant AT variant as compared to wild-type TT variant with an odds ratio of 3.51 (P value = 0.017). This finding is in agreement with the findings in previous studies that explored the role of sodium channel mutations, including D188V, in epilepsy. A functional study by Cossette et al[9] revealed that compared to wild-type sodium channels, in vitro expression of channels with the D188V mutation demonstrated greater resistance to the amplitude decline during high-frequency pulses. However, the precise mechanism causing intractable seizures in these patients still remains unclear. SCN1A rs121917953 T/A* polymorphism causes the substitution of aspartate with valine at a conserved site within the coding region of the SCN1A gene, potentially impacting the function of the inactivation gate that regulates the influx and efflux of sodium ions. As a result, this amino acid change may alter the function of SCN1A, a key target of AEDs[6]. Overall, these observations indicate that D188V mutation can alter drug binding and, this could therefore result in variations in epilepsy treatment efficacy among individuals with different genotypes.
In our study, we also investigated the role of the SCN1A rs121918623 C/T* polymorphism in anti-epileptic drug response. On comparing the distribution of SCN1A rs121918623 C/T* polymorphism, no significant difference was observed between the drug-resistant and drug-sensitive groups. A similar study conducted by Kahlig et al[14] evaluated ranolazine's ability to reduce the magnitude of persistent current in mutant channels to levels comparable to those seen in wild-type NaV1.1 channels. The authors reported that for T875M mutant channels, the persistent current in the presence of ranolazine was similar to that observed in wild-type NaV1.1 channels without the drug. Although there is paucity of data on SCN1A rs121918623 C/T* polymorphism and drug responsiveness in human subjects, some functional studies suggest that this polymorphism may be associated with susceptibility to epilepsy and variations in drug effectiveness. For example, in oocyte expression studies, T875M enhanced slow inactivation of sodium channels[19]. It is suggested that SCN1A rs121918623 C/T* polymorphism may cause loss of function of the SCN1A α1-subunit. In the absence of a functioning α1-subunit, sodium-channel α-subunits are less likely to be inactivated, leading to enhanced sodium influx and hyperexcitability[10].
The association between SCN1A mutations and drug resistance is well-documented for many other SCN1A genetic variants. Missense or in-frame deletion SCN1A mutations (L986F, delF1289, R1648C, F1661S, G1674R, and G1979E) exhibited reduced cell surface expression relative to wild-type NaV1.1 in pediatric epileptic patients as observed by Thompson et al[12]. Lossin et al[6] revealed that the persistent current was found to be associated with T1875M in tsA201 cells. In addition, phenytoin and lamotrigine promoted greater persistent current density in cells expressing R1648C channels. In contrast, an earlier study showed no association between another SCN1A genotype rs3812718 and phenytoin response in epileptic patients[20]. However, due to the analyzed variability in polymorphisms across the available literature this raises difficulty in deriving broad conclusions regarding their significant impact. Nonetheless, identifying a common influence on treatment response could potentially allow for more effective drug selection and personalized treatment.
Our findings support the hypothesis that the SCN1A gene polymorphism plays a significant role in epilepsy. The current analysis is an expansion of our prior report, in which we selected more genes controlling distinct sodium voltage-gated channel alpha receptor subunits associated with epilepsy risk. In our previous study we investigated the asso
The present study is, to our knowledge, the first to explore the relationship between SCN1A rs121917953 T/A and SCN1A rs121918623 C/T polymorphisms and AED responsiveness in patients with epilepsy. Our data revealed that SCN1A rs121917953 T/A* D188V mutation exhibited a higher risk of drug resistance as compared to wild-type. To conclude, our results suggest that alterations in the structure and function of voltage-gated sodium channels subunits may impact seizure susceptibility, which could, in turn, affect the response to anti-epileptic medications. Further research is needed at the functional level and in larger cohorts to tailor more effective treatments for epilepsy patients, particularly those unresponsive to AEDs.
Thanks to the Multidisciplinary Research Unit (MRU), Maulana Azad Medical College, New Delhi for the provision of molecular analysis research facilities and support.
| 1. | Depondt C. The potential of pharmacogenetics in the treatment of epilepsy. Eur J Paediatr Neurol. 2006;10:57-65. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 40] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 2. | Łukawski K, Czuczwar SJ. Understanding mechanisms of drug resistance in epilepsy and strategies for overcoming it. Expert Opin Drug Metab Toxicol. 2021;17:1075-1090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 20] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 3. | Hauser WA. The prevalence and incidence of convulsive disorders in children. Epilepsia. 1994;35 Suppl 2:S1-S6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 443] [Cited by in RCA: 430] [Article Influence: 13.9] [Reference Citation Analysis (0)] |
| 4. | Hauser WA, Annegers JF, Kurland LT. Incidence of epilepsy and unprovoked seizures in Rochester, Minnesota: 1935-1984. Epilepsia. 1993;34:453-468. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1415] [Cited by in RCA: 1357] [Article Influence: 42.4] [Reference Citation Analysis (0)] |
| 5. | Lu Y, Wang X. Genes associated with idiopathic epilepsies: a current overview. Neurol Res. 2009;31:135-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 27] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 6. | Lossin C, Wang DW, Rhodes TH, Vanoye CG, George AL Jr. Molecular basis of an inherited epilepsy. Neuron. 2002;34:877-884. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 270] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 7. | Wallace RH, Scheffer IE, Barnett S, Richards M, Dibbens L, Desai RR, Lerman-Sagie T, Lev D, Mazarib A, Brand N, Ben-Zeev B, Goikhman I, Singh R, Kremmidiotis G, Gardner A, Sutherland GR, George AL Jr, Mulley JC, Berkovic SF. Neuronal sodium-channel alpha1-subunit mutations in generalized epilepsy with febrile seizures plus. Am J Hum Genet. 2001;68:859-865. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 281] [Cited by in RCA: 244] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 8. | Lakhan R, Kumari R, Misra UK, Kalita J, Pradhan S, Mittal B. Differential role of sodium channels SCN1A and SCN2A gene polymorphisms with epilepsy and multiple drug resistance in the north Indian population. Br J Clin Pharmacol. 2009;68:214-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 63] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 9. | Cossette P, Loukas A, Lafrenière RG, Rochefort D, Harvey-Girard E, Ragsdale DS, Dunn RJ, Rouleau GA. Functional characterization of the D188V mutation in neuronal voltage-gated sodium channel causing generalized epilepsy with febrile seizures plus (GEFS). Epilepsy Res. 2003;53:107-117. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 10. | Escayg A, MacDonald BT, Meisler MH, Baulac S, Huberfeld G, An-Gourfinkel I, Brice A, LeGuern E, Moulard B, Chaigne D, Buresi C, Malafosse A. Mutations of SCN1A, encoding a neuronal sodium channel, in two families with GEFS+2. Nat Genet. 2000;24:343-345. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 689] [Cited by in RCA: 679] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 11. | Margari L, Legrottaglie AR, Vincenti A, Coppola G, Operto FF, Buttiglione M, Cassano A, Bartolomeo N, Mariggiò MA. Association between SCN1A gene polymorphisms and drug resistant epilepsy in pediatric patients. Seizure. 2018;55:30-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 12. | Thompson CH, Porter JC, Kahlig KM, Daniels MA, George AL Jr. Nontruncating SCN1A mutations associated with severe myoclonic epilepsy of infancy impair cell surface expression. J Biol Chem. 2012;287:42001-42008. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 13. | Viswas A, Dabla PK, Gupta S, Yadav M, Tanwar A, Upreti K, Koner BC. SCN1A Genetic Alterations and Oxidative Stress in Idiopathic Generalized Epilepsy Patients: A Causative Analysis in Refractory Cases. Ind J Clin Biochem. 2023. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Reference Citation Analysis (0)] |
| 14. | Kahlig KM, Lepist I, Leung K, Rajamani S, George AL. Ranolazine selectively blocks persistent current evoked by epilepsy-associated Naν1.1 mutations. Br J Pharmacol. 2010;161:1414-1426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 15. | Tang HX, Ho MD, Vu NP, Cao HV, Ngo VA, Nguyen VT, Nguyen TD, Nguyen TD. Association between Genetic Polymorphism of SCN1A, GABRA1 and ABCB1 and Drug Responsiveness in Vietnamese Epileptic Children. Medicina (Kaunas). 2024;60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 4] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 16. | Li M, Zhong R, Lu Y, Zhao Q, Li G, Lin W. Association Between SCN1A rs2298771, SCN1A rs10188577, SCN2A rs17183814, and SCN2A rs2304016 Polymorphisms and Responsiveness to Antiepileptic Drugs: A Meta-Analysis. Front Neurol. 2020;11:591828. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 17. | Wang ZJ, Chen J, Chen HL, Zhang LY, Xu D, Jiang WT. Association between SCN1A polymorphism rs3812718 and valproic acid resistance in epilepsy children: a case-control study and meta-analysis. Biosci Rep. 2018;38. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 18. | Abe T, Seo T, Ishitsu T, Nakagawa T, Hori M, Nakagawa K. Association between SCN1A polymorphism and carbamazepine-resistant epilepsy. Br J Clin Pharmacol. 2008;66:304-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 19. | Spampanato J, Escayg A, Meisler MH, Goldin AL. Functional effects of two voltage-gated sodium channel mutations that cause generalized epilepsy with febrile seizures plus type 2. J Neurosci. 2001;21:7481-7490. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 126] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 20. | Khairani AF, Sutarni S, Sholikhah EN, Malueka RG, Luthffia A, Vidyanti AN. Association of SCN1A Gene Polymorphism with Phenytoin Response in Patients with Epilepsy: Relevance of Stratification by the History of Febrile Seizure. Open Access Maced J Med Sci. 2022;10:1676-1681. [RCA] [DOI] [Full Text] [Reference Citation Analysis (0)] |