Published online Jun 19, 2023. doi: 10.5498/wjp.v13.i6.319
Peer-review started: December 27, 2022
First decision: January 17, 2023
Revised: January 31, 2023
Accepted: April 25, 2023
Article in press: April 25, 2023
Published online: June 19, 2023
Processing time: 174 Days and 3 Hours
The global burden of psychopathologies appears to be underestimated, since the global psychiatric disorder burden is exceeding other medical burdens. To be able to address this problem more effectively, we need to better understand the etiology of psychiatric disorders. One of the hallmarks of psychiatric disorders appears to be epigenetic dysregulation. While some epigenetic modifications (such as DNA methylation) are well known and studied, the roles of others have been investigated much less. DNA hydroxymethylation is a rarely studied epigenetic modification, which as well as being an intermediate stage in the DNA demethylation cycle is also an independent steady cell state involved in neuro
Core Tip: DNA hydroxymethylation is one of the least investigated epigenetic mechanisms but based on currently available results it is abundant in the brain, thus potentially influencing the development of psychiatric disorders through modulation of gene expression. Currently no particular genetic locus or gene can be linked to hydroxymethylation in psychiatric disorders. However, the dynamics in gene expression of the ten-eleven translocation enzymes that catalyze hydroxymethylation have been demonstrated, and changes in hydroxymethylation levels when comparing healthy subjects with psychiatric disorders can be determined using genome-wide and candidate gene approaches.
- Citation: Kouter K, Šalamon Arčan I, Videtič Paska A. Epigenetics in psychiatry: Beyond DNA methylation. World J Psychiatry 2023; 13(6): 319-330
- URL: https://www.wjgnet.com/2220-3206/full/v13/i6/319.htm
- DOI: https://dx.doi.org/10.5498/wjp.v13.i6.319
Psychiatric disorders comprise different types of mental disorder, including anxiety disorder, major depressive disorder (MDD), bipolar disorder (BP), post-traumatic stress disorder, schizophrenia (SZ), eating disorders, neurodevelopmental disorders, disruptive behavior, and dissocial disorders. All disorders have in common a clinically significant disturbance in an individual’s cognition, emotional regulation, or behavior and are normally associated with distress or impairment in important areas of functioning. In 2019, there were 970 million people around the world living with a psychiatric disorder. More than half of those people struggle with anxiety disorder (301 million) or depression (280 million), both of which are also common in children and adolescents. BP was experienced by 40 million people, and SZ by approximately 24 million people[1]. Obsessive-compulsive disorder (OCD) affects 1%-3% of the worldwide population[2]. Suicide is strongly connected with psychiatric disorders (in particular, depression and BP). However, many suicides happen impulsively in moments of crisis, such as breakdown in the ability to deal with life stressors, such as financial or relationship problems, chronic pain, or illness. More than 700000 people die due to suicide and there are many more people attempting it[3]. 13% of the global population is living with a psychiatric disorder which has a substantial effect on all areas of life, such as school or work performance, relationships with family and friends, and the ability to participate in the community[4]. Besides social impact, there is also an economic burden that costs the global economy US$ 1 trillion each year[5].
A biomarker is a measurable indicator of some biological state or condition. Diagnostic biomarkers serve for narrowing down diagnoses specific to an individual patient[6]. In the field of psychiatry, diagnoses are made based on psychiatric examination using the Diagnostic and Statistical Manual or Mental Disorders or International Classification of Diseases[7,8]. As clinical presentations of the same psychiatric disorders are heterogeneous, identifying biomarkers is essential step to facilitate diagnosis through markers that allow stratification of groups within the syndrome, early detection of the disease, prescribing the right medications, and improving treatment outcome[6]. Several factors, including genetic, neurobiological, cultural, and life experiences and their interplay contribute to psychiatric disorders. Heritability plays one part in understanding pathogenesis. For example, for the development of SZ and BP there is an approximately 80% contribution of heritability, while this drops to only around 40% for depression and suicidality[9]. Scientists have tried to understand the association of genetic polymorphisms on bigger cohorts by way of genome-wide association studies (GWAS), but there still remain many unanswered questions. This indicates that environmental factors also need to be taken into account[10]. Epigenetic mechanisms such as DNA methylation and hydroxymethylation, histone tail modifications and non-coding RNAs, respond to environmental factors and cause changes in gene expression or translation without changing DNA sequence[9]. The final biological outcome results from the interplay between different epigenetic mechanisms. In recent years hydroxymethylation has received increasing attention in the “neuroepigenome”[10]. There is particular interest in hydroxymethylation because it is highly enriched in brain and synapse-related genes, and exhibits dynamic regulation during development[11].
Mental disorders are influenced by many biological and external factors that can be linked together by epigenetics[9,12]. The term epigenetics describes innate (hereditary) or lifetime-acquired changes in gene expression that do not originate from differences in DNA sequence[13]. Epigenetics can thus help us understand the delicate interplay of environmental and genetic influences within the cell. One of the commonly studied mechanisms of epigenetic regulation is DNA methylation and the changes in its pattern. During the DNA methylation process a single methyl group is attached to the 5’ C site of the cytosine, making the cytosine either methylated or, when removed, unmethylated. DNA methylation can have a significant effect on the regulation of gene expression[14]. While DNA methylation is still the best researched and understood epigenetic modification, other types (such as noncoding RNAs) are getting more recognition[15,16].
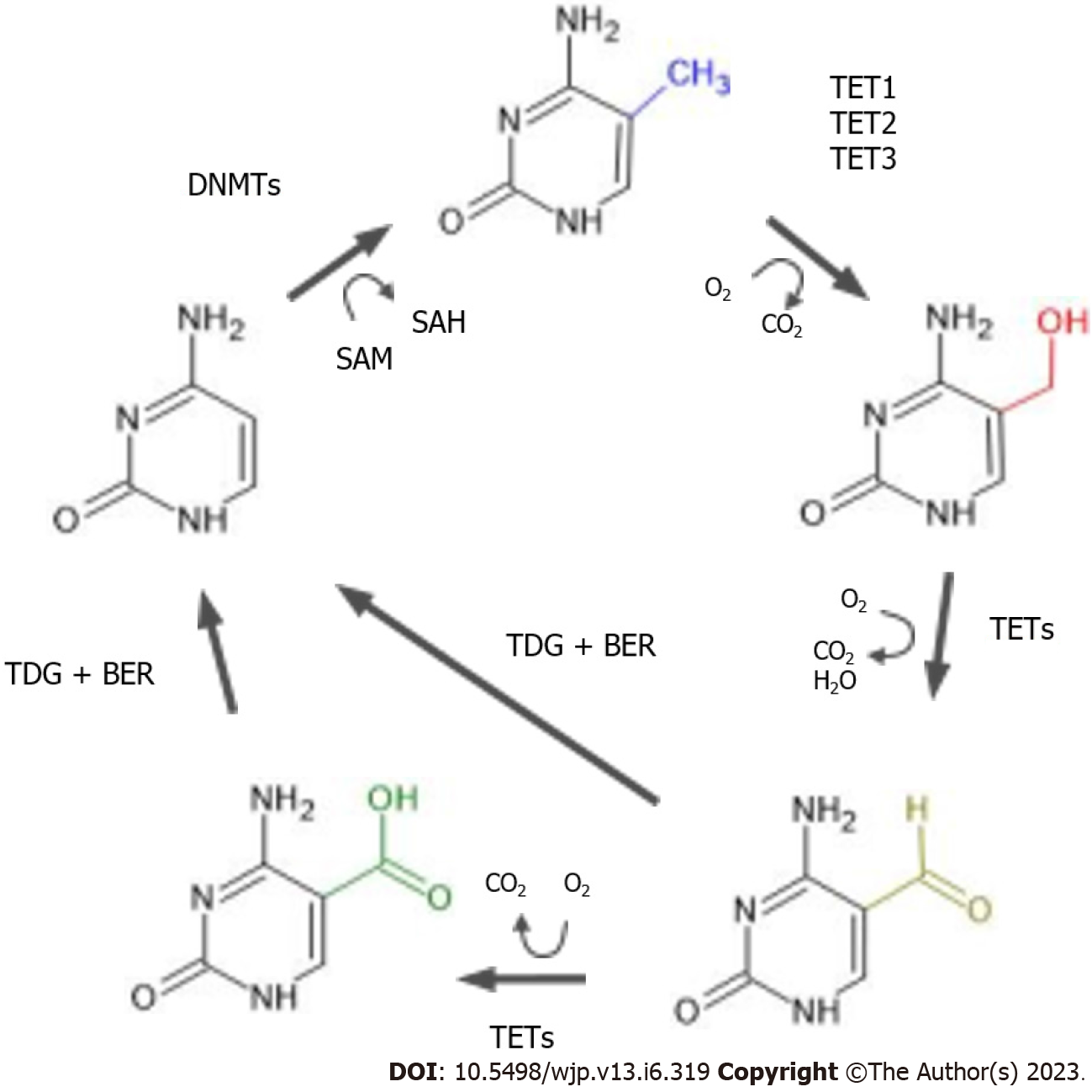
DNA methylation was long thought of as a binary state; whether the switch was turned on (resulting in a methylated cytosine) or off (resulting in an unmethylated cytosine). However more recently it has been discovered that methylation can have stable intermediate stages, which can happen during the process of demethylation. Active demethylation is catalyzed by enzymes of the ten-eleven translocation (TET) family and the enzyme thymine-DNA glycosylase mediated base excision. TET family proteins (the enzymes TET1, TET2 and TET3) are 2-oxoglutarate and Fe(II)-dependent dioxygenases, which catalyze 5-methylcytosine (5mC) conversion to 5-hydroxymethycytosine (5hmC) (illustrated in Figure 1). DNA hydroxymethylation was first described in 1972 but it was only in 2009 that 5hmC was identified as an oxidation metabolite of DNA methylation[17,18]. 5hmCs are often localized in gene bodies and untranslated regions. Compared to methylation, levels of 5hmC are lower (roughly 10% of methylation levels), but a high abundance of 5hmC is observed in the central nervous system. Animal studies demonstrate that DNA hydroxymethylation appears to be important during neurodevelopment as levels of 5hmC in mice embryos increase in the absence of demethylation, making it a stable cell state. This is confirmed by many animal studies[19]. 5hmC can be recognized by specific binding proteins, and can be maintained through cell division[20]. While its exact role in adult brain is not known, it appears that DNA hydroxymethylation can affect neuroplasticity and neurotransmission. 5hmC as such serves a dual role, not only as an intermediate of active demethylation but also as an important mark of epigenetic regulation. As with DNA methylation, numerous environmental factors such as medication, stress and pollutants can affect levels of 5hmC[19].
Like DNA methylation, DNA hydroxymethylation also affects gene expression, but the manner and direction (upregulation or downregulation of gene expression) are not as well characterized as for DNA methylation. Nevertheless, research indicates that DNA hydroxymethylation of the gene body leads to increased gene expression. Enrichment of DNA hydroxymethylation is also often observed at 5’ splicing sites, enhancers and CpG island borders[21].
Several methods are available for the analysis of DNA hydroxymethylation, following in the footsteps of DNA methylation methods. For DNA methylation, bisulfite conversion of DNA is the gold standard. During the conversion, unmethylated cytosines are deaminated, allowing methylated cytosines to be identified. However, the classical bisulfite conversion approach does not work for DNA hydroxymethylation because it is not possible to distinguish between methylated and hydroxymethylated cytosines[22]. Since the ratio of methylation to hydroxymethylation is approximately 9 to 1 in favor of methylation other approaches are needed to measure hydroxymethylation[23]. When the number of target genes is small, the candidate gene approach is most appropriate. When the number of target genes increases, a genome-wide approach is usually better in terms of finances, time, and information. Therefore, the selection of the appropriate method for DNA hydroxymethylation analysis should be based on the broader experimental design. Since genome-wide methods are more commonly used for 5hmC assessment, this section focuses only on such methods.
Genome-wide approaches are often better than candidate gene approaches when studying lesser-known epigenetic states such as hydroxymethylation. In addition, psychiatric disorders are complex behaviors that likely involve multiple genes. Currently, there are several methods. Some methods use restriction enzymes and glucosylation (the process by which β-glucosyltransferase specifically transfers a glucose unit to 5hmC, resulting in glucosylated 5hmC). One example is Aba-seq, which uses a restriction endonuclease AbaSI that selectively recognizes glucosylated 5hmC and cleaves DNA in its vicinity (within approximately 10 base pairs)[24]. Another example is Reduced Representation Hydroxymethylation Profiling, in which DNA is first digested with the enzyme MspI, which recognizes and cleaves the 5’-CCGG sequence. 5hmCs are later glucosylated, which prevents MspI from further cleavage, so that only fragments containing 5hmC are prevented from enzymatic digestion[25].
Tet-assisted bisulfite sequencing (TAB-seq) also takes advantage of glucosylation of 5hmC. After the glucosylation step, the DNA is oxidized using Tet proteins. The glucosylated 5hmC is retained, while 5mC converts to 5-carboxylcytosine (5caC). This oxidized DNA is then treated with bisulfite conversion and polymerase chain reaction (PCR) amplification, where unmethylated cytosines and 5mC (now 5caC) are converted to thymine, while glucosylated 5hmCs become cytosines. TAB-seq is more appropriate when only the 5hmC content is of interest[26].
A more complex method is a variant of bisulfite sequencing called oxidative bisulfite sequencing (oxBs-seq). In oxBs-seq, the sample of interest is divided into two parts; one part is subjected to bisulfite treatment, and after PCR amplification both 5hmC and 5mC are observed as cytosines. The second part of the sample is oxidized before bisulfite treatment, converting 5hmC to 5fC and after PCR amplification to thymine, with 5mC seen as cytosines. This parallel sequencing of the same sample allows accurate detection of 5hmC but increases the price because very high coverage is required[27].
Immuno-dot blot methods use antibodies that can recognize 5-hydroxymethylcytosine. A major advantage of these methods is their relative simplicity in terms of wet-lab work. They provide information on global DNA hydroxymethylation throughout the genome and provide a single numerical measure. The results do not provide detailed information on the state of hydroxymethylation in specific genomic regions, but can be used to provide a comprehensive overview of the state of hydroxymethylation in the genome[28]. A more complex immune approach uses the combination of hydroxymethylation-specific antibodies with next generation sequencing. In antibody-based methods such as hydroxymethylated DNA immunoprecipitation sequencing, DNA is cut into short fragments. Using antibodies against hydroxymethylation, only the hydroxymethylated DNA is isolated from the mixture of DNA fragments. A library is created from the purified DNA fragments and sequenced using next-generation sequencing[29]. Compared with oxBs-seq, this is a less costly option. It does not provide base pair resolution, but a resolution of a few hundred base pairs is usually sufficient for regional overview and gene identification[23,30].
Nanopore technologies is a third-generation (long-read) sequencing approach. Compared with the commonly used Illumina approach of “sequencing by synthesis”, sequencing with Nanopore technologies is based on small changes in current. During sequencing, DNA strands pass through protein nanopores about 1.8 nanometers in diameter embedded in a polymer membrane. As DNA strands enter the pore, the current changes according to the DNA bases that are inside the pore (about 6 DNA bases at a time). This also allows detection of epigenetic changes[31]. The advantage of this method is that it provides genome-wide information, while long reads (up to several Mb = 106 base pairs) allow detection of rare variants/deletions/insertions/repetitive regions as well as epigenetic DNA changes (both DNA methylation and hydroxymethylation), all in a single sample. This reduces the possibility of biological and technical errors. Although the price per sample is high (the total cost is nearly € 1000 per sample), it allows a much more detailed examination of a sample[32].
As mentioned above, psychiatric disorders are multifactorial disorders influenced by both genetic and epigenetic factors. As epigenetic mechanisms show a high degree of tissue specificity, the best biological source (tissue or organ) for studying psychiatric disorders is therefore the brain. Studies often focus on brain regions related to emotional and behavioral responses, and cognitive and executive control functions. These often-studied regions include the limbic system (hippocampus) and the prefrontal cortex (PFC) (Brodmann area 9 in the dorsolateral PFC)[33,34]. Since the brain is not accessible during life, other peripheral, more easily accessible tissues, such as blood, are often included in studies of psychiatric disorders as well. In the following paragraphs the available studies on DNA hydroxymethylation are summarized, with more technical data presented in Table 1.
| Ref. | Studied disorder | Subjects | Tissue | Method | Main results |
| Gross et al[35], 2017 | Suicidal behavior/major depressive disorder | Subjects with major depressive disorder; n = 19 and psychiatrically healthy controls; n = 19 | Prefrontal cortex | AbaSI sequencing | No differences in 5hmC density across the genome. Site-specific differential hydroxymethylation in the depressed brain |
| Lutz et al[36], 2018 | Suicidal behavior | Suicide completers (severe child abuse history); n = 30, suicide completers (no child abuse history); n = 30 and control group; n = 34 | Anterior insula | Targeted oxidative bisulfite sequencing | Childhood abuse in the past is associated with decreased hydroxymethylation of Kappa Intron 2 |
| Jiang et al[41], 2017 | Schizophrenia | Schizophrenic subjects; n = 264 and controls; n = 221 | Peripheral blood | Dot blot-based measurement | 5hmC levels increased in male SZ patients but decreased levels in female SZ patients |
| Zong et al[42], 2017 | Schizophrenia | Schizophrenic subjects; n = 279 and controls; n = 256 | Peripheral white blood cells | HpaII/MspI restriction-based EpiMark 5hmC/5mC Analysis Kit followed by qPCR | 5hmC increased in the promoter of the GABRB2 gene; correlation between heterozygous genotype rs72815526 (C/A) and 5hmC levels increased |
| Gavin et al[44], 2012 | Schizophrenia and bipolar disorder | Schizophrenic subjects; n = 15, subjects with bipolar disorder; n = 11, and controls; n = 15 | Parietal cortex (Brodmann area 39-40) | hMeDIP | 5hmC increased at the promoter IXabcd of the BDNF gene |
| Dong et al[45], 2012 | Schizophrenia and bipolar disorder | Schizophrenic subjects; n = 10, subjects with bipolar disorder; n = 9, and controls; n = 11 | Inferior parietal lobule | Immuno-dot-blot analysis | 5hmC increased at the GAD67 gene at -537 to -415, and -145 to +21; 5hmC increased at the BDNF IXabcd gene at -60 to +50 not within BDNF exon IXd at +1185 to +1305 |
| D'Addario et al[57], 2019 | Obsessive compulsive disorder | Obsessive compulsive disorder; n = 35, and controls; n = 32 | Peripheral blood mononuclear cells | Bisulfite conversion or oxidative bisulfite conversion, followed by pyrosequencing | BDNF hydroxymethlation decreased at promoter exon I; no changes in promoter exon IV and IX |
| Clark et al[60], 2022 | Substance use disorder | Alcohol use disorder; n = 25, and controls; n = 25 | Prefrontal cortex (Brodmann area 10) | Selective chemical labelling and enrichment of hmC (hMe-Seal) | BAIAP2 hydroxymethylation increased |
In the field of suicidal behavior only two studies have been published that examined the pattern of DNA hydroxymethylation in suicide completers. Gross et al[35] examined DNA hydroxymethylation levels in PFC of male suicide completers with MDD. Compared to the control group, no global changes in DNA hydroxymethylation distribution were observed. They did however observe 550 differentially hydroxymethylated CpG sites in the genome. Lutz et al[36] included DNA hydroxymethylation analysis of opioid related genes in three brain regions (anterior cingulate cortex, thalamus and anterior insula). Suicide completers who experienced child abuse showed decreased level of DNA hydroxymethylation, while suicide completers with no history of child abuse did not. The decrease in DNA hydroxymethylation may therefore be a mark of child abuse.
SZ is a complex, multifactorial disease that causes psychosis. Psychotic symptoms include changes that affect personal and occupational life, e.g., positive symptoms such as hallucinations, delusions, thought and movement disorder, and negative and cognitive symptoms[37]. SZ manifests in late adolescence or early adulthood. Due to psychotic symptoms, SZ is also accompanied by an elevated risk of suicide, which reduces life expectancy for people developing it[38].
Factors that contribute to the development of SZ are both genetic and environmental[39]. Large-scale GWAS has highlighted a large number of variants with small effects which are highly heterogeneous between individuals. The importance of environmental factors is studied through epigenetics. Genome-wide approach and specific candidate gene studies have shown differences in DNA methylation and histone modifications in schizophrenic patients. Associated genes are involved in axon and dendrite extension, oligodendrocyte differentiation, regulation of GABAergic transmission, metabolism of dopamine, and glutamate and serotonin receptors[40].
Candidate gene studies have shown hyper- and hypo-methylated genes in brain tissue in concordance with studies on peripheral tissue (whole blood, peripheral leukocytes, or saliva). However, the results of DNA methylation studies are not always similar between male and female patients. Moreover, many interesting results from DNA methylation studies have pushed interest toward DNA hydroxymethylation. Even though there are not as many studies investigating hydroxymethylation, and much more work needs to be done, some interesting results have been obtained[40].
The level of 5mC is increased in both male and female SZ patients, but 5hmC level in peripheral blood is increased in males and decreased in females compared to controls. Furthermore, 5mC globally increased with age in all participants (schizophrenic and controls), but there was no significant correlation with age for the 5hmC level[41]. An increased level of 5hmC was also reported in the promoter of the GABRB2 gene in peripheral white blood cells of patients with SZ compared with controls. Furthermore, they found a correlation between promoter 5mC and 5hmC levels, with single nucleotide polymorphisms in patients with SZ. Heterozygous (C/A) genotypes of rs72815526 were correlated with increased 5hmC levels whereas heterozygous (C/T) genotypes of rs3811997 were correlated with decreased 5mC levels[42].
BP is a chronic psychiatric disorder characterized by unusual shifts in mood, energy, activity levels, and concentration. Impulsive reckless behavior leads to an increased risk of suicide[43]. Since SZ and BP are both psychoses, some studies have been made on a combined group of people with SZ and BP[44,45].
A number of studies address DNA methylation in patients with BP but just two have investigated hydroxymethylation where patients with BP were included in the study alongside patients with SZ, forming a combined group of psychotic illnesses. In a study made on parietal cortical samples from psychotic patients (Brodmann area[39,40]), an increase in 5mC and 5hmC was observed at the promoter IXabcd of the brain-derived neurotrophic factor (BDNF) gene. This result was also associated with reduced transcription of the BDNF gene[44]. In another study on the inferior parietal lobule of psychotic patients increased expression of the TET1 gene was detected. Consistent with the increase of expression the TET1 gene, there was an increased level of 5hmC. Specifically, higher 5hmC levels were observed at the glutamate decarboxylase 67 gene at positions -537 to -415 and -145 to +21 in psychosis patients vs control. There was also an increase in 5hmC at the BDNF IXabcd gene at position -60 to +50 in psychosis patients vs control, but not within BDNF exon IXd at +1185 to +1305[45].
MDD is the psychiatric disorder that contributes the most to the global health burden, and is the primary cause of disability globally. The environmental factors that increase the risk of MDD the most are stress and exposure to adverse life events, meta-analyses and genome-wide studies have shown that genetic factors also contribute to its development, contributing about 40%[46].
The DNA methylation studies were summarized in a recent systematic review[47]. Out of 67 studies 61 were performed on blood samples and thus lacked information on the status of DNA methylation in the central nervous system. Among the already well-established genes, hypermethylation of BDNF and the serotonin transporter gene showed association with depression on general or MDD. Results of other genes showed mixed results[47].
Only two studies on hydroxymethylation in MDD have been reported so far the studies by Gross et al[35] and Lutz et al[36] mentioned previously. In both studies the brain tissue of subjects with MDD who died due to suicide was used, making it difficult to link the observed differences to MDD alone.
Although studies in human are lacking, three studies on mice strain C57BL/6 have been reported, all investigating the TET enzymes. Feng et al[48] applied chronic stress, and determined decreased levels of expression of TET1 in the nucleus accumbens in stress-susceptible mice. In a TET1 knockout genes related to the immune system were demonstrated to be the most highly dysregulated. Zhang et al[49] examined the TET2 knockout animals, and identified some hydroxymethylated regions that overlapped with known depression-associated loci. They also showed abnormal translocation of TET2 protein from the cytosol to the nucleus in chronic stress induced mice. Cheng et al[50] showed that chronic restraint stress induced depression-like behavior in mice and reduced hydroxymethylation of the PFC. Knocking out TET1 resulted in resistance to chronic restraint stress, while a knockout of TET2 increased the susceptibility to chronic restraint stress[50]. The above results indicate that hydroxymethylation exhibits dynamic changes in response to stress-induced stimuli, resulting in changes of epigenetically regulated gene expression.
OCD is a psychiatric disorder characterized by obsessions and compulsions of various natures and degrees, and is in the majority of cases accompanied by at least one comorbid psychiatric disorder, most commonly MDD[51]. OCD is a heritable disorder with a polygenic background. Candidate gene and genome-wide studies suggest that the serotonergic, dopaminergic, and glutamatergic systems, and the interaction between them, contribute to the risk of the OCD[52]. Important contributing environmental factors are perinatal complications, childhood trauma, reproductive cycle events, and stressful life events[51].
Epigenetic studies of OCD have mostly investigated DNA methylation. Using the genome-wide approach, covering over 485000 CpG sites, genes previously associated with OCD were detected[53]. With the candidate gene approach the oxytocin receptor and serotonin transporter genes were interrogated. The results were mixed, showing both higher and lower level of methylation in OCD patients compared to controls[54-56].
In the context of the candidate gene approach BDNF methylation and hydroxymethylation have also been analyzed. Expression of BDNF in peripheral blood mononuclear cells demonstrated broad association with OCD, showing an increase in gene expression, as well as significant correlation of both lower methylation and higher hydroxymethylation at promoter exon I[57]. The results were replicated on saliva samples, showing lower levels of methylation in OCD compared to controls[2].
Substance use disorder (SUD) is a mental disorder with a range of symptoms from moderate to severe. The most severe SUD is addiction. SUD affects a person’s brain and behavior, which leads to the uncontrollable use of substances, such as drugs, alcohol, and medications[58]. It is common that people with SUD are also diagnosed with mental disorders and vice versa. Common mental disorders that co-occur with SUD are depression, BP, attention-deficit hyperactivity disorder, psychotic illness, borderline personality disorder, and antisocial personality disorder[59]. There are three suggestions for their joint occurrence. Firstly, common risk factors might contribute to SUD and other mental disorders; secondly, mental disorders can contribute to substance use and SUD; thirdly, substance use and SUD contribute to the development of other mental disorders[58]. It is estimated that there is a 40%-60% genetic contribution to vulnerability for developing SUD. For example, genetic vulnerability can be seen through changes in the degradation of the consumed, inhaled, or injected substance[59]. Moreover, there are also environmental factors that work through the epigenome[60].
There are several studies investigating 5hmC changes in the brains of people with SUD[61]. Clark et al[60] showed a difference in 5hmC of the BAIAP2 gene in the PFC from people with alcohol use disorder (AUD)[60]. One study showed differential expression of TET1 in the cerebellum of people with AUD, but no differences in 5hmC in the promoter of GABAergic genes[62]. More studies of SUD were made on model organisms, where they found 5hmC alteration after cocaine and methamphetamine administration in the nucleus accumbens[48,63,64], and a higher global 5hmC level was observed in nucles accumbens after ethanol administration[65].
In treatment of psychiatric disorders there is currently only a limited selection of validated biomarkers in use (e.g., electrophysiological biomarkers, neuropsychological biomarkers, brain imaging biomarkers, blood biomarkers), while molecular-genetic markers particularly have not yet entered the routine practice. The selection of treatment is based on clinical interview. In many cases patients do not respond to the medication prescribed or their treatment is changed too quickly due to side effects, since the response to the same medication can be highly variable among patients. Thus, the screening of a patient’s genetic background could improve the selection of a suitable medicine(s) based on an individual’s genetic background, taking into account pharmacokinetic and pharmacodynamics parameters. These specific differences in gene makeup have been tested in genes responsible for drug metabolism, however other genes also show changes that can significantly affect the development of disease. Shortly after the beginning of the millennium the human genome was sequenced, initiating exponential growth of genome-wide studies that continues until today. It has become evident that psychiatric disorders are polygenic, and that changes in gene sequences (e.g., single nucleotide polymorhpisms, copy number variations) contribute small effects which result in the development of the disorder, also through genetic pleiotropy and epistasis. However, the results of these studies have been mixed, identifying different genes, so no particular genetic locus can be linked to a given psychiatric disorder and used as a biomarker. Since psychiatric disorders show a significant proportion of environmental risk factors, the study of epigenetics has become an important approach to linking genetics with environmental effects, and might therefore provide applicable biomarkers for diagnosis and treatment.
The majority of epigenetic studies have interrogated DNA methylation as the prime epigenetic mark, but again these have produced mixed results at the level of genome-wide analysis, while the candidate gene approach has confirmed the importance of genes involved in neurotransmission and neurotrophic growth factors. Since the development of psychiatric disorders stems from disrupted signaling in the brain DNA hydroxymethylation, which can affect neuroplasticity and neurotransmission, has become an interesting research field. To date not many studies are available, as this area develops, and joint studies on transcriptional regulation are carried out, we expect to be able to improve effects evoked by pharmacotherapy with beneficial effects on patients’ health.
As is the case with genetic biomarkers, epigenetic biomarkers are not currently useful in a clinical setting, and many gaps remain in the understanding of their function. To advance our understanding of potential biomarkers, more studies simultaneously interrogating the status of the brain and peripheral tissue such as blood or even saliva are needed. Biomarkers showing good overlap between different tissue types could become key elements of validation studies on bigger, clinically well-defined cohorts, helping to discern the molecular mechanisms of psychiatric disorder development, diagnosis and treatment.
The authors would like to thank Dr. John Hancock for critical appraisal and scientific English editing of the manuscript.
Provenance and peer review: Invited article; Externally peer reviewed.
Peer-review model: Single blind
Corresponding Author’s Membership in Professional Societies: Slovenian Biochemical Society; Slovenian Neuroscience Association.
Specialty type: Psychiatry
Country/Territory of origin: Slovenia
Peer-review report’s scientific quality classification
Grade A (Excellent): A
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Hosak L, Czech Republic; Stevovic LI, Montenegro S-Editor: Wang JJ L-Editor: A P-Editor: Liu JH
| 1. | World Health Organization. Mental disorders. [cited 18 November 2022]. Available from: https://www.who.int/news-room/fact-sheets/detail/mental-disorders. |
| 2. | D’Addario C, Macellaro M, Bellia F, Benatti B, Annunzi E, Palumbo R, Conti D, Fasciana F, Vismara M, Varinelli A, Ferrara L, Celebre L, Viganò C, Dell'Osso B. In Search for Biomarkers in Obsessive-Compulsive Disorder: New Evidence on Saliva as a Practical Source of DNA to Assess Epigenetic Regulation. Curr Med Chem. 2022;29:5782-5791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 3. | World Health Organization. Suicide. [cited 18 November 2022]. Available from: https://www.who.int/news-room/fact-sheets/detail/suicide. |
| 4. | World Health Organization. World mental health report: transforming mental health for all. [cited 18 November 2022]. Available from: https://www.who.int/publications/i/item/9789240049338. |
| 5. | The Lancet Global Health. Mental health matters. Lancet Glob Health. 2020;8:e1352. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 61] [Cited by in RCA: 177] [Article Influence: 35.4] [Reference Citation Analysis (0)] |
| 6. | García-Gutiérrez MS, Navarrete F, Sala F, Gasparyan A, Austrich-Olivares A, Manzanares J. Biomarkers in Psychiatry: Concept, Definition, Types and Relevance to the Clinical Reality. Front Psychiatry. 2020;11:432. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 159] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 7. | American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-5™, 5th ed. Washington: American Psychiatric Publishing, 2013. |
| 8. | World Health Organization. International Classification of Diseases 11th Revision. [cited 15 November 2022]. Available from: https://icd.who.int/en. |
| 9. | Nestler EJ, Peña CJ, Kundakovic M, Mitchell A, Akbarian S. Epigenetic Basis of Mental Illness. Neuroscientist. 2016;22:447-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 222] [Article Influence: 22.2] [Reference Citation Analysis (0)] |
| 10. | Cariaga-Martinez A, Alelú-Paz R. Rethinking the Epigenetic Framework to Unravel the Molecular Pathology of Schizophrenia. Int J Mol Sci. 2017;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 11. | Kato T, Iwamoto K. Comprehensive DNA methylation and hydroxymethylation analysis in the human brain and its implication in mental disorders. Neuropharmacology. 2014;80:133-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 64] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 12. | Turecki G, Brent DA, Gunnell D, O'Connor RC, Oquendo MA, Pirkis J, Stanley BH. Suicide and suicide risk. Nat Rev Dis Primers. 2019;5:74. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 509] [Article Influence: 84.8] [Reference Citation Analysis (0)] |
| 13. | Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17:487-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1401] [Cited by in RCA: 1790] [Article Influence: 198.9] [Reference Citation Analysis (0)] |
| 14. | Deaton AM, Bird A. CpG islands and the regulation of transcription. Genes Dev. 2011;25:1010-1022. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2107] [Cited by in RCA: 2226] [Article Influence: 159.0] [Reference Citation Analysis (0)] |
| 15. | Cheung S, Woo J, Maes MS, Zai CC. Suicide epigenetics, a review of recent progress. J Affect Disord. 2020;265:423-438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 16. | Girdhar K, Rahman S, Dong P, Fullard JF, Roussos P. The Neuroepigenome: Implications of Chemical and Physical Modifications of Genomic DNA in Schizophrenia. Biol Psychiatry. 2022;92:443-449. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 17. | Kriaucionis S, Heintz N. The nuclear DNA base 5-hydroxymethylcytosine is present in Purkinje neurons and the brain. Science. 2009;324:929-930. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2321] [Cited by in RCA: 2054] [Article Influence: 128.4] [Reference Citation Analysis (0)] |
| 18. | Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930-935. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4860] [Cited by in RCA: 4387] [Article Influence: 274.2] [Reference Citation Analysis (0)] |
| 19. | Nikolac Perkovic M, Videtic Paska A, Konjevod M, Kouter K, Svob Strac D, Nedic Erjavec G, Pivac N. Epigenetics of Alzheimer's Disease. Biomolecules. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 117] [Article Influence: 29.3] [Reference Citation Analysis (0)] |
| 20. | Lunnon K, Hannon E, Smith RG, Dempster E, Wong C, Burrage J, Troakes C, Al-Sarraj S, Kepa A, Schalkwyk L, Mill J. Variation in 5-hydroxymethylcytosine across human cortex and cerebellum. Genome Biol. 2016;17:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 65] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 21. | Wen L, Tang F. Genomic distribution and possible functions of DNA hydroxymethylation in the brain. Genomics. 2014;104:341-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 70] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 22. | Hong SR, Shin KJ. Bisulfite-Converted DNA Quantity Evaluation: A Multiplex Quantitative Real-Time PCR System for Evaluation of Bisulfite Conversion. Front Genet. 2021;12:618955. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 24] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 23. | Kochmanski J, Bernstein AI. The Impact of Environmental Factors on 5-Hydroxymethylcytosine in the Brain. Curr Environ Health Rep. 2020;7:109-120. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 24. | Sun Z, Terragni J, Borgaro JG, Liu Y, Yu L, Guan S, Wang H, Sun D, Cheng X, Zhu Z, Pradhan S, Zheng Y. High-resolution enzymatic mapping of genomic 5-hydroxymethylcytosine in mouse embryonic stem cells. Cell Rep. 2013;3:567-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 130] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 25. | Petterson A, Chung TH, Tan D, Sun X, Jia XY. RRHP: a tag-based approach for 5-hydroxymethylcytosine mapping at single-site resolution. Genome Biol. 2014;15:456. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 26. | Yu M, Hon GC, Szulwach KE, Song CX, Zhang L, Kim A, Li X, Dai Q, Shen Y, Park B, Min JH, Jin P, Ren B, He C. Base-resolution analysis of 5-hydroxymethylcytosine in the mammalian genome. Cell. 2012;149:1368-1380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 848] [Cited by in RCA: 812] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 27. | Booth MJ, Ost TW, Beraldi D, Bell NM, Branco MR, Reik W, Balasubramanian S. Oxidative bisulfite sequencing of 5-methylcytosine and 5-hydroxymethylcytosine. Nat Protoc. 2013;8:1841-1851. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 282] [Cited by in RCA: 257] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 28. | de Dieuleveult M. Analysis of DNA methylation and hydroxymethylation: Easy and fast Dot Blot protocol. 2019. [DOI] [Full Text] |
| 29. | Tan L, Xiong L, Xu W, Wu F, Huang N, Xu Y, Kong L, Zheng L, Schwartz L, Shi Y, Shi YG. Genome-wide comparison of DNA hydroxymethylation in mouse embryonic stem cells and neural progenitor cells by a new comparative hMeDIP-seq method. Nucleic Acids Res. 2013;41:e84. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 79] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 30. | Ficz G, Branco MR, Seisenberger S, Santos F, Krueger F, Hore TA, Marques CJ, Andrews S, Reik W. Dynamic regulation of 5-hydroxymethylcytosine in mouse ES cells and during differentiation. Nature. 2011;473:398-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 958] [Cited by in RCA: 874] [Article Influence: 62.4] [Reference Citation Analysis (0)] |
| 31. | Steinbock LJ, Radenovic A. The emergence of nanopores in next-generation sequencing. Nanotechnology. 2015;26:074003. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 52] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 32. | Liu Y, Cheng J, Siejka-Zielińska P, Weldon C, Roberts H, Lopopolo M, Magri A, D'Arienzo V, Harris JM, McKeating JA, Song CX. Accurate targeted long-read DNA methylation and hydroxymethylation sequencing with TAPS. Genome Biol. 2020;21:54. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 57] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 33. | Mirza S, Docherty AR, Bakian A, Coon H, Soares JC, Walss-Bass C, Fries GR. Genetics and epigenetics of self-injurious thoughts and behaviors: Systematic review of the suicide literature and methodological considerations. Am J Med Genet B Neuropsychiatr Genet. 2022;189:221-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 10] [Reference Citation Analysis (0)] |
| 34. | Šalamon Arčan I, Kouter K, Videtič Paska A. Depressive disorder and antidepressants from an epigenetic point of view. World J Psychiatry. 2022;12:1150-1168. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 4] [Cited by in RCA: 9] [Article Influence: 3.0] [Reference Citation Analysis (3)] |
| 35. | Gross JA, Pacis A, Chen GG, Drupals M, Lutz PE, Barreiro LB, Turecki G. Gene-body 5-hydroxymethylation is associated with gene expression changes in the prefrontal cortex of depressed individuals. Transl Psychiatry. 2017;7:e1119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 43] [Cited by in RCA: 48] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 36. | Lutz PE, Gross JA, Dhir SK, Maussion G, Yang J, Bramoulle A, Meaney MJ, Turecki G. Epigenetic Regulation of the Kappa Opioid Receptor by Child Abuse. Biol Psychiatry. 2018;84:751-761. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 37. | World Health Organization. Schizophrenia. [cited 16 November 2022]. Available from: https://www.who.int/news-room/fact-sheets/detail/schizophrenia. |
| 38. | Trubetskoy V, Pardiñas AF, Qi T, Panagiotaropoulou G, Awasthi S, Bigdeli TB, Bryois J, Chen CY, Dennison CA, Hall LS, Lam M, Watanabe K, Frei O, Ge T, Harwood JC, Koopmans F, Magnusson S, Richards AL, Sidorenko J, Wu Y, Zeng J, Grove J, Kim M, Li Z, Voloudakis G, Zhang W, Adams M, Agartz I, Atkinson EG, Agerbo E, Al Eissa M, Albus M, Alexander M, Alizadeh BZ, Alptekin K, Als TD, Amin F, Arolt V, Arrojo M, Athanasiu L, Azevedo MH, Bacanu SA, Bass NJ, Begemann M, Belliveau RA, Bene J, Benyamin B, Bergen SE, Blasi G, Bobes J, Bonassi S, Braun A, Bressan RA, Bromet EJ, Bruggeman R, Buckley PF, Buckner RL, Bybjerg-Grauholm J, Cahn W, Cairns MJ, Calkins ME, Carr VJ, Castle D, Catts SV, Chambert KD, Chan RCK, Chaumette B, Cheng W, Cheung EFC, Chong SA, Cohen D, Consoli A, Cordeiro Q, Costas J, Curtis C, Davidson M, Davis KL, de Haan L, Degenhardt F, DeLisi LE, Demontis D, Dickerson F, Dikeos D, Dinan T, Djurovic S, Duan J, Ducci G, Dudbridge F, Eriksson JG, Fañanás L, Faraone SV, Fiorentino A, Forstner A, Frank J, Freimer NB, Fromer M, Frustaci A, Gadelha A, Genovese G, Gershon ES, Giannitelli M, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, González Peñas J, González-Pinto A, Gopal S, Gratten J, Green MF, Greenwood TA, Guillin O, Gülöksüz S, Gur RE, Gur RC, Gutiérrez B, Hahn E, Hakonarson H, Haroutunian V, Hartmann AM, Harvey C, Hayward C, Henskens FA, Herms S, Hoffmann P, Howrigan DP, Ikeda M, Iyegbe C, Joa I, Julià A, Kähler AK, Kam-Thong T, Kamatani Y, Karachanak-Yankova S, Kebir O, Keller MC, Kelly BJ, Khrunin A, Kim SW, Klovins J, Kondratiev N, Konte B, Kraft J, Kubo M, Kučinskas V, Kučinskiene ZA, Kusumawardhani A, Kuzelova-Ptackova H, Landi S, Lazzeroni LC, Lee PH, Legge SE, Lehrer DS, Lencer R, Lerer B, Li M, Lieberman J, Light GA, Limborska S, Liu CM, Lönnqvist J, Loughland CM, Lubinski J, Luykx JJ, Lynham A, Macek M Jr, Mackinnon A, Magnusson PKE, Maher BS, Maier W, Malaspina D, Mallet J, Marder SR, Marsal S, Martin AR, Martorell L, Mattheisen M, McCarley RW, McDonald C, McGrath JJ, Medeiros H, Meier S, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mitjans M, Molden E, Molina E, Molto MD, Mondelli V, Moreno C, Morley CP, Muntané G, Murphy KC, Myin-Germeys I, Nenadić I, Nestadt G, Nikitina-Zake L, Noto C, Nuechterlein KH, O'Brien NL, O'Neill FA, Oh SY, Olincy A, Ota VK, Pantelis C, Papadimitriou GN, Parellada M, Paunio T, Pellegrino R, Periyasamy S, Perkins DO, Pfuhlmann B, Pietiläinen O, Pimm J, Porteous D, Powell J, Quattrone D, Quested D, Radant AD, Rampino A, Rapaport MH, Rautanen A, Reichenberg A, Roe C, Roffman JL, Roth J, Rothermundt M, Rutten BPF, Saker-Delye S, Salomaa V, Sanjuan J, Santoro ML, Savitz A, Schall U, Scott RJ, Seidman LJ, Sharp SI, Shi J, Siever LJ, Sigurdsson E, Sim K, Skarabis N, Slominsky P, So HC, Sobell JL, Söderman E, Stain HJ, Steen NE, Steixner-Kumar AA, Stögmann E, Stone WS, Straub RE, Streit F, Strengman E, Stroup TS, Subramaniam M, Sugar CA, Suvisaari J, Svrakic DM, Swerdlow NR, Szatkiewicz JP, Ta TMT, Takahashi A, Terao C, Thibaut F, Toncheva D, Tooney PA, Torretta S, Tosato S, Tura GB, Turetsky BI, Üçok A, Vaaler A, van Amelsvoort T, van Winkel R, Veijola J, Waddington J, Walter H, Waterreus A, Webb BT, Weiser M, Williams NM, Witt SH, Wormley BK, Wu JQ, Xu Z, Yolken R, Zai CC, Zhou W, Zhu F, Zimprich F, Atbaşoğlu EC, Ayub M, Benner C, Bertolino A, Black DW, Bray NJ, Breen G, Buccola NG, Byerley WF, Chen WJ, Cloninger CR, Crespo-Facorro B, Donohoe G, Freedman R, Galletly C, Gandal MJ, Gennarelli M, Hougaard DM, Hwu HG, Jablensky AV, McCarroll SA, Moran JL, Mors O, Mortensen PB, Müller-Myhsok B, Neil AL, Nordentoft M, Pato MT, Petryshen TL, Pirinen M, Pulver AE, Schulze TG, Silverman JM, Smoller JW, Stahl EA, Tsuang DW, Vilella E, Wang SH, Xu S; Indonesia Schizophrenia Consortium; PsychENCODE; Psychosis Endophenotypes International Consortium; SynGO Consortium, Adolfsson R, Arango C, Baune BT, Belangero SI, Børglum AD, Braff D, Bramon E, Buxbaum JD, Campion D, Cervilla JA, Cichon S, Collier DA, Corvin A, Curtis D, Forti MD, Domenici E, Ehrenreich H, Escott-Price V, Esko T, Fanous AH, Gareeva A, Gawlik M, Gejman PV, Gill M, Glatt SJ, Golimbet V, Hong KS, Hultman CM, Hyman SE, Iwata N, Jönsson EG, Kahn RS, Kennedy JL, Khusnutdinova E, Kirov G, Knowles JA, Krebs MO, Laurent-Levinson C, Lee J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, Malhotra D, McIntosh A, McQuillin A, Menezes PR, Morgan VA, Morris DW, Mowry BJ, Murray RM, Nimgaonkar V, Nöthen MM, Ophoff RA, Paciga SA, Palotie A, Pato CN, Qin S, Rietschel M, Riley BP, Rivera M, Rujescu D, Saka MC, Sanders AR, Schwab SG, Serretti A, Sham PC, Shi Y, St Clair D, Stefánsson H, Stefansson K, Tsuang MT, van Os J, Vawter MP, Weinberger DR, Werge T, Wildenauer DB, Yu X, Yue W, Holmans PA, Pocklington AJ, Roussos P, Vassos E, Verhage M, Visscher PM, Yang J, Posthuma D, Andreassen OA, Kendler KS, Owen MJ, Wray NR, Daly MJ, Huang H, Neale BM, Sullivan PF, Ripke S, Walters JTR, O'Donovan MC; Schizophrenia Working Group of the Psychiatric Genomics Consortium. Mapping genomic loci implicates genes and synaptic biology in schizophrenia. Nature. 2022;604:502-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1733] [Cited by in RCA: 1461] [Article Influence: 487.0] [Reference Citation Analysis (0)] |
| 39. | National Institute of Mental Health. Schizophrenia. [cited 20 November 2022]. Available from: https://www.nimh.nih.gov/health/topics/schizophrenia. |
| 40. | Khavari B, Cairns MJ. Epigenomic Dysregulation in Schizophrenia: In Search of Disease Etiology and Biomarkers. Cells. 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 65] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 41. | Jiang T, Zong L, Zhou L, Hou Y, Zhang L, Zheng X, Han H, Li S, Zhang W, Zhang J, Deng C, Jia Y, Zhao C. Variation in global DNA hydroxymethylation with age associated with schizophrenia. Psychiatry Res. 2017;257:497-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 19] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 42. | Zong L, Zhou L, Hou Y, Zhang L, Jiang W, Zhang W, Wang L, Luo X, Wang S, Deng C, Peng Z, Li S, Hu J, Zhao H, Zhao C. Genetic and epigenetic regulation on the transcription of GABRB2: Genotype-dependent hydroxymethylation and methylation alterations in schizophrenia. J Psychiatr Res. 2017;88:9-17. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 43. | National Institute of Mental Health. Bipolar Disorder. [cited 20 November 2022]. Available from: https://www.nimh.nih.gov/health/topics/bipolar-disorder. |
| 44. | Gavin DP, Sharma RP, Chase KA, Matrisciano F, Dong E, Guidotti A. Growth arrest and DNA-damage-inducible, beta (GADD45b)-mediated DNA demethylation in major psychosis. Neuropsychopharmacology. 2012;37:531-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 91] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 45. | Dong E, Gavin DP, Chen Y, Davis J. Upregulation of TET1 and downregulation of APOBEC3A and APOBEC3C in the parietal cortex of psychotic patients. Transl Psychiatry. 2012;2:e159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 95] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 46. | Penner-Goeke S, Binder EB. Epigenetics and depression. Dialogues Clin Neurosci. 2019;21:397-405. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 150] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 47. | Li M, D'Arcy C, Li X, Zhang T, Joober R, Meng X. What do DNA methylation studies tell us about depression? Transl Psychiatry. 2019;9:68. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 79] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 48. | Feng J, Pena CJ, Purushothaman I, Engmann O, Walker D, Brown AN, Issler O, Doyle M, Harrigan E, Mouzon E, Vialou V, Shen L, Dawlaty MM, Jaenisch R, Nestler EJ. Tet1 in Nucleus Accumbens Opposes Depression- and Anxiety-Like Behaviors. Neuropsychopharmacology. 2017;42:1657-1669. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 50] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 49. | Zhang Q, Hu Q, Wang J, Miao Z, Li Z, Zhao Y, Wan B, Allen EG, Sun M, Jin P, Xu X. Stress modulates Ahi1-dependent nuclear localization of ten-eleven translocation protein 2. Hum Mol Genet. 2021;30:2149-2160. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |
| 50. | Cheng Y, Sun M, Chen L, Li Y, Lin L, Yao B, Li Z, Wang Z, Chen J, Miao Z, Xin N, Huang L, Allen EG, Wu H, Xu X, Jin P. Ten-Eleven Translocation Proteins Modulate the Response to Environmental Stress in Mice. Cell Rep. 2018;25:3194-3203.e4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 53] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 51. | Mahjani B, Bey K, Boberg J, Burton C. Genetics of obsessive-compulsive disorder. Psychol Med. 2021;51:2247-2259. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 56] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 52. | Pauls DL, Abramovitch A, Rauch SL, Geller DA. Obsessive-compulsive disorder: an integrative genetic and neurobiological perspective. Nat Rev Neurosci. 2014;15:410-424. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 442] [Cited by in RCA: 496] [Article Influence: 45.1] [Reference Citation Analysis (0)] |
| 53. | Yue W, Cheng W, Liu Z, Tang Y, Lu T, Zhang D, Tang M, Huang Y. Genome-wide DNA methylation analysis in obsessive-compulsive disorder patients. Sci Rep. 2016;6:31333. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 34] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 54. | Cappi C, Diniz JB, Requena GL, Lourenço T, Lisboa BC, Batistuzzo MC, Marques AH, Hoexter MQ, Pereira CA, Miguel EC, Brentani H. Epigenetic evidence for involvement of the oxytocin receptor gene in obsessive-compulsive disorder. BMC Neurosci. 2016;17:79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 51] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 55. | Grünblatt E, Marinova Z, Roth A, Gardini E, Ball J, Geissler J, Wojdacz TK, Romanos M, Walitza S. Combining genetic and epigenetic parameters of the serotonin transporter gene in obsessive-compulsive disorder. J Psychiatr Res. 2018;96:209-217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 56. | Park CI, Kim HW, Jeon S, Kang JI, Kim SJ. Reduced DNA methylation of the oxytocin receptor gene is associated with obsessive-compulsive disorder. Clin Epigenetics. 2020;12:101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 57. | D'Addario C, Bellia F, Benatti B, Grancini B, Vismara M, Pucci M, De Carlo V, Viganò C, Galimberti D, Fenoglio C, Scarpini E, Maccarrone M, Dell'Osso B. Exploring the role of BDNF DNA methylation and hydroxymethylation in patients with obsessive compulsive disorder. J Psychiatr Res. 2019;114:17-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 21] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 58. | National Institute of Mental Health. Substance Use and Co-Occurring Mental Disorders. [cited 20 November 2022]. Available from: https://www.nimh.nih.gov/health/topics/substance-use-and-mental-health. |
| 59. | National Institute on Drug Abuse. Common Comorbidities with Substance Use Disorders Research Report. [cited 20 November 2022]. Available from: https://nida.nih.gov/publications/research-reports/common-comorbidities-substance-use-disorders/introduction. |
| 60. | Clark SL, Chan RF, Zhao M, Xie LY, Copeland WE, Penninx BWJH, Aberg KA, van den Oord EJCG. Dual methylation and hydroxymethylation study of alcohol use disorder. Addict Biol. 2022;27:e13114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 61. | Cadet JL, Jayanthi S. Epigenetics of addiction. Neurochem Int. 2021;147:105069. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 21] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 62. | Gatta E, Auta J, Gavin DP, Bhaumik DK, Grayson DR, Pandey SC, Guidotti A. Emerging Role of One-Carbon Metabolism and DNA Methylation Enrichment on δ-Containing GABAA Receptor Expression in the Cerebellum of Subjects with Alcohol Use Disorders (AUD). Int J Neuropsychopharmacol. 2017;20:1013-1026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 63. | Jayanthi S, Gonzalez B, McCoy MT, Ladenheim B, Bisagno V, Cadet JL. Methamphetamine Induces TET1- and TET3-Dependent DNA Hydroxymethylation of Crh and Avp Genes in the Rat Nucleus Accumbens. Mol Neurobiol. 2018;55:5154-5166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 39] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 64. | Anier K, Urb M, Kipper K, Herodes K, Timmusk T, Zharkovsky A, Kalda A. Cocaine-induced epigenetic DNA modification in mouse addiction-specific and non-specific tissues. Neuropharmacology. 2018;139:13-25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 23] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 65. | Niinep K, Anier K, Eteläinen T, Piepponen P, Kalda A. Repeated Ethanol Exposure Alters DNA Methylation Status and Dynorphin/Kappa-Opioid Receptor Expression in Nucleus Accumbens of Alcohol-Preferring AA Rats. Front Genet. 2021;12:750142. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 11] [Reference Citation Analysis (0)] |