Published online Jan 19, 2024. doi: 10.5498/wjp.v14.i1.88
Peer-review started: July 31, 2023
First decision: November 1, 2023
Revised: November 8, 2023
Accepted: December 7, 2023
Article in press: December 7, 2023
Published online: January 19, 2024
Processing time: 172 Days and 7.5 Hours
Early diagnosis and therapeutic interventions can greatly enhance the developmental trajectory of children with autism spectrum disorder (ASD). However, the etiology of ASD is not completely understood. The presence of confounding factors from environment and genetics has increased the difficulty of the identification of diagnostic biomarkers for ASD.
To estimate and interpret the causal relationship between ASD and metabolite profile, taking into consideration both genetic and environmental influences.
A two-sample Mendelian randomization (MR) analysis was conducted using summarized data from large-scale genome-wide association studies (GWAS) including a metabolite GWAS dataset covering 453 metabolites from 7824 European and an ASD GWAS dataset comprising 18381 ASD cases and 27969 healthy controls. Metabolites in plasma were set as exposures with ASD as the main outcome. The causal relationships were estimated using the inverse variant weight (IVW) algorithm. We also performed leave-one-out sensitivity tests to validate the robustness of the results. Based on the drafted metabolites, enrichment analysis was conducted to interpret the association via constructing a protein-protein interaction network with multi-scale evidence from databases including Infinome, SwissTargetPrediction, STRING, and Metascape.
Des-Arg(9)-bradykinin was identified as a causal metabolite that increases the risk of ASD (β = 0.262, SE = 0.064,
Through the application of MR, this study provides practical insights into the potential causal association between plasma metabolites and ASD. These findings offer perspectives for the discovery of diagnostic or predictive biomarkers to support clinical practice in treating ASD.
Core Tip: This study employs Mendelian randomization to uncover a potential causal relationship between plasma metabolites and autism spectrum disorder (ASD), emphasizing the role of Des-Arg(9)-bradykinin in increasing the risk of ASD. The findings underscore the significance of neuroinflammation and the response to stimulus as possible mediating factors, offering new directions for the development of diagnostic and predictive biomarkers in the clinical management of ASD.
- Citation: Huang ZY, Lyu ZP, Li HG, You HZ, Yang XN, Cha CH. Des-Arg(9) bradykinin as a causal metabolite for autism spectrum disorder. World J Psychiatry 2024; 14(1): 88-101
- URL: https://www.wjgnet.com/2220-3206/full/v14/i1/88.htm
- DOI: https://dx.doi.org/10.5498/wjp.v14.i1.88
Autism spectrum disorder (ASD) is an early-onset neurodevelopmental disorder characterized by persistent deficits in social communication and social interaction, coupled with restricted and repetitive patterns of behavior and interests[1]. The negative impacts of ASD extend across biological, social, and cognitive functions, significantly influencing the developmental trajectory of affected children[2]. Achieving an accurate early diagnosis of ASD is crucial for enhancing the quality of life in adulthood. However, current ASD diagnosis criteria rely on behavioral pattern analysis and assessments. The early-stage clinical presentations are heterogeneous and inconspicuous, making a definitive ASD diagnosis challenging. There is a need to optimize diagnostic approaches for ASD to better align with clinical practice[3].
There is growing interest in identifying diagnostic biomarkers to support early diagnosis and effective treatments for ASD[4]. To unravel the etiology and pathogenesis of ASD, interdisciplinary approaches have been employed, gathering multi-scale evidence from genomics, metabolomics, transcriptomics, and beyond. Accumulating evidence suggests that ASD development is influenced by both genetic and environmental factors[5,6]. Risk genes such as CHD8, DYRK1A and SHANK3 have been identified. These genes formed a functional network that indicates potential biological processes involved in the development of ASD. Moreover, genetic models have integrated risk genes such as NRXN2 and DYRK1A, which are involved in neurodevelopment, to explain the inherited patterns and heterogeneity observed in ASD[7,8]. Maternal environmental factors, including inflammation and the immune response, have also been recognized as crucial contributors to ASD development[9,10]. These findings provide essential information for developing diagnostic instruments for ASD. However, due to the complexity of interactions among biological processes and the existence of undetectable confounding factors, it is still challenging to identify biomarkers on an appropriate scale, ensuring effective implementation with sufficient biological validity[11].
The utilization of metabolomics technology for estimating metabolic profiles has advanced the development of workflows in biomarker research[12]. As detectable variables reflecting both genomic and environmental effects, metabolites that participate in mitochondrial dysfunction were identified as potential biomarkers of ASD[13-15]. However, the limitations stemming from the availability of large biological samples and technical barriers in sample preparation have impeded the attainment of standardized and reproducible results in metabolomics studies focused on ASD[16]. Additionally, confounding factors should be well handled to identify a reliable biomarker from the association between metabolites and ASD. Therefore, it is imperative to explore practical methods for identifying metabolic biomarkers that can address the shortcomings of current paradigms.
Mendelian randomization (MR) analysis utilizes genetic variants as instrumental variables to investigate the causal relationships between exposures and outcomes in observational studies[17]. Two-sample MR offers increased statistical power and enhanced flexibility by gathering instrumental variable-exposure association and instrumental variable outcome association from two different sets of participants to examine the causal relationship between an exposure and an outcome[18]. This approach eliminates the need to measure both exposure and outcome variables in the same sample, making it particularly useful in cases where the exposure or outcome is rare, challenging to measure, or when there is limited sample overlap between exposure and outcome studies[19]. Therefore, aiming at identifying candidate biomarkers to assist early diagnosis of ASD, this study attempted to uncover potential associations between metabolites and ASD taking advantage of the two sample MR. To strengthen the interpretation of the potential association, enrichment analysis was carried out with the integration of multi-scale evidence.
We implemented a two-sample MR design using datasets of European ancestry, chosen due to their public availability, ample sample size, and population consistency. Specifically, we selected two datasets for this analysis. Firstly, we utilized the metabolite genome-wide association study (mGWAS) published by Shin et al[20] in 2014. This dataset provided genetic association evidence for 486 metabolites and included data from 7824 individuals across two European studies. Additionally, we employed the meta-analysis of ASD conducted by the Lundbeck Foundation Initiative for Integrative Psychiatric Research (iPSYCH) and the Psychiatric Genomics Consortium (PGC) in 2017[21]. This ASD dataset consisted of a total of 18381 ASD cases and 27969 healthy controls as shown in Table 1. These datasets were publicly available, and any missing frequency data in the ASD GWAS dataset were acquired by contacting the authors via email in July 2022.
| Variable | Ancestry | Ref. | Sample size | Sex | Data download |
| Metabolite | European | Shin et al[20], 2014 | 7824 | Mixed | http://metabolomics.helmholtz-muenchen.de/gwas/ |
| Autism spectrum disorder | European | Grove et al[21], 2017 | Cases = 18381; Controls = 27969 | Mixed | https://doi.org/10.6084/m9.figshare.14671989 |
The two-sample MR analysis aimed to estimate the associations between metabolite traits (from the mGWAS) and ASD (from the iPSYCH-PGC) using genetic variants as instrumental variables. The effect size was calculated as the association between a 1-standard deviation change in metabolite trait levels and ASD, with the odds ratio (OR) as the main result. This approach provided quantified potential directional associations between ASD and metabolite traits.
Quality control: To ensure the reliability of the association between metabolite and ASD, the genetic variances that exhibited a significant association (P < 5 × 10-8) with the metabolite from the mGWAS dataset were selected. Moreover, acknowledging the potential no-random association of alleles at two or more loci, especially those in close genomic proximity, a linkage disequilibrium (LD) analysis was performed using the European 1000G reference panel to exclude single nucleotide polymorphisms (SNPs) with a correlation coefficient (r2) less than 0.01 within a 500-kb window. The clumping procedure was conducted using the TwoSampleMR R package[22].
Assumptions: Firstly, the relevance assumption postulated that the genetic variables used as instrumental variables were associated with the metabolite of interest. Secondly, the independence assumption stated that the genetic variables were independent of potential confounding factors. Thirdly, the exclusion restriction assumption proposed that the disease outcome was solely influenced by the genetic instrument through the metabolites and not through alternative pathways. Additionally, as MR-Egger regression was employed to obtain pleiotropy-corrected causal estimates, a fourth assumption was made that the association between an SNP and the exposure variable was independent of its direct effects on the outcome.
In the two-sample MR analyses, the causal effects of metabolites on the disease were assessed using the inverse variance weighted (IVW) method. The analyses were conducted using the TwoSampleMR and MR-PRESSO packages in R 3.6.2[22,23] with statistical significance defined as P < 0.05. The weighted median, Wald ratio, weighted mode, and MR-Egger algorithm were also employed to evaluate the consistency of the causal inferences, thereby safeguarding false positive results. Sensitivity analysis was performed to investigate the robustness of the estimated significant MR causal associations. To test the robustness of the results and investigate the influence of individual SNPs, a global test and the leave-one-out method were employed. Forest plots were used to show the consistency of the effect of each SNP. Horizontal pleiotropy, which refers to the presence of genetic variants affecting both the exposure and the outcome through different pathways, was assessed using the MR pleiotropy residual sum and outlier global test.
Genes near the SNPs were searched and collected on the Infinome website. DrugBank provided information on the structure, SMILE representation, and validated target genes of the quantified causal metabolite. SwissTargetPrediction was used to predict target genes based on structural similarity and molecular affinity. In this study, we collected the top 10 predicted target genes of the metabolites for further interpretation of the causal relationship between metabolite and ASD. Meanwhile, previously reported ASD-related genes were retrieved from the Malacard dataset. To further discover potential biological clues for interpreting the causal association between metabolites and ASD, a protein-protein interaction network was constructed using the STRING database. Then enrichment analysis was performed using GO and KEGG. Cluster modules of the interaction network were identified using Metascape. Maximal cliques are identified and genes with high clique centrality in the network were considered as hub genes through the maximal clique centrality (MCC) algorithm in Cystoscape 3.9.1, utilizing the interaction network.
The ASD GWAS dataset covered 9112386 SNPs from the iPSYCH-PGC consortium, and there were 207856 SNPs of 453 metabolites in the mGWAS dataset. After filtering based on the significance of the correlation between SNPs and metabolites (P < 5 × 10-8), a subset of 18824 SNPs was used for LD clumping, resulting in 609 SNPs as representative genetic variants for the MR analysis.
The association between each metabolite and ASD was estimated using five different methods. Among the evaluated associations, 11 were deemed significant (P < 0.05) with the IVW method, as shown in Table 2. Three metabolites, including M32847, M33137, and M34306, were excluded from further analysis as neither their structures nor their identification were available in the original study.
| ID | Metabolite | Outcome | SNP, n | β | SE | P value |
| M02137 | Biliverdin | ASD | 3 | -0.255 | 0.122 | 0.038 |
| M20675 | 1,5-Anhydroglucitol | ASD | 5 | -0.881 | 0.407 | 0.031 |
| M32412 | Butyrylcarnitine | ASD | 12 | -0.219 | 0.111 | 0.049 |
| M32497 | 10-undecenoate (11:1n1) | ASD | 3 | 0.546 | 0.258 | 0.034 |
| M32847 | X-11530 | ASD | 3 | -0.302 | 0.125 | 0.016 |
| M33137 | X-11792 | ASD | 3 | 0.388 | 0.093 | 3.03 × 10-5 |
| M33138 | Oxidized bilirubin | ASD | 3 | -0.318 | 0.148 | 0.032 |
| M34306 | X-12696 | ASD | 4 | -0.831 | 0.325 | 0.011 |
| M34407 | Isovalerylcarnitine | ASD | 4 | -0.976 | 0.393 | 0.013 |
| M34420 | Des-Arg(9) bradykinin | ASD | 3 | 0.262 | 0.064 | 4.64 × 10-5 |
| M36131 | Alpha-glutamyltyrosine | ASD | 3 | -0.619 | 0.213 | 0.004 |
Among the remaining metabolites, two were found to be positively associated with ASD: M34420: Des-Arg(9) bradykinin (β = 0.262, SE = 0.064, P = 4.64 × 10-5) and M32497: 10-undecenoate (11:1n1) (β = 0.546, SE = 0.258, P = 0.034). Additionally, six metabolites were identified as protective factors for ASD: M02137: Biliverdin (β = -0.255, SE = 0.122, P = 0.038), M20675: 1,5-Anhydroglucitol (β = -0.881, SE = 0.407, P = 0.031), M32412: Butyrylcarnitine (β = -0.219, SE = 0.111, P = 0.0497), M33138: Oxidized bilirubin (β = -0.318, SE = 0.148, P = 0.032), M34407: Isovalerylcarnitine (β = -0.976, SE = 0.393, P = 0.013), and M36131: Alpha-glutamyltyrosine (β = -0.619, SE = 0.213, P = 0.004).
Scatterplots illustrating the relationship between these eight metabolites and ASD are presented in Figure 1. The consistency across the SNPs of each metabolite, except M20675: 1,5-Anhydroglucitol, M32412: Butyrylcarnitine, and M34407: Isovalerylcarnitine, was evaluated as adequate with heterogeneity P > 0.05 and pleiotropy assumption was satisfied with P > 0.05 as shown in Table 3.
| Metabolite | Heterogeneity, MR Egger P value | Heterogeneity, IVW P value | Pleiotropy, P value |
| Biliverdin | 0.392 | 0.415 | 0.496 |
| 1,5-anhydroglucitol | 0.482 | 0.034 | 0.067 |
| Butyrylcarnitine | 0.174 | 0.007 | 0.016 |
| 10-Undecenoate (11:1n1) | 0.844 | 0.689 | 0.556 |
| Oxidized bilirubin | 0.271 | 0.385 | 0.586 |
| Isovalerylcarnitine | 0.030 | 0.058 | 0.744 |
| Des-Arg(9) bradykinin | 0.663 | 0.906 | 0.949 |
| Alpha-glutamyltyrosine | 0.785 | 0.923 | 0.817 |
However, since the heterogeneity and pleiotropy tests were not satisfied, it suggests that the assumption of instrumental variance was violated. This may be due to inconsistent estimates from the genetic variants of the metabolites or the presence of potential confounding affecting both the exposure and outcome. Consequently, these three metabolites were excluded at this stage.
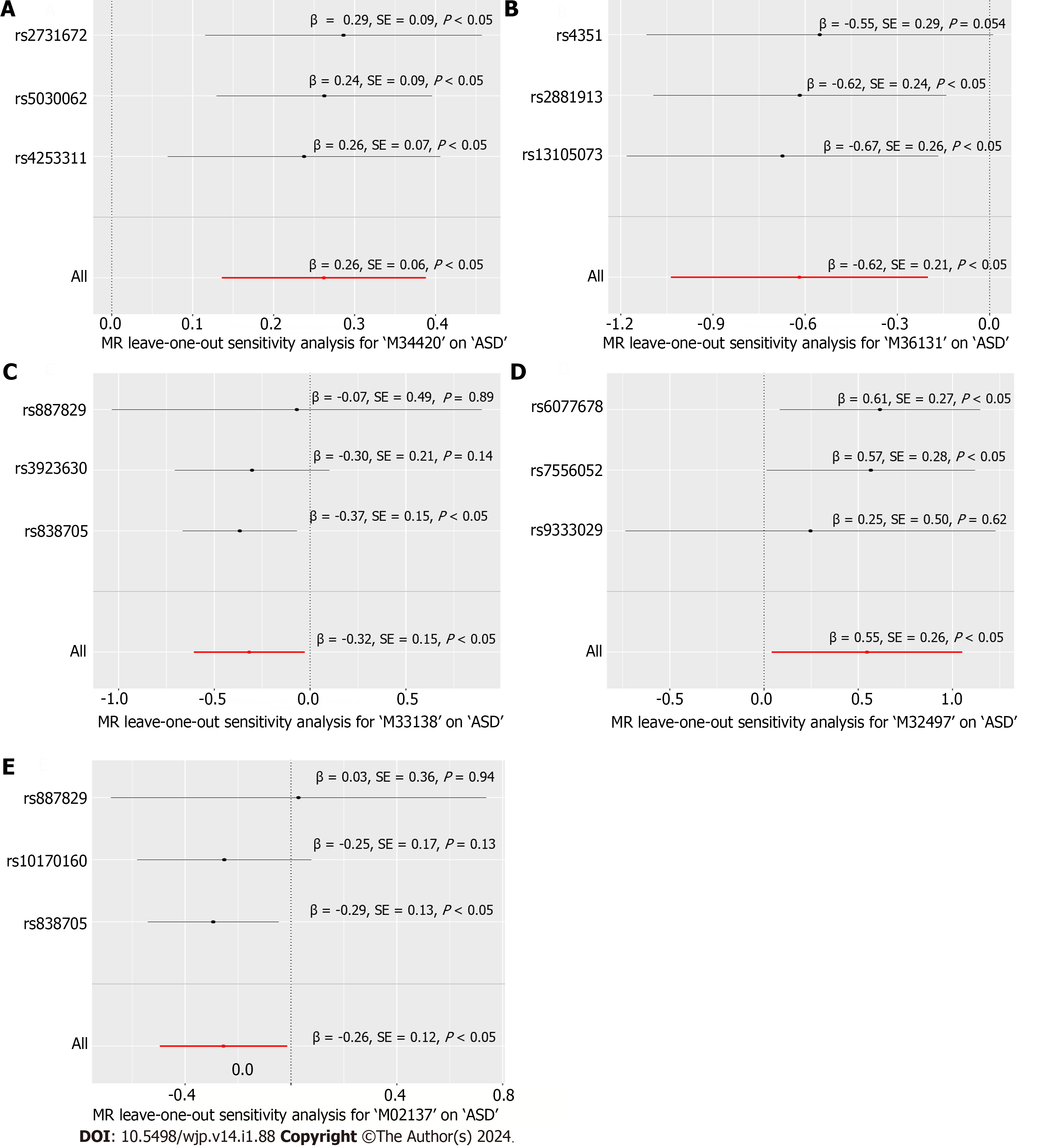
We conducted a leave-one-out sensitivity analysis for the remaining five metabolites, and the corresponding forest plots are presented in Figure 2. The consistency of the estimated effect size for Des-Arg(9) bradykinin about ASD was assessed as adequate through this leave-one-out sensitivity analysis, which involved systematically excluding each SNP from the analysis. As a result, Des-Arg(9) bradykinin was ultimately identified as a causal metabolite for ASD.
The assumption regarding the consistency among SNPs influencing ASD through M02137: Biliverdin, M32497: 10-undecenoate (11:1n1), M33138: Oxidized bilirubin, and M36131: Alpha-glutamyltyrosine was found to be unsatisfactory, as indicated by the p-value of the SNP. To put it more intuitively, certain SNPs were observed to cross the vertical null value line in the horizontal line plot.
To interpret the potential biological processes and pathways mediating the association between Des-Arg(9) bradykinin and ASD, evidence on gene-metabolite-disease interactions is collected and estimated. Firstly, nearby genes of the location of the SNPs were gathered from the Infinome database, revealing that F12, GRK6 and PFN3 were in proximity to rs2731672, while KNG1 was near rs5030062 and KLKB1 near rs4253311. Secondly, supported by the SwissTargetPrediction database, 10 predicted targets ranking on top of the predicted list of Des-Arg(9) bradykinin were retrieved and listed in Table 4. Among the predicted targets, the B1 receptor of bradykinin ranked the 1st which is consistent with the fact that the B1 receptor performed as a specific receptor for Des-Arg(9) bradykinin. Other markers were predicted as potential targets of Des-Arg(9)-bradykinin including NTSR1, NTSR2, OPRM1, OPRD1, OPRK1, F2, CCNA1, CCNA2, CDK2, HLA-A and HLA-DRB3. These targets are used as clues to discover potential biological interaction between Des-Arg(9)-bradykinin and ASD.
| Target | Common name | Target class | Probability |
| Bradykinin B1 receptor | BDKRB1 | Family A G protein-coupled receptor | 0.420 |
| Neurotensin receptor 1 | NTSR1 | Family A G protein-coupled receptor | 0.189 |
| Thrombin | F2 | Protease | 0.169 |
| HLA class I histocompatibility antigen A-3 | HLA-A | Surface antigen | 0.140 |
| Mu opioid receptor | OPRM1 | Family A G protein-coupled receptor | 0.131 |
| Delta opioid receptor | OPRD1 | Family A G protein-coupled receptor | 0.131 |
| Kappa opioid receptor | OPRK1 | Family A G protein-coupled receptor | 0.122 |
| Neurotensin receptor 2 | NTSR2 | Family A G protein-coupled receptor | 0.122 |
| HLA class II histocompatibility antigen DRB3-1 | HLA-DRB3 | Surface antigen | 0.122 |
| Cyclin-dependent kinase 2/cyclin A | CDK2, CCNA1, CCNA2 | Other cytosolic protein | 0.122 |
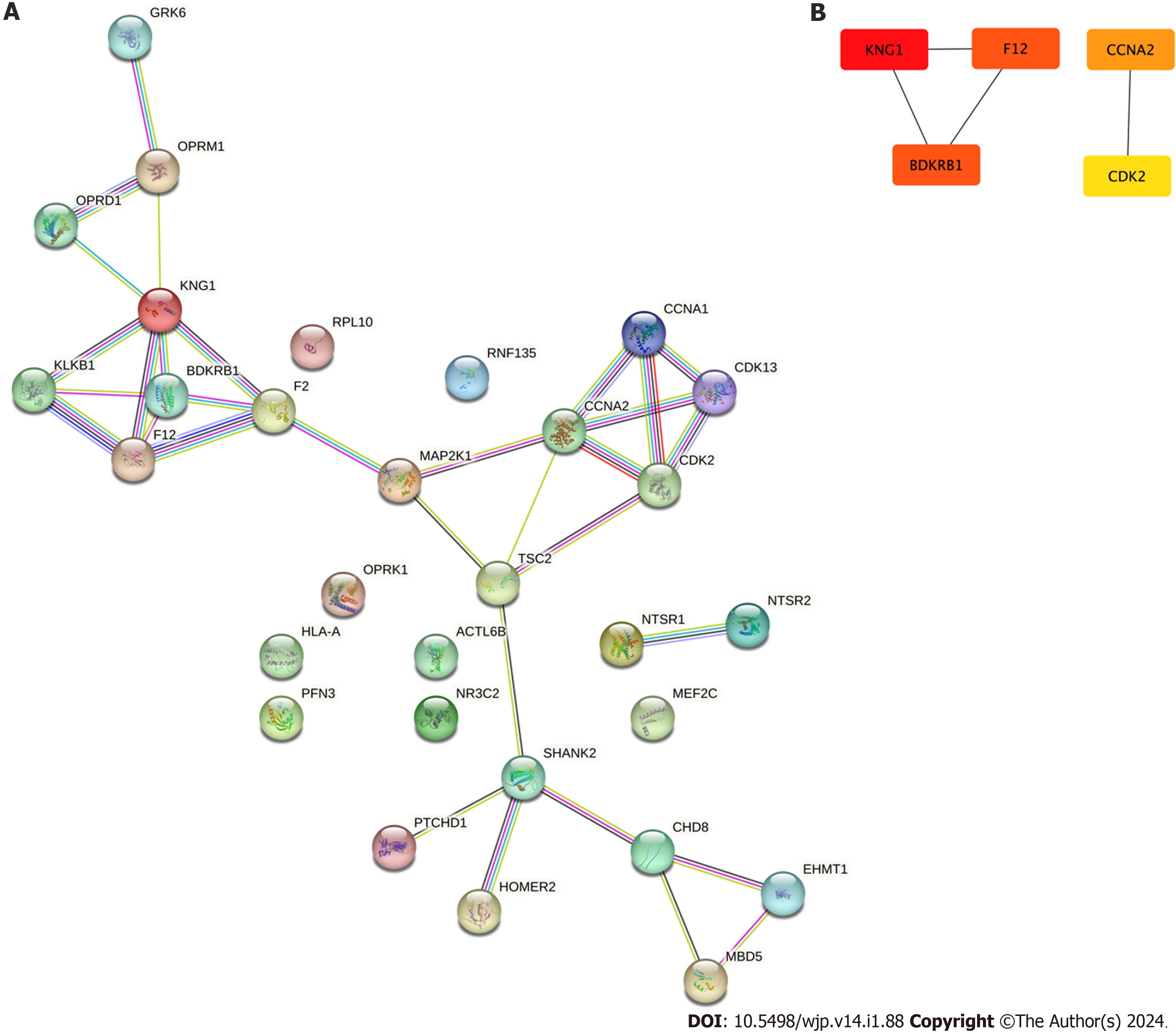
In the STRING database, a protein-protein interaction network was constructed based on a union set of the ASD-related gene set and the Des-Arg(9) bradykinin-related gene set as presented in Figure 3A. Pathways in the network represented potential interaction processes between Des-Arg(9) bradykinin and ASD. Furthermore, as shown in Figure 3B, five hub genes including KNG1, F12, BDKRB1, CCNA2 and CDK2 were identified with the MCC algorithm in the interaction network. These genes could play important direct or indirect roles in modulating the biological activity of other makers of the network.
To further explore the potential biological processes bridging Des-Arg(9) bradykinin and ASD, enrichment analysis was performed and the result is shown in Figure 4. Genes in the interaction network were enriched in biological processes including Peptide GPCRs, behavior, cellular senescence, multicellular organismal processes, response to stimuli, metabolic processes, immune system processes, and developmental processes. These findings provide valuable insights into the potential mechanisms underlying the association between Des-Arg(9)-bradykinin and ASD, highlighting key genes and biological processes involved in this interaction.
This study used two-sample MR to investigate the causal relationship between metabolites and ASD. The analysis identified Des-Arg(9)-bradykinin as a potential risk factor for ASD(β = 0.262, SE = 0.064, P = 4.64 × 10-5). Furthermore, an interaction network analysis revealed five hub genes associated with Des-Arg(9)-bradykinin and ASD. Enrichment analysis indicated that this association may be involved in complex biological processes, including behavior, response to stimulus, and interaction between organisms. These findings provide valuable insights into the potential mechanisms underlying the association between Des-Arg(9)-bradykinin and ASD, highlighting potential key genes and biological processes involved in the development of ASD.
The pathophysiology of ASD is believed to result from intricate interactions between environmental and genetic factors[24]. Studies have found that elevated levels of inflammatory markers, such as cytokines, were detected in the blood and brains of some individuals with autism[25]. Dysregulated neuroinflammation has been proposed as a potential contributor to ASD development, emphasizing the importance of immune-neuronal crosstalk in neuroinflammation research[26]. There is also a suggestion of possible interaction between innate immunity and neuronal activity in the etiology of autism[27]. Increased microglial activation and density, and increased proinflammatory cytokines were observed in several brain regions among ASD individuals[28]. Furthermore, certain environmental factors associated with autism, such as prenatal infection, are known to trigger immune responses and inflammation. The enrichment of behavior-related processes aligns with the core symptoms of ASD, including impaired social interaction and repetitive behaviors. Additionally, the association with the biological process of response to stimulus suggests potential sensitivities or altered sensory processing in individuals with ASD.
Current research revealing the role of Des-Arg(9)-bradykinin in the development of ASD is limited. However, there is evidence suggesting that the Des-Arg(9)-bradykinin is involved in inflammatory and neurobiological processes. As a member of the kallikrein–kinin system, bradykinin participates in the modulation of important biological activities including vasocontraction, nitric oxide release, and anti-ischemic effects via interaction with the renin-angiotensin system[29]. Contributing to fluid retention within cerebral tissue, bradykinin was also considered to have a pathophysiological role in the mammalian brain[30]. The BDKRB1 and BDKRB2 were two of the main targets of bradykinin and its metabolites. The expression of B1R is induced during injury by cytokines such as interleukin-1beta while B2R is ubiquitously expressed. As a metabolite of bradykinin mediated by carboxypeptidase N, Des-Arg(9)-bradykinin was reported as a B1 receptor agonist that increases intracellular Ca2+ and induces vasocontraction, cell proliferation, and collagen synthesis[31,32]. The activation of the B1 receptor was reported to participate in the inflammatory response by inducing the release of pro-inflammatory cytokines and promoting leukocyte recruitment to the site of inflammation[33]. In light of the regulatory approval of drugs targeting bradykinin receptors, notably lcatibant, there arises an interesting topic for the in-depth examination and exploration of their potential effects on neurodevelopmental diseases. This prospect warrants further observation and comprehensive investigation.
Enrichment analysis results of this study also indicated the complexity of the interactions between the metabolism of Des-Arg(9) bradykinin and ASD. Given the potential role of the B1 receptor activation in immune cell infiltration, microglia activation, and cytokine production within the central nervous system, B1 receptor-mediated signaling cascades might result in elevated neuroinflammation[31]. As predicted targets of Des-Arg(9)-bradykinin, NTSR1, and NTSR2 participates in the modulation of neurotensin signaling that has been reported to stimulate microglia to secrete IL-1beta and CXCL8[34] and affect social behavior and repetitive behaviors, which are core features of ASD[35-37]. The Mu opioid receptors have also been reported as a critical neurobiological substrate of social behavior that participate in stress response and immune processes[38]. Therefore, immune-inflammation pathways could be the potential factors mediating Des-Arg(9)-bradykinin and ASD. The exact mechanism should be further explored and validated.
Setting genetic variances as instrumental variables, the causal relationship between metabolites and diseases estimated by MR provides practical clues for exploring biomarkers for neurodevelopmental disorders, such as ASD. The bioinformatic approaches integrating multi-scale evidence also provide a novel perspective for interpreting the association between metabolite and disease. Even if the causal relationship between Des-Arg(9)-bradykinin and ASD is validated pathogenetically, further research on the pharmacological treatment targeting the metabolic pathway of bradykinin or repurposing the drug target B1R would promote the discovery of a novel approach to treating ASD.
This study identified Des-Arg(9)-bradykinin as a causal metabolite for ASD using MR analysis. However, there are several limitations to be considered in this study. Firstly, linear algorithms were used in MR analysis hypothesizing a simple interaction mode between metabolites and disease. However, the modulation of biological processes is complex and dynamic. It should be also noticed that other metabolites, though evaluated as non-significant in this study, could also participate in the modulation of inflammation condition and immune status, and further affect the development of disease. Therefore, the pathogenetic interaction between the metabolite profile and ASD needs to be further validated using GWAS data from a larger sample size over more adaptive subgrouping. This would enhance the robustness and generalizability of the findings. Secondly, while genetic variables were utilized as reliable instrument variables for causal inference between metabolic exposure and ASD, the presence of pleiotropy in SNPs can disrupt their reliability. It is important to acknowledge that the strength of the instrument variables and the sample size employed in this study may introduce potential biases in the results. Thirdly, the study is based on the hypothesis that altered gene expression leads to changes in metabolite levels, ultimately contributing to the development of ASD. However, it is important to note that not all cases of ASD can be attributed to gene expression alterations alone. Other factors and mechanisms may also play a role in the disorder. Additionally, the practicality of the analytical results may be limited as the study did not comprehensively consider actual conditions such as the dosage, duration of exposure, interindividual variation in metabolism, and the ability of metabolites to reach the target tissues. Lastly, the study predominantly involved individuals of European ancestry to mitigate potential biases stemming from population variations. However, this selection may limit the generalizability and applicability of the findings to other demographic groups. Considering the genetic and environmental heterogeneity prevalent in various ethnic groups, it is imperative to replicate these findings in diverse populations to confirm their broader applicability. The journey toward identifying valid biomarkers to aid in clinical disease diagnosis is undeniably lengthy[39]. Although this study offers valuable insights into the potential role of Des-Arg(9)-bradykinin in ASD development, further research addressing these limitations is essential to bolster the validity, reliability, and practicality of the results.
Our study suggests a potential role for Des-Arg(9)-bradykinin in the development of ASD, mediated through metabolic and immune processes. The identification of hub genes and the enrichment of relevant biological processes provide additional evidence to support the interpretation of this association. Understanding the underlying pathological mechanisms and exploring the metabolic pathway of bradykinin, including the B1 receptor, could pave the way for novel therapeutic approaches for ASD. Further research, involving larger and more diverse cohorts, is warranted to validate and extend our findings.
This study delves into the complex landscape of autism spectrum disorder (ASD), an early-onset neurodevelopmental condition characterized by social communication deficits and repetitive behaviors. The etiology of ASD remains enigmatic, prompting a crucial need for robust diagnostic biomarkers. The study explores the potential interplay of genetic and environmental factors in ASD development, with a focus on plasma metabolites and their causal associations, providing a foundation for future advancements in diagnosis and intervention.
In the realm of ASD research, the pressing motivation lies in early diagnosis and effective therapeutic interventions. The study aims to tackle the pivotal challenge of identifying diagnostic biomarkers, given the intricate interplay of genetic and environmental factors in ASD etiology. A successful exploration of these causal associations can pave the way for innovative diagnostic tools and targeted interventions, offering hope for improved outcomes and quality of life for individuals with ASD.
This study sought to uncover the causal connections between plasma metabolites and ASD while accounting for genetic and environmental factors. The realized objective of identifying these associations carries profound implications for advancing diagnostic biomarkers and guiding future research in the field of ASD.
This study employed a two-sample Mendelian randomization (MR) analysis, a robust method that harnessed data from large-scale genome-wide association studies on metabolites and ASD. Novelty lies in the integration of genetic variants as instrumental variables to estimate causal associations, and the innovative use of the inverse variant weight algorithm. These methods unveiled the potential role of plasma metabolites in ASD etiology, shedding new light on diagnostic biomarkers and therapeutic avenues.
Des-Arg(9)-bradykinin emerged as a compelling causal metabolite associated with an increased risk of ASD. The sensitivity analysis underscored the robustness of this association. Furthermore, the identification of five hub genes, including KNG1, F12, BDKRB1, CCNA2 and CDK2, signifies the potential involvement of these genes in the ASD-Des-Arg(9)-bradykinin association. Enrichment analysis shed light on a multitude of biological processes, from peptide GPCRs to immune system functions, offering a comprehensive insight into the potential mechanisms linking Des-Arg(9)-bradykinin with ASD.
This study contributes novel insights by proposing Des-Arg(9)-bradykinin as a potential causal metabolite for ASD. The study set metabolites as proxy of genetic and environmental factors, and leveraged two-sample MR methods to elucidate the associations. These findings introduce new diagnostic and predictive biomarker for ASD, offering a promising pathway for future research and clinical practice.
The direction of future research should focus on comprehensive investigations into the complex interplay of genetic and environmental factors in ASD etiology. Expanding datasets to include diverse populations and incorporating multi-omics approaches can provide a more nuanced understanding of ASD development. Additionally, future research should explore the identification of additional biomarkers and the underlying mechanisms, potentially paving the way for innovative diagnostic tools and personalized interventions to enhance the lives of individuals with ASD.
Acknowledgment to Guangzhou Library for providing space and digital resources for the research.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Psychiatry
Country/Territory of origin: China
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Dauyey K, Kazakhstan; Moshref RH, Saudi Arabia S-Editor: Gao CC L-Editor: A P-Editor: Cai YX
| 1. | Yates K, Le Couteur A. Diagnosing autism/autism spectrum disorders. Paediatr Child Health. 2016;26:513-518. [RCA] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 20] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Hus Y, Segal O. Challenges Surrounding the Diagnosis of Autism in Children. Neuropsychiatr Dis Treat. 2021;17:3509-3529. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 3. | Dawson G, Sapiro G. Potential for Digital Behavioral Measurement Tools to Transform the Detection and Diagnosis of Autism Spectrum Disorder. JAMA Pediatr. 2019;173:305-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 36] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 4. | Frye RE, Vassall S, Kaur G, Lewis C, Karim M, Rossignol D. Emerging biomarkers in autism spectrum disorder: a systematic review. Ann Transl Med. 2019;7:792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 5. | Bölte S, Girdler S, Marschik PB. The contribution of environmental exposure to the etiology of autism spectrum disorder. Cell Mol Life Sci. 2019;76:1275-1297. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 276] [Cited by in RCA: 294] [Article Influence: 49.0] [Reference Citation Analysis (0)] |
| 6. | Kim YS, Leventhal BL. Genetic epidemiology and insights into interactive genetic and environmental effects in autism spectrum disorders. Biol Psychiatry. 2015;77:66-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 158] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 7. | Leblond CS, Le TL, Malesys S, Cliquet F, Tabet AC, Delorme R, Rolland T, Bourgeron T. Operative list of genes associated with autism and neurodevelopmental disorders based on database review. Mol Cell Neurosci. 2021;113:103623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 60] [Article Influence: 15.0] [Reference Citation Analysis (0)] |
| 8. | Li J, Wang L, Guo H, Shi L, Zhang K, Tang M, Hu S, Dong S, Liu Y, Wang T, Yu P, He X, Hu Z, Zhao J, Liu C, Sun ZS, Xia K. Targeted sequencing and functional analysis reveal brain-size-related genes and their networks in autism spectrum disorders. Mol Psychiatry. 2017;22:1282-1290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 72] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 9. | Madore C, Leyrolle Q, Lacabanne C, Benmamar-Badel A, Joffre C, Nadjar A, Layé S. Neuroinflammation in Autism: Plausible Role of Maternal Inflammation, Dietary Omega 3, and Microbiota. Neural Plast. 2016;2016:3597209. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 90] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 10. | Kwon HK, Choi GB, Huh JR. Maternal inflammation and its ramifications on fetal neurodevelopment. Trends Immunol. 2022;43:230-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 76] [Article Influence: 25.3] [Reference Citation Analysis (0)] |
| 11. | de la Torre-Ubieta L, Won H, Stein JL, Geschwind DH. Advancing the understanding of autism disease mechanisms through genetics. Nat Med. 2016;22:345-361. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 521] [Cited by in RCA: 567] [Article Influence: 63.0] [Reference Citation Analysis (0)] |
| 12. | Liu Y, Qing H, Deng Y. Biomarkers in Alzheimer's disease analysis by mass spectrometry-based proteomics. Int J Mol Sci. 2014;15:7865-7882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 13. | Glinton KE, Elsea SH. Untargeted Metabolomics for Autism Spectrum Disorders: Current Status and Future Directions. Front Psychiatry. 2019;10:647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 14. | Ritz B, Yan Q, Uppal K, Liew Z, Cui X, Ling C, Inoue K, von Ehrenstein O, Walker DI, Jones DP. Untargeted Metabolomics Screen of Mid-pregnancy Maternal Serum and Autism in Offspring. Autism Res. 2020;13:1258-1269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 15. | Liang Y, Ke X, Xiao Z, Zhang Y, Chen Y, Li Y, Wang Z, Lin L, Yao P, Lu J. Untargeted Metabolomic Profiling Using UHPLC-QTOF/MS Reveals Metabolic Alterations Associated with Autism. Biomed Res Int. 2020;2020:6105608. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 16. | Likhitweerawong N, Thonusin C, Boonchooduang N, Louthrenoo O, Nookaew I, Chattipakorn N, Chattipakorn SC. Profiles of urine and blood metabolomics in autism spectrum disorders. Metab Brain Dis. 2021;36:1641-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 17. | Sanderson E, Glymour MM, Holmes MV, Kang H, Morrison J, Munafò MR, Palmer T, Schooling CM, Wallace C, Zhao Q, Smith GD. Mendelian randomization. Nat Rev Methods Primers. 2022;2. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 644] [Cited by in RCA: 890] [Article Influence: 296.7] [Reference Citation Analysis (0)] |
| 18. | Lawlor DA. Commentary: Two-sample Mendelian randomization: opportunities and challenges. Int J Epidemiol. 2016;45:908-915. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 480] [Cited by in RCA: 523] [Article Influence: 58.1] [Reference Citation Analysis (0)] |
| 19. | Grover S, Del Greco M F, Stein CM, Ziegler A. Mendelian Randomization. Methods Mol Biol. 2017;1666:581-628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 86] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 20. | Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, Walter K, Menni C, Chen L, Vasquez L, Valdes AM, Hyde CL, Wang V, Ziemek D, Roberts P, Xi L, Grundberg E; Multiple Tissue Human Expression Resource (MuTHER) Consortium, Waldenberger M, Richards JB, Mohney RP, Milburn MV, John SL, Trimmer J, Theis FJ, Overington JP, Suhre K, Brosnan MJ, Gieger C, Kastenmüller G, Spector TD, Soranzo N. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46:543-550. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1097] [Cited by in RCA: 1096] [Article Influence: 99.6] [Reference Citation Analysis (1)] |
| 21. | Grove J, Ripke S, Als T, Mattheisen M, Walters R, Won H, Pallesen J, Agerbo E, Andreasssen O, Anney R, Belliveau R, Bettella F, Buxbaum J, Grauholm J, Bækved-Hansen M, Cerrato F, Chambert K, Christensen J, Churchhouse C, Børglum A. Common risk variants identified in autism spectrum disorder. 2017 Preprint. Available from: bioRxiv:224774. [DOI] [Full Text] |
| 22. | Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4747] [Cited by in RCA: 4756] [Article Influence: 679.4] [Reference Citation Analysis (0)] |
| 23. | Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50:693-698. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2495] [Cited by in RCA: 5300] [Article Influence: 757.1] [Reference Citation Analysis (0)] |
| 24. | Gevezova M, Sarafian V, Anderson G, Maes M. Inflammation and Mitochondrial Dysfunction in Autism Spectrum Disorder. CNS Neurol Disord Drug Targets. 2020;19:320-333. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 37] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 25. | Stavridou T, Driga A M, Drigas A. Blood Markers in Detection of Autism. Int J Recent Contributions Eng Sci IT. 2021;9:79-86. [DOI] [Full Text] |
| 26. | Matta SM, Hill-Yardin EL, Crack PJ. The influence of neuroinflammation in Autism Spectrum Disorder. Brain Behav Immun. 2019;79:75-90. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 250] [Article Influence: 41.7] [Reference Citation Analysis (0)] |
| 27. | Gupta S, Ellis SE, Ashar FN, Moes A, Bader JS, Zhan J, West AB, Arking DE. Transcriptome analysis reveals dysregulation of innate immune response genes and neuronal activity-dependent genes in autism. Nat Commun. 2014;5:5748. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 342] [Cited by in RCA: 377] [Article Influence: 34.3] [Reference Citation Analysis (0)] |
| 28. | Toscano CVA, Barros L, Lima AB, Nunes T, Carvalho HM, Gaspar JM. Neuroinflammation in autism spectrum disorders: Exercise as a "pharmacological" tool. Neurosci Biobehav Rev. 2021;129:63-74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 29. | Bekassy Z, Lopatko Fagerström I, Bader M, Karpman D. Crosstalk between the renin-angiotensin, complement and kallikrein-kinin systems in inflammation. Nat Rev Immunol. 2022;22:411-428. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 96] [Article Influence: 24.0] [Reference Citation Analysis (0)] |
| 30. | Dobrivojević M, Špiranec K, Sinđić A. Involvement of bradykinin in brain edema development after ischemic stroke. Pflugers Arch. 2015;467:201-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 31. | Sriramula S. Kinin B1 receptor: A target for neuroinflammation in hypertension. Pharmacol Res. 2020;155:104715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 25] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 32. | Sharma JN. Does the kinin system mediate in cardiovascular abnormalities? An overview. J Clin Pharmacol. 2003;43:1187-1195. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 17] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 33. | Mugisho OO, Robilliard LD, Nicholson LFB, Graham ES, O'Carroll SJ. Bradykinin receptor-1 activation induces inflammation and increases the permeability of human brain microvascular endothelial cells. Cell Biol Int. 2020;44:343-351. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 23] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 34. | Patel AB, Tsilioni I, Leeman SE, Theoharides TC. Neurotensin stimulates sortilin and mTOR in human microglia inhibitable by methoxyluteolin, a potential therapeutic target for autism. Proc Natl Acad Sci U S A. 2016;113:E7049-E7058. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 97] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 35. | Boules MM, Fredrickson P, Muehlmann AM, Richelson E. Elucidating the role of neurotensin in the pathophysiology and management of major mental disorders. Behav Sci (Basel). 2014;4:125-153. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 36. | Angelidou A, Francis K, Vasiadi M, Alysandratos KD, Zhang B, Theoharides A, Lykouras L, Sideri K, Kalogeromitros D, Theoharides TC. Neurotensin is increased in serum of young children with autistic disorder. J Neuroinflammation. 2010;7:48. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 60] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 37. | Tsilioni I, Dodman N, Petra AI, Taliou A, Francis K, Moon-Fanelli A, Shuster L, Theoharides TC. Elevated serum neurotensin and CRH levels in children with autistic spectrum disorders and tail-chasing Bull Terriers with a phenotype similar to autism. Transl Psychiatry. 2014;4:e466. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 34] [Cited by in RCA: 37] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 38. | Pellissier LP, Gandía J, Laboute T, Becker JAJ, Le Merrer J. μ opioid receptor, social behaviour and autism spectrum disorder: reward matters. Br J Pharmacol. 2018;175:2750-2769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 70] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 39. | Cortese S, Solmi M, Michelini G, Bellato A, Blanner C, Canozzi A, Eudave L, Farhat LC, Højlund M, Köhler-Forsberg O, Leffa DT, Rohde C, de Pablo GS, Vita G, Wesselhoeft R, Martin J, Baumeister S, Bozhilova NS, Carlisi CO, Leno VC, Floris DL, Holz NE, Kraaijenvanger EJ, Sacu S, Vainieri I, Ostuzzi G, Barbui C, Correll CU. Candidate diagnostic biomarkers for neurodevelopmental disorders in children and adolescents: a systematic review. World Psychiatry. 2023;22:129-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 56] [Article Influence: 28.0] [Reference Citation Analysis (0)] |