Published online Mar 19, 2024. doi: 10.5498/wjp.v14.i3.445
Peer-review started: November 30, 2023
First decision: December 18, 2023
Revised: December 31, 2023
Accepted: February 1, 2024
Article in press: February 1, 2024
Published online: March 19, 2024
Processing time: 110 Days and 2.3 Hours
Epidemiological studies have revealed a correlation between Alzheimer’s disease (AD) and type 2 diabetes mellitus (T2D). Insulin resistance in the brain is a common feature in patients with T2D and AD. KAT7 is a histone acetyltransferase that participates in the modulation of various genes.
To determine the effects of KAT7 on insulin patients with AD.
APPswe/PS1-dE9 double-transgenic and db/db mice were used to mimic AD and diabetes, respectively. An in vitro model of AD was established by Aβ stimulation. Insulin resistance was induced by chronic stimulation with high insulin levels. The expression of microtubule-associated protein 2 (MAP2) was assessed using immunofluorescence. The protein levels of MAP2, Aβ, dual-specificity tyrosine phosphorylation-regulated kinase-1A (DYRK1A), IRS-1, p-AKT, total AKT, p-GSK3β, total GSK3β, DYRK1A, and KAT7 were measured via western blotting. Accumulation of reactive oxygen species (ROS), malondialdehyde (MDA), and SOD activity was measured to determine cellular oxidative stress. Flow cytometry and CCK-8 assay were performed to evaluate neuronal cell death and proliferation, respectively. Relative RNA levels of KAT7 and DYRK1A were examined using quantitative PCR. A chromatin immunoprecipitation assay was conducted to detect H3K14ac in DYRK1A.
KAT7 expression was suppressed in the AD mice. Overexpression of KAT7 decreased Aβ accumulation and MAP2 expression in AD brains. KAT7 overexpression decreased ROS and MDA levels, elevated SOD activity in brain tissues and neurons, and simultaneously suppressed neuronal apoptosis. KAT7 upregulated levels of p-AKT and p-GSK3β to alleviate insulin resistance, along with elevated expression of DYRK1A. KAT7 depletion suppressed DYRK1A expression and impaired H3K14ac of DYRK1A. HMGN1 overexpression recovered DYRK1A levels and reversed insulin resistance caused by KAT7 depletion.
We determined that KAT7 overexpression recovered insulin sensitivity in AD by recruiting HMGN1 to enhance DYRK1A acetylation. Our findings suggest that KAT7 is a novel and promising therapeutic target for the resistance in AD.
Core Tip: Type 2 diabetes mellitus (T2D) is closely associated with neurodegenerative diseases, such as Alzheimer’s disease (AD), in which insulin resistance dysfunction plays a critical role. However, the pathological mechanisms underlying diabetes mellitus-related atopic dermatitis remain unclear. Our study demonstrated that the histone acetyltransferase KAT7 ameliorated neuronal death and oxidative stress in AD and restored insulin sensitivity in insulin-resistant neurons by recruiting HMGN1 to enhance the acetylation of the dual-specificity tyrosine phosphorylation-regulated kinase-1A gene, suggesting the promising therapeutic potential of KAT7 in diabetes mellitus-associated AD.
- Citation: Lu QS, Ma L, Jiang WJ, Wang XB, Lu M. KAT7/HMGN1 signaling epigenetically induces tyrosine phosphorylation-regulated kinase 1A expression to ameliorate insulin resistance in Alzheimer’s disease. World J Psychiatry 2024; 14(3): 445-455
- URL: https://www.wjgnet.com/2220-3206/full/v14/i3/445.htm
- DOI: https://dx.doi.org/10.5498/wjp.v14.i3.445
Alzheimer's disease (AD) is a complicated and prevalent neurodegenerative disease that commonly occurs among older adults globally[1,2]. It is characterized by a progressive decline in cognitive ability and memory loss[3]. The deposition of Aβ-comprised extracellular plaques and neurofibrillary tangles are the main pathological hallmarks of AD[3]. Moreover, most patients with AD have cerebrovascular diseases, including impaired integrity of the blood-brain barrier[4]. An increasing number of epidemiological studies have shown a strong association between AD and type 2 diabetes mellitus (T2D), in which insulin resistance is a common and critical pathological feature[5,6]. However, the pathological mechanisms underlying the association between insulin resistance and AD remain unclear.
Histone acetyltransferases (HATs) are divided into different families according to their structure and sequence homology, including the P300/CBP, MYST, and GCN5 families[7]. The HATs play a central role in transcriptional regulation by catalyzing the transfer of acetyl from acetyl CoA to ε-amino of histone lysine residues[8]. Abnormal HAT function is closely correlated with various diseases, including developmental disorders and cancers[9-11]. HATs of the MYST family are characterized by conserved MYST catalytic domains, which include the KAT5 (TIP60), KAT6A (MOZ and MYST3), KAT6B (MORF and MYST4), KAT7 (HBO1 and MYST), and KAT8 (MOF)[12]. KAT7 acetylates the K14 and K23 on histone H3 by interacting with scaffolding protein BRPF and acetylates K5, K8, and K12 on histone H4 via scaffolding protein JADE[13,14]. During tissue development, depletion of KAT7 Leads to significantly decreased H3K14ac levels in erythrocytes of the fetal liver and mouse embryos[15].
Dual-specificity tyrosine phosphorylation-regulated kinase-1A (DYRK1A) is a highly conserved protein kinase that phosphorylates tyrosine and silk/threonine residues on exogenous substrates[16]. DYRK1A catalyzes multiple critical proteins, such as NOTCH, CREB, STAT3, eIF2B, and caspase-9[17]. Transgenic mice with high DYRK1A levels exhibit impaired motor and spatial learning abilities[18]. DYRK1A knockout mice died at the embryonic stage, and heterozygous mice exhibited low survival rates and abnormal neurological behavior[19]. DYRK1A has also been reported to participate in the development of AD, Down syndrome, diabetes, and cancer[20,21].
In this study, we explored the mechanisms underlying insulin resistance in AD and determined that KAT7 epigenetically upregulates the acetylation and expression of DYRK1A to reduce insulin resistance during AD. Our study identified novel therapeutic targets for AD.
Eight-month-old APPswe/PS1-dE9 double-transgenic mice were brought from Vital River Laboratory (China). The mice were randomly divided into experimental groups; the KAT7 overexpressing lentivirus (1 × 109 IU/mL) was stereotactically injected (3 µL/min) into the CA1 area of the hippocampus. All experiments were approved by the Animal Ethics Committee of the Qilu Hospital of Shandong University.
Twelve-week-old db/db and control mice were purchased from Vital River Laboratory (China). Brain tissues were collected from these mice, and protein expression was assessed via western blotting.
Primary neurons were isolated from mice and maintained in a specific culture medium at 37 °C in a humidified atmosphere containing 5% CO2[22]. To mimic insulin resistance, the cells were stimulated with culture medium containing insulin (3 μM), no foetal bovine serum, and no B27 for 24 h, followed by insulin deprivation for 30 min. The cells were stimulated with or without insulin (10 nM) for 15 min and collected for subsequent experiments.
A lentiviral system for KAT7 and HMGN1 overexpression and siRNAs targeting KAT7 (siKAT7) and HMGN1 (siHMGN1) were synthesized by GenePharma (Shanghai, China). Oligonucleotides were transfected into cells using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, United States), following the manufacturer’s instructions.
Cell viability was assessed using cell counting kit-8 (CCK-8) (Beyotime, China). Briefly, 5000 cells were seeded in each well of a 96-well plate and incubated for 24 h. Then, 20 μL CCK-8 reagent was added and hatched for another 2 h at 37 °C. Absorbance was measured at 450 nm using a microplate detector (Thermo Fisher Scientific). Apoptosis was assessed via flow cytometry using an Annexin V/PI Apoptosis Detection Kit (Beyotime, China).
For immunofluorescence staining, brain tissues were fixed and coated with optimal cutting temperature compound, made into 5 μm slices, and then probed with primary antibodies against microtubule-associated protein 2 (MAP2) overnight at 4 °C. The next day, samples were incubated with Alexa Fluor 633-conjugated secondary antibodies (Thermo Fisher Scientific) for 1 h at room temperature. Nuclei were labeled with DAPI (Thermo Fisher Scientific). Five random images were captured using a microscope (Leica, Germany).
Brain tissues and cells were homogenized using TRIzol reagent (Thermo Fisher Scientific) to extract total RNA, followed by reverse transcription to cDNA using the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific). Gene expression levels were quantified using the SYBR Green system (Thermo Fisher Scientific). Relative gene expression was normalized to that of GAPDH.
Total protein was obtained from brain tissues and cells using ice-cold RIPA lysis buffer (Thermo, United States) containing protease inhibitors (Sigma, United States). Equal amounts of proteins were separated via SDS-PAGE, blotted onto the PVDF membranes (Millipore, United States), blocked with 5% non-fat milk, and then hatched with anti-Aβ, anti-MAP2, anti-KAT7, anti-DYRK1A, anti-AKT, anti-pAKT, anti-GSK3β, and ani-β-actin for one night at 4 °C. The blots were visualized after incubation with secondary antibodies and ECL reagent (Millipore, United States). All the antibodies were purchased from Abcam and used according to the manufacturer’s instructions.
The levels of reactive oxygen species (ROS) were evaluated by staining with 2',7'-dichlorodihydrofluorescein diacetate (Sigma, United States) according to the manufacturer’s protocol. Samples were hatched with DCF-DA (25 μM) at 37 °C incubator in dark for 30 min. Relative fluorescence at 485 nm was measured using a microplate detector (Thermo, United States).
The levels of malondialdehyde (MDA) and superoxide dismutase[23] activity were assessed using MDA and SOD kits (Beyotime, China), according to the manufacturer’s instructions.
The chromatin immunoprecipitation (ChIP) assay was performed using the EZ-ChIP kit (Millipore, United States) according to the manufacturer’s instructions. Briefly, neurons were treated with formaldehyde for 10 min to obtain a crosslink between DNA and protein. Chromatin fragments were obtained after sonication of the cell lysates and incubation with an antibody targeting H3K27me3. The precipitated DNA was evaluated using quantitative PCR.
All data are presented as mean ± SD and were analyzed using SPSS software (SPSS, United States). Data comparisons between two groups or among multiple groups were conducted using Student’s t-test or one-way analysis of variance[24]. Statistical significance was set at P < 0.05, significant.
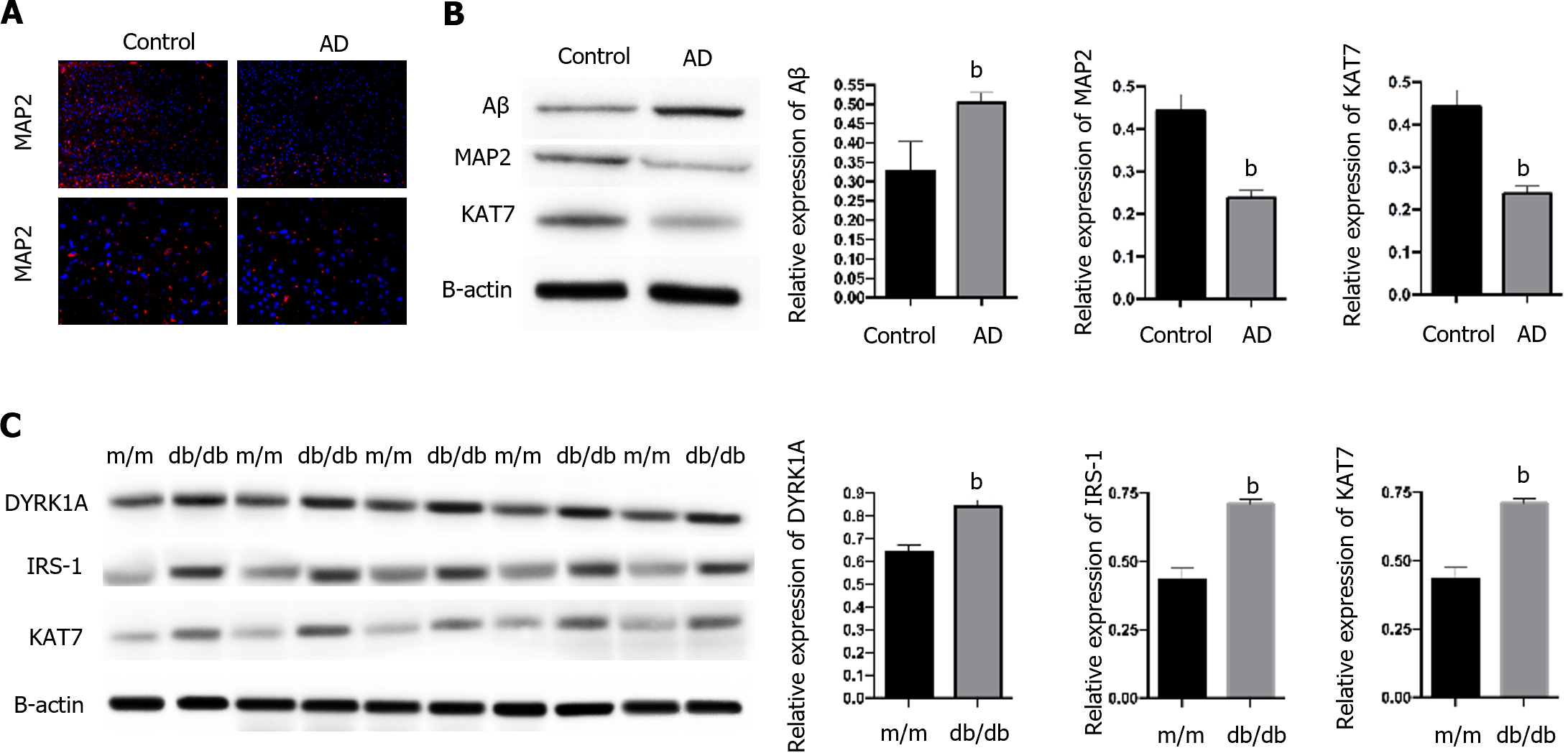
To determine the role of KAT7 in IR-induced AD, we established an in vivo AD model. We observed a notable accumulation of Aβ and decreased expression of MAP2, the biomarker of neuron generation (Figure 1A and B) in brain tissues from AD mice, compared with control mice, which suggested the successful establishment of the AD model. In contrast, we observed decreased KAT7 expression in the AD group (Figure 1A and B). In addition, KAT7 was coordinately overexpressed with IRS-1 and DYPK1A in diabetic mice (db/db) compared to that in normal mice (m/m), as shown in Figure 1C. The insulin receptor substrate-1 is an important regulator of insulin homeostasis, and its downregulation promotes insulin resistance[25,26]. Recent studies have indicated that DYPK1A/IRS-1 signaling represses insulin resistance[27]. Hence, we speculate that KAT7 may modulate insulin resistance in AD.
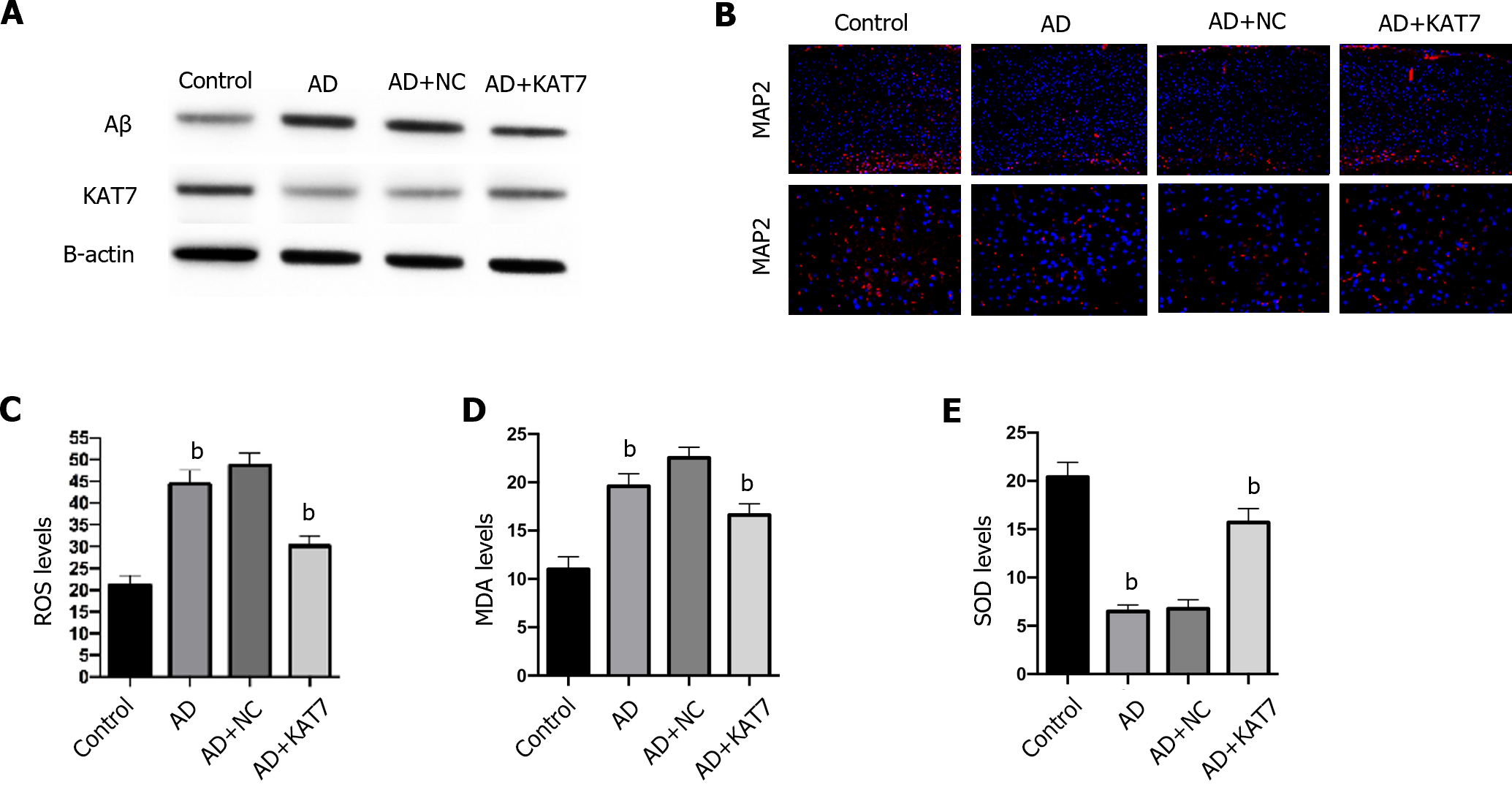
Next, we determined how KAT7 overexpression affected damage and oxidative stress in the brain. As shown in Figure 2A, treatment with KAT7 overexpression vectors led to significant elevation of KAT7 in brain tissues, along with decreased Aβ accumulation, which revered the phenotype of AD brains. KAT7 treatment also enhanced the proportion of MAP2-positive neurons compared to that in AD brains (Figure 2B). Moreover, AD brains exhibited elevated ROS accumulation, enhanced MDA levels, and decreased SOD activity, whereas KAT7 overexpression reversed these effects (Figure 2C-E).
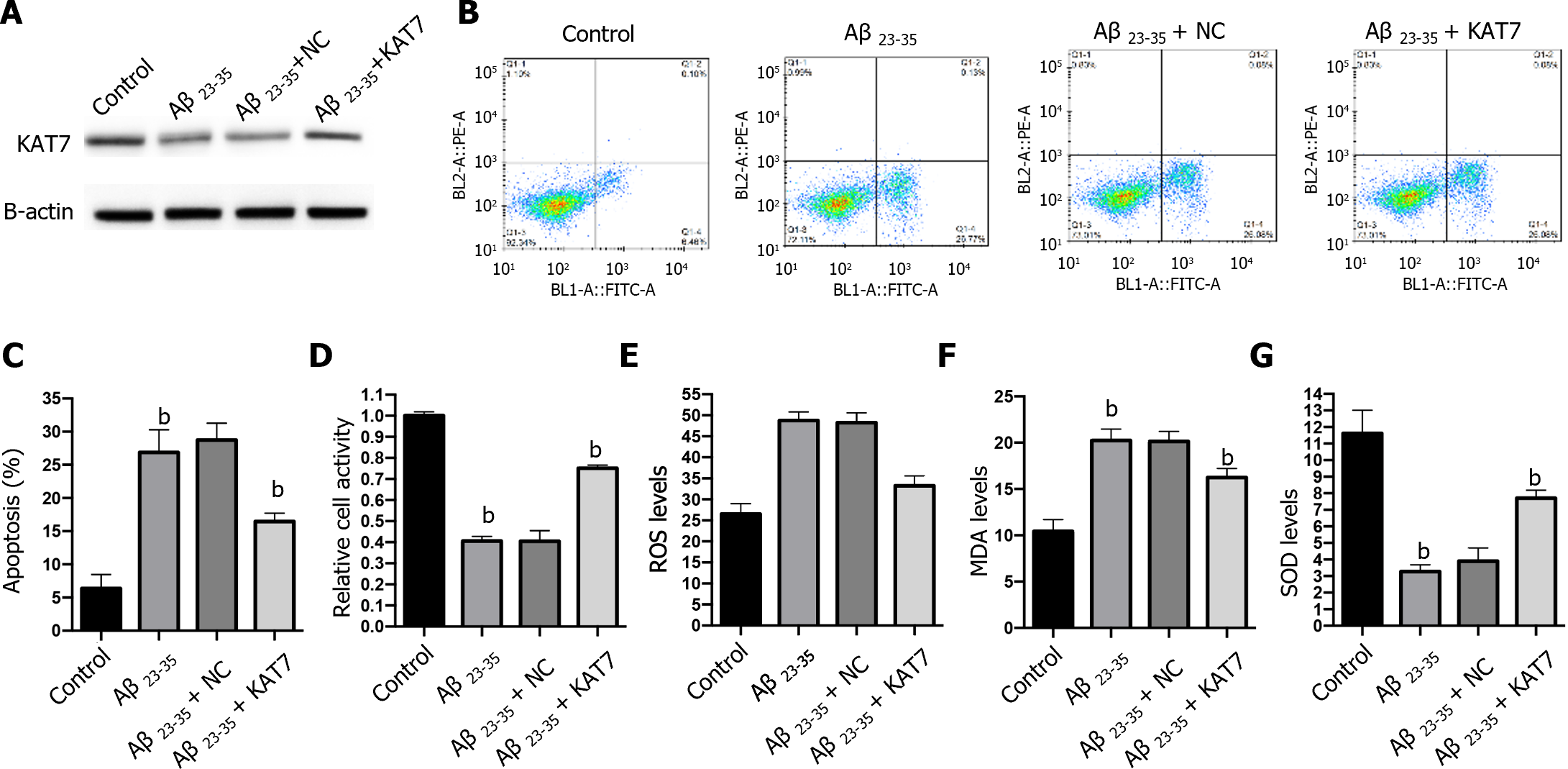
We also adopted an in vitro model to assess the effects of KAT7 overexpression on Aβ-induced neuron cell damage. Stimulation with Aβ repressed the expression of KAT7, and transfection with KAT7 vectors enhanced its protein levels in neurons (Figure 3A). Results from flow cytometry and CCK-8 demonstrated suppressed cell viability and increased apoptosis of neurons in the Aβ-stimulated cell model, whereas KAT7 overexpression recovered cell viability and alleviated cell apoptosis (Figure 3B-D). In contrast with the in vivo model, KAT7 also alleviated oxidative stress induced by Aβ (Figure 3E-G). These data indicated that KAT7 alleviated AD-induced neuronal cell death and oxidative stress.
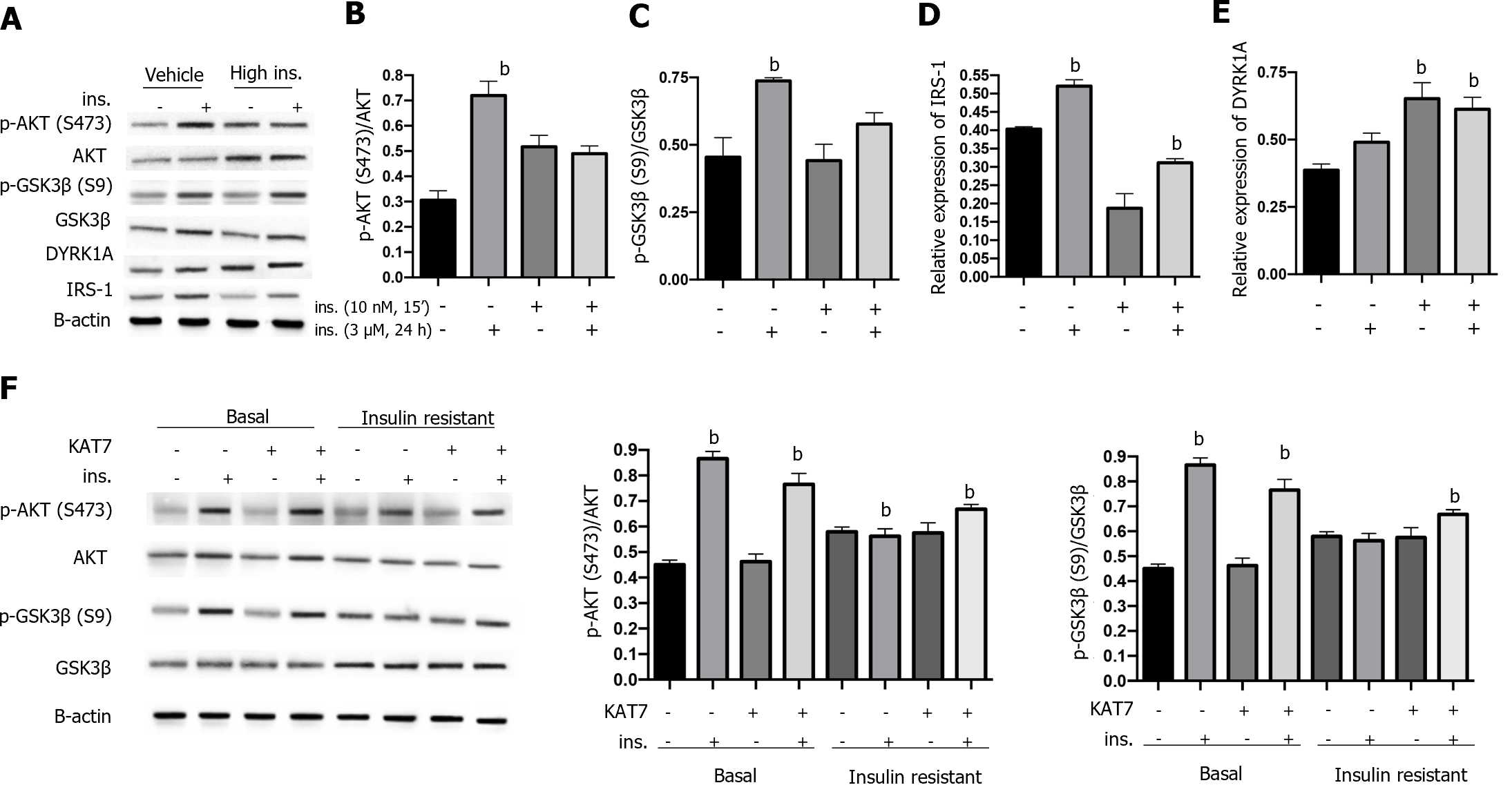
Insulin resistance can be caused by the sustained stimulation of high levels of insulin. Here, we first treated neurons with insulin (3 μM) for 24 h to achieve insulin resistance, and treatment with serum-free medium reached a basal status, followed by acute stimulation with 10 nM insulin for 15 min. As shown in Figure 4A-C, acute stimulation by insulin caused an elevated ratio of p-AKT and p-GSK3β in control neurons, indicating insulin sensitivity. In contrast, neurons pre-treated with insulin (3 μM) for 24 h presented no significant alteration of p-AKT and p-GSK3β ratio (Figure 4A-C), indicating the acquired insulin resistance. We also found that IRS-1 expression was decreased by pre-stimulation with insulin and was increased by acute stimulation (Figure 4D), consistent with previously reported findings. Notably, chronic stimulation with insulin caused increased expression of DYRK1A with or without insulin pre-stimulation (Figure 4E). Overexpression of KAT7 upregulated the sensitivity to insulin in both stimulated and basal neurons, manifested by elevated levels of p-AKT and p-GSK3β ratio (Figure 4F). These data suggest that KAT7 ameliorates chronic insulin-induced insulin resistance.
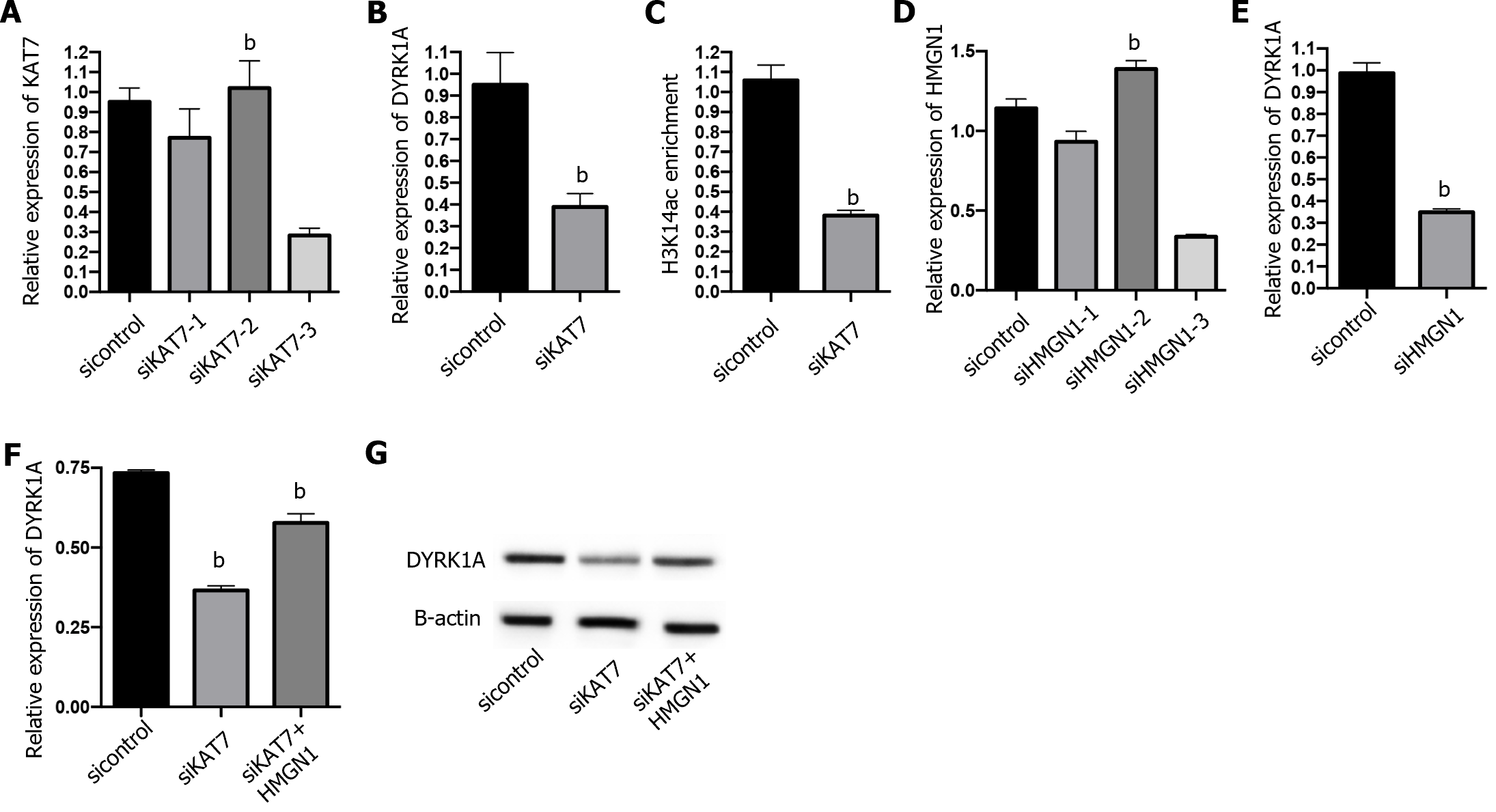
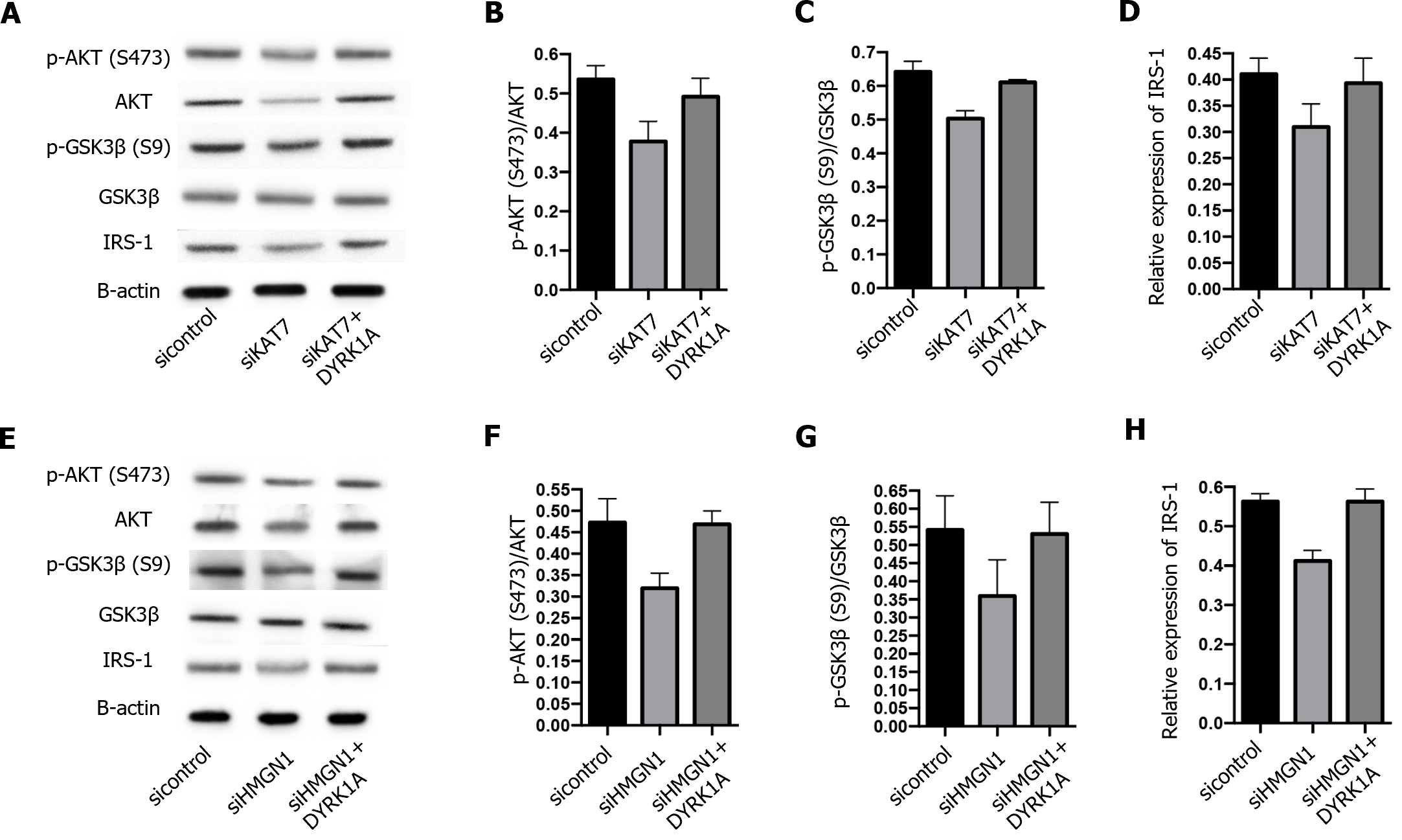
Next, we explored the downstream regulation of KAT7 during insulin resistance in AD. We depleted KAT7 in the neurons and evaluated the expression of DYRK1A. Transfection with siKAT7-3 effectively downregulated KAT7 and DYRK1A levels (Figure 5A and B). ChIP results revealed that the depletion of KAT7 alleviated the acetylation of K14 on histone 3 of DYRK1A (Figure 5C). Moreover, HMGN1 binds to the nucleosome and facilitates H4K14 acetylation[28]. We observed that siHMGN1-3 effectively suppressed HMGN1 and DYRK1A expression in neurons (Figure 5D and E). HMGN1 overexpression reversed both the RNA and protein levels of DYRK1A (Figure 5F and G). We used insulin-resistant neurons to evaluate the function of KAT7/HMGN1/DYRK1A. We observed that p-AKT, p-GSK3β, and IRS-1 expression were decreased by KAT7 knockdown (Figure 6A-D) or HMGN1 (Figure 6E-H), whereas overexpression of DYRK1A reversed this phenomenon. These findings indicate that KAT7 modulates DYRK1A expression by recruiting HMGN1 and ameliorating neuronal insulin resistance via DYRK1A/HMGN1 signaling.
Epidemiological and basic research studies have revealed a correlation between AD and T2D[4,29]. Diabetes is a novel risk factor for AD[5]. However, mechanisms underlying the correlation between AD and T2D remain unclear. Insulin resistance in the brain is a common feature in both T2D and AD[30]. Studies have reported that diabetic mice with cognitive disorders exhibit notable insulin resistance in the brain[31]. Accumulating evidence demonstrates that insulin resistance promotes Tau phosphorylation and Aβ plaques accumulation in AD brains[31]. Here, we established an in vivo AD model and determined a notable decrease in KAT7 expression in AD brains compared to control mice. KAT7 overexpression alleviated the accumulation of Aβ and increased MAP2 positive neurons, simultaneously suppressing oxidative stress and apoptosis of neurons, suggesting the protective function of KAT7 against AD.
DYRK1A is a protein kinase that phosphorylates serine and tyrosine residues of target proteins[18]. It has been reported that the dosage of DYRK1A is critical in the central nervous system during development and aging, and abnormal DYRK1A levels occur in neurodegenerative diseases, such as AD and Parkinson's disease[18]. Previous studies have reported that DYRK1A interacts with IRS-1 via serine phosphorylation[27]. In addition, DYRK1A inhibitors have been proposed as potential therapeutic agents for diabetes[32-34]. Consistently, we showed that both DYRK1A and IRS-1 were elevated in the brain tissue of diabetic mice, along with elevated KAT7 expression. IRS-1 is a critical factor that mediates insulin signal transduction, and decreased IRS-1 Levels are a feature of insulin resistance[35]. Studies have revealed that drugs that upregulate IRS-1 expression alleviate insulin resistance[36]. In this study, we established an insulin-resistant neuronal model by chronic stimulation with high levels of insulin. The levels of p-AKT and pGSK3β in established insulin-resistant neurons did not change under insulin stimulation, indicating the successful establishment of the model. Subsequently, we found that overexpression of KAT7 Led to elevated p-AKT and p-GSK3β levels.
KAT7 is a histone acetyltransferase that acetylates the K14 and K23 on histone H3 by interacting with scaffolding protein[13,14]. Here, we evaluated the acetylation of DYRK1A in neurons and determined the decreased enrichment of H3K14ac on DYRK1A upon depletion of KAT7. HMGN1 is a DNA-binding protein[37,38]. A recent study reported that HMGN1 could increase the acetylation H3K14 by enhancing the function of HATs[28]. Hence, we investigated whether KAT7 modulated DYRK1A expression by recruiting HMGN1. As expected, the depletion of HMGN1 downregulated DYRK1A and H3K14ac enrichment in DYRK1A cells. HMGN1 knockdown also recovered the phosphorylation of AKT and GSK3β in insulin-resistant neurons. However, the current study did not identify any direct interactions among KAT7, HMGN1, and DYRK1A. Verification of the KAT7–HMGN1–DYRK1A axis in an in vivo model requires further experiments.
In summary, we observed decreased KAT7 Levels in AD. Overexpression of KAT7 ameliorates neuronal death and oxidative stress in AD and restores insulin sensitivity in insulin-resistant neurons by recruiting HMGN1 to enhance DYRK1A acetylation. Our findings suggest that KAT7 is a potential therapeutic target for the treatment of insulin resistance in AD.
Epidemiological studies increasingly suggest a significant connection between Alzheimer's disease (AD) and type 2 diabetes mellitus, primarily attributed to insulin resistance, a prominent and pivotal pathological characteristic.
The precise pathological mechanisms that underlie the correlation between insulin resistance and AD remain elusive.
This study aims to investigate the impact of KAT7, a histone acetyltransferase involved in regulating multiple genes, on insulin resistance in AD.
APPswe/PS1-dE9 transgenic mice were employed to study AD, while db/db mice were utilized as a model for diabetes. An in vitro AD model was established through Aβ stimulation.
Overexpression of KAT7 decreased Aβ accumulation, alleviated ferroptosis and apoptosis in brain tissues and neurons. KAT7 epigenetically regulated the expression of DYRK1A via recruiting the HMGN1 and activated AKT and GSK3β to alleviate insulin resistance.
Our study revealed that upregulation of KAT7 restored insulin sensitivity in AD by recruiting HMGN1 to augment acetylation of the DYRK1A gene.
Our findings highlight KAT7 as a novel and promising therapeutic target for addressing insulin resistance in AD.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Modrego P, Spain S-Editor: Li L L-Editor: A P-Editor: Chen YX
| 1. | Lorente-Gea L, García B, Martín C, Quirós LM, Fernández-Vega I. Heparan sulfate proteoglycans and heparanases in Alzheimer's disease: current outlook and potential therapeutic targets. Neural Regen Res. 2017;12:914-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 13] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 2. | Unschuld PG. Novel Translational Research Methodology and the Prospect to a Better Understanding of Neurodegenerative Disease. Neurodegener Dis. 2018;18:1-4. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 3. | Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete NV, Coté S, De Simone A, Doig AJ, Faller P, Garcia A, Laio A, Li MS, Melchionna S, Mousseau N, Mu Y, Paravastu A, Pasquali S, Rosenman DJ, Strodel B, Tarus B, Viles JH, Zhang T, Wang C, Derreumaux P. Amyloid β Protein and Alzheimer's Disease: When Computer Simulations Complement Experimental Studies. Chem Rev. 2015;115:3518-3563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 463] [Cited by in RCA: 505] [Article Influence: 50.5] [Reference Citation Analysis (0)] |
| 4. | Pugazhenthi S, Qin L, Reddy PH. Common neurodegenerative pathways in obesity, diabetes, and Alzheimer's disease. Biochim Biophys Acta Mol Basis Dis. 2017;1863:1037-1045. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 476] [Article Influence: 52.9] [Reference Citation Analysis (0)] |
| 5. | Arnold SE, Arvanitakis Z, Macauley-Rambach SL, Koenig AM, Wang HY, Ahima RS, Craft S, Gandy S, Buettner C, Stoeckel LE, Holtzman DM, Nathan DM. Brain insulin resistance in type 2 diabetes and Alzheimer disease: concepts and conundrums. Nat Rev Neurol. 2018;14:168-181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 1039] [Article Influence: 148.4] [Reference Citation Analysis (0)] |
| 6. | Akhtar A, Sah SP. Insulin signaling pathway and related molecules: Role in neurodegeneration and Alzheimer's disease. Neurochem Int. 2020;135:104707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 169] [Article Influence: 33.8] [Reference Citation Analysis (0)] |
| 7. | Zhao M, Tao Y, Peng GH. The Role of Histone Acetyltransferases and Histone Deacetylases in Photoreceptor Differentiation and Degeneration. Int J Med Sci. 2020;17:1307-1314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 8. | Voss AK, Thomas T. Histone Lysine and Genomic Targets of Histone Acetyltransferases in Mammals. Bioessays. 2018;40:e1800078. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 92] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 9. | Baell JB, Leaver DJ, Hermans SJ, Kelly GL, Brennan MS, Downer NL, Nguyen N, Wichmann J, McRae HM, Yang Y, Cleary B, Lagiakos HR, Mieruszynski S, Pacini G, Vanyai HK, Bergamasco MI, May RE, Davey BK, Morgan KJ, Sealey AJ, Wang B, Zamudio N, Wilcox S, Garnham AL, Sheikh BN, Aubrey BJ, Doggett K, Chung MC, de Silva M, Bentley J, Pilling P, Hattarki M, Dolezal O, Dennis ML, Falk H, Ren B, Charman SA, White KL, Rautela J, Newbold A, Hawkins ED, Johnstone RW, Huntington ND, Peat TS, Heath JK, Strasser A, Parker MW, Smyth GK, Street IP, Monahan BJ, Voss AK, Thomas T. Inhibitors of histone acetyltransferases KAT6A/B induce senescence and arrest tumour growth. Nature. 2018;560:253-257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 115] [Cited by in RCA: 205] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 10. | Gomathi K, Akshaya N, Srinaath N, Rohini M, Selvamurugan N. Histone acetyl transferases and their epigenetic impact on bone remodeling. Int J Biol Macromol. 2021;170:326-335. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 11. | Sharma S, Sarathlal KC, Taliyan R. Epigenetics in Neurodegenerative Diseases: The Role of Histone Deacetylases. CNS Neurol Disord Drug Targets. 2019;18:11-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 12. | Wiesel-Motiuk N, Assaraf YG. The key roles of the lysine acetyltransferases KAT6A and KAT6B in physiology and pathology. Drug Resist Updat. 2020;53:100729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 58] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 13. | Yan MS, Turgeon PJ, Man HJ, Dubinsky MK, Ho JJD, El-Rass S, Wang YD, Wen XY, Marsden PA. Histone acetyltransferase 7 (KAT7)-dependent intragenic histone acetylation regulates endothelial cell gene regulation. J Biol Chem. 2018;293:4381-4402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 14. | Newman DM, Voss AK, Thomas T, Allan RS. Essential role for the histone acetyltransferase KAT7 in T cell development, fitness, and survival. J Leukoc Biol. 2017;101:887-892. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 22] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 15. | Yang Y, Kueh AJ, Grant ZL, Abeysekera W, Garnham AL, Wilcox S, Hyland CD, Di Rago L, Metcalf D, Alexander WS, Coultas L, Smyth GK, Voss AK, Thomas T. The histone lysine acetyltransferase HBO1 (KAT7) regulates hematopoietic stem cell quiescence and self-renewal. Blood. 2022;139:845-858. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 46] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 16. | Bhansali RS, Rammohan M, Lee P, Laurent AP, Wen Q, Suraneni P, Yip BH, Tsai YC, Jenni S, Bornhauser B, Siret A, Fruit C, Pacheco-Benichou A, Harris E, Besson T, Thompson BJ, Goo YA, Hijiya N, Vilenchik M, Izraeli S, Bourquin JP, Malinge S, Crispino JD. DYRK1A regulates B cell acute lymphoblastic leukemia through phosphorylation of FOXO1 and STAT3. J Clin Invest. 2021;131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 61] [Article Influence: 15.3] [Reference Citation Analysis (0)] |
| 17. | Bellmaine SF, Ovchinnikov DA, Manallack DT, Cuddy CE, Elefanty AG, Stanley EG, Wolvetang EJ, Williams SJ, Pera M. Inhibition of DYRK1A disrupts neural lineage specificationin human pluripotent stem cells. Elife. 2017;6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 18. | Arbones ML, Thomazeau A, Nakano-Kobayashi A, Hagiwara M, Delabar JM. DYRK1A and cognition: A lifelong relationship. Pharmacol Ther. 2019;194:199-221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 113] [Article Influence: 18.8] [Reference Citation Analysis (0)] |
| 19. | Watson-Scales S, Kalmar B, Lana-Elola E, Gibbins D, La Russa F, Wiseman F, Williamson M, Saccon R, Slender A, Olerinyova A, Mahmood R, Nye E, Cater H, Wells S, Yu YE, Bennett DLH, Greensmith L, Fisher EMC, Tybulewicz VLJ. Analysis of motor dysfunction in Down Syndrome reveals motor neuron degeneration. PLoS Genet. 2018;14:e1007383. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 20. | Laham AJ, Saber-Ayad M, El-Awady R. DYRK1A: a down syndrome-related dual protein kinase with a versatile role in tumorigenesis. Cell Mol Life Sci. 2021;78:603-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 12.8] [Reference Citation Analysis (0)] |
| 21. | García-Cerro S, Rueda N, Vidal V, Lantigua S, Martínez-Cué C. Normalizing the gene dosage of Dyrk1A in a mouse model of Down syndrome rescues several Alzheimer's disease phenotypes. Neurobiol Dis. 2017;106:76-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 69] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 22. | Zhang X, Wei M, Fan J, Yan W, Zha X, Song H, Wan R, Yin Y, Wang W. Ischemia-induced upregulation of autophagy preludes dysfunctional lysosomal storage and associated synaptic impairments in neurons. Autophagy. 2021;17:1519-1542. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 154] [Article Influence: 38.5] [Reference Citation Analysis (0)] |
| 23. | Matsushita M, Hasegawa S, Kitoh H, Mori K, Ohkawara B, Yasoda A, Masuda A, Ishiguro N, Ohno K. Meclozine promotes longitudinal skeletal growth in transgenic mice with achondroplasia carrying a gain-of-function mutation in the FGFR3 gene. Endocrinology. 2015;156:548-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 46] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 24. | Ruiz de Galarreta M, Bresnahan E, Molina-Sánchez P, Lindblad KE, Maier B, Sia D, Puigvehi M, Miguela V, Casanova-Acebes M, Dhainaut M, Villacorta-Martin C, Singhi AD, Moghe A, von Felden J, Tal Grinspan L, Wang S, Kamphorst AO, Monga SP, Brown BD, Villanueva A, Llovet JM, Merad M, Lujambio A. β-Catenin Activation Promotes Immune Escape and Resistance to Anti-PD-1 Therapy in Hepatocellular Carcinoma. Cancer Discov. 2019;9:1124-1141. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 620] [Article Influence: 103.3] [Reference Citation Analysis (0)] |
| 25. | Guo S. Insulin signaling, resistance, and the metabolic syndrome: insights from mouse models into disease mechanisms. J Endocrinol. 2014;220:T1-T23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 235] [Cited by in RCA: 347] [Article Influence: 31.5] [Reference Citation Analysis (0)] |
| 26. | Xuguang H, Aofei T, Tao L, Longyan Z, Weijian B, Jiao G. Hesperidin ameliorates insulin resistance by regulating the IRS1-GLUT2 pathway via TLR4 in HepG2 cells. Phytother Res. 2019;33:1697-1705. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 35] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 27. | Tian S, Jia W, Lu M, Zhao J, Sun X. Dual-specificity tyrosine phosphorylation-regulated kinase 1A ameliorates insulin resistance in neurons by up-regulating IRS-1 expression. J Biol Chem. 2019;294:20164-20176. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 28. | Lim JH, West KL, Rubinstein Y, Bergel M, Postnikov YV, Bustin M. Chromosomal protein HMGN1 enhances the acetylation of lysine 14 in histone H3. EMBO J. 2005;24:3038-3048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 29. | Burillo J, Marqués P, Jiménez B, González-Blanco C, Benito M, Guillén C. Insulin Resistance and Diabetes Mellitus in Alzheimer's Disease. Cells. 2021;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 119] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 30. | Tumminia A, Vinciguerra F, Parisi M, Frittitta L. Type 2 Diabetes Mellitus and Alzheimer's Disease: Role of Insulin Signalling and Therapeutic Implications. Int J Mol Sci. 2018;19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 106] [Cited by in RCA: 175] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 31. | Shieh JC, Huang PT, Lin YF. Alzheimer's Disease and Diabetes: Insulin Signaling as the Bridge Linking Two Pathologies. Mol Neurobiol. 2020;57:1966-1977. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 79] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 32. | Guo Y, Li L, Yao Y, Li H. Regeneration of Pancreatic β-Cells for Diabetes Therapeutics by Natural DYRK1A Inhibitors. Metabolites. 2022;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 33. | Kumar K, Suebsuwong C, Wang P, Garcia-Ocana A, Stewart AF, DeVita RJ. DYRK1A Inhibitors as Potential Therapeutics for β-Cell Regeneration for Diabetes. J Med Chem. 2021;64:2901-2922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 34. | Liu YA, Jin Q, Zou Y, Ding Q, Yan S, Wang Z, Hao X, Nguyen B, Zhang X, Pan J, Mo T, Jacobsen K, Lam T, Wu TY, Petrassi HM, Bursulaya B, DiDonato M, Gordon WP, Liu B, Baaten J, Hill R, Nguyen-Tran V, Qiu M, Zhang YQ, Kamireddy A, Espinola S, Deaton L, Ha S, Harb G, Jia Y, Li J, Shen W, Schumacher AM, Colman K, Glynne R, Pan S, McNamara P, Laffitte B, Meeusen S, Molteni V, Loren J. Selective DYRK1A Inhibitor for the Treatment of Type 1 Diabetes: Discovery of 6-Azaindole Derivative GNF2133. J Med Chem. 2020;63:2958-2973. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 53] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 35. | Ardestani A, Maedler K. mTORC1 and IRS1: Another Deadly Kiss. Trends Endocrinol Metab. 2018;29:737-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 36. | Wang W, Tanokashira D, Fukui Y, Maruyama M, Kuroiwa C, Saito T, Saido TC, Taguchi A. Serine Phosphorylation of IRS1 Correlates with Aβ-Unrelated Memory Deficits and Elevation in Aβ Level Prior to the Onset of Memory Decline in AD. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 37. | Yang D, Han Z, Alam MM, Oppenheim JJ. High-mobility group nucleosome binding domain 1 (HMGN1) functions as a Th1-polarizing alarmin. Semin Immunol. 2018;38:49-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 38. | Zhu N, Hansen U. HMGN1 modulates estrogen-mediated transcriptional activation through interactions with specific DNA-binding transcription factors. Mol Cell Biol. 2007;27:8859-8873. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 21] [Article Influence: 1.2] [Reference Citation Analysis (0)] |