Published online Jun 2, 2023. doi: 10.5496/wjmg.v11.i2.8
Peer-review started: February 8, 2023
First decision: April 28, 2023
Revised: April 28, 2023
Accepted: May 22, 2023
Article in press: May 22, 2023
Published online: June 2, 2023
Processing time: 113 Days and 11.9 Hours
John Henryism (JH) is a strategy for dealing with chronic psychological stress characterized by high levels of physical effort and work. Cynicism is a belief that people are motivated primarily by self-interest. High scores on the JH scale and cynicism measures correlate with an increased risk of cardiovascular disease. High cynicism is also a hallmark of burnout syndrome, another known risk factor for heart disease.
To evaluate possible interactions between JH and cynicism hoping to clarify risk factors of burnout.
We analyzed genetic and psychological data available from the Database of Genotypes and Phenotypes for genome-wide associations with these traits. We split the total available samples and used plink to perform the association studies on the discovery set (n = 1852, 80%) and tested for replication using the validation set (n = 465). We used scikit-learn to perform supervised machine learning for developing genetic risk algorithms.
We identified 2, 727, and 204 genetic associations for scores on the JH, cynicism and cynical distrust (CD) scales, respectively. We also found 173 associations with high cynicism, 109 with high CD, but no associations with high JH. We also produced polygenic classifiers for high cynicism using machine learning with areas under the receiver operator characteristics curve greater than 0.7.
We found significant genetic components to these traits but no evidence of an interaction. Therefore, while there may be a genetic risk, JH is not likely a burnout risk factor.
Core Tip: This study evaluates the interaction of a job-related cardiovascular disease risk factor (John Henryism) and the development of occupational burnout (specifically the cynicism and cynical distrust components). Genome-wide associations and statistical genetics revealed that while John Henryism is not a risk factor for burnout syndrome, there are independent genetic risk factors for both John Henryism and cynicism. These new results provide additional tools to industrial and occupational psychologists, as well as cardiologists, to help reduce burnout incidence.
- Citation: Chapleau RR. Genome-wide associations, polygenic risk, and Mendelian randomization reveal limited interactions between John Henryism and cynicism. World J Med Genet 2023; 11(2): 8-20
- URL: https://www.wjgnet.com/2220-3184/full/v11/i2/8.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v11.i2.8
First described formally by Freudenberger[1] and later expanded upon by Maslach and Jackson[2], burnout syndrome is generally considered to be a response to long-term occupational stress[3]. The most widely used definition of burnout syndrome is a 3-component syndrome comprised of emotional exhaustion, depersonalization or cynicism, and feelings of low professional efficacy or personal[4]. High levels of burnout syndrome components have been associated with increased disease prevalence including a heightened risk of cardiovascular disease[5,6], impaired cognitive function[7], increased sleep disorders[8,9], and even type II diabetes and hyperlipidemia[10,11]. Clearly, the impacts of burnout syndrome extend beyond just performance in the workplace and can adversely affect health, well-being, and quality of life.
Another response to chronic social stressors is John Henryism (JH), a coping mechanism by which an individual exerts increased effort to overcome stress[12]. As with burnout syndrome, the effects of JH affect both the individual’s health and the workplace. Also, like the effects of burnout syndrome, JH can have a negative impact on cardiovascular health, hypertension, and increased rates of alcoholism[13-16]. Paradoxically, high levels of JH have been associated with lower levels of depressive symptoms in African Americans of varying socioeconomic status, suggesting a protective effect on mental health[17]. These findings suggest a nuanced interaction between JH and overall health and well-being.
The literature on the genetic contribution to burnout syndrome and JH is limited. Preliminary work regarding the heritability of cynicism (one of the three components of burnout syndrome) was reported in the late 1980’s to early 1990’s and revealed mixed results. One twin study of cynical hostility revealed a non-significant genetic component[18]. In contrast, three other twin studies using the Cook Medley Hostility Scale and the MMPI showed significant heritability[19-21]. The genetic heritability of JH also has limited reports, with one estimate suggesting up to 35% of the variability is explained by genetic factors[22,23]. To the best of our knowledge, there have not been any genome-wide or candidate gene studies performed to assess the genetic contribution to these traits.
Here we report the results of our study investigating the relationship between JH and cynicism. We hypothesized that JH would exert a causal influence on cynicism. We came to this hypothesis because of the nature of JH as a coping mechanism for dealing with discriminatory acts and the skepticism and negative view of others inherent to cynicism. We tested this hypothesis through the statistical approach called Mendelian randomization[24], where genetic associations with JH are considered for their independent influence on cynicism. In order to take this approach, we first performed a genome-wide association study (GWAS) to identify genetic variables for JH and cynicism independently. We also extended those GWAS results to develop polygenic risk scores (risk scores considering multiple genetic variants) for each trait and assessed if the higher genetic risk in one trait correlated to higher levels of the other trait. To our knowledge, this study is the first to report genetic associations with any of the three outcomes.
This study was reviewed and approved by WCG IRB (Study number 1332892) for human subjects research oversight. All data were obtained from the National Institutes of Health’s Database of Genotypes and Phenotypes (dbGAP). We used data from the Coronary Artery Risk Development in Young Adults (CARDIA) Study (dbGAP study accession phs000285)[25]. The CARDIA cohort study design was a longitudinal study, but we used the data in a retrospective fashion. We followed the STREGA guidelines available from the EQUATOR network[26]. The guideline table with annotations regarding how we addressed each point is available as Supplementary material.
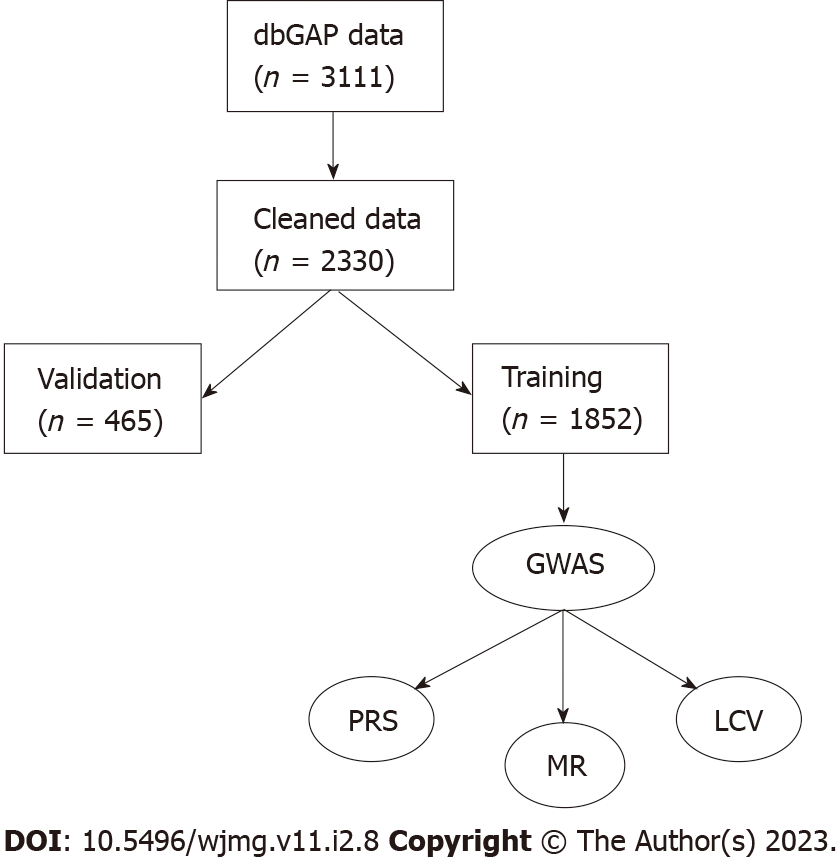
JH was measured by the 12-item John Henryism Active Coping Scale, and responses were reverse-coded. JH was calculated as a mean of the responses. As our goal was to observe the effect of having above-average JH, we defined a dichotomous variable of JH as individuals with scores above the median (49) as having high JH[13,15]. Individuals with scores at or below the median were considered average or below average JH. We also performed tests using the JH score as a continuous, mean-centered variable. The JH data were obtained from dbGAP accession number phv001133534.v2.p2.c1. Cynicism and cynical distrust (CD) were measured as 12- and 8-item subscales of the Cook-Medley Hostility Scale (CMHS), respectively[27-29]. Mean-centered continuous variables and median-adjusted dichotomous variables were created for cynicism and CD in the same manner as for JH, resulting in four CMHS-derived variables (continuous cynicism, high/Low cynicism, continuous CD, and high/Low CD). The CMHS data were obtained from dbGAP accession number phv00113478.v2.p2.c1. As not all participants completed all questionnaires, missing data were filled in as the mean. Processed data were then split into a validation set (20%, n = 623) and the remainder were used for training (Figure 1). Statistical analyses were performed in R version 4.2.0.
We performed a power analysis for our study to determine if the sample sizes available in the CARDIA cohort were sufficient to identify significant genetic associations. We assumed a 10% effect allele frequency (EAF) in the control population, 20% type-II error rate (beta) and 5% type-I error rate (alpha). We used the observed case-control ratios for each condition to calculate the statistical power. We report the results of the analysis as the minimum EAF in the case population to achieve 80% power in Table 1 alongside the cohort characteristics.
| John Henryism | Cynicism | Cynical distrust | |
| Training high | 877 (47.4) | 724 (39.1) | 680 (36.7) |
| Training not high | 975 | 1128 | 1172 |
| Case:Control ratio | 1:1.1 | 1:1.6 | 1:1.7 |
| Minimum EAF, % | 14.5 | 15.7 | 16.0 |
| Validation high | 246 (52.9) | 180 (38.7) | 171 (36.8) |
| Validation not high | 219 | 285 | 294 |
Microarray (Affymetrix Genome-Wide Human SNP 6.0 Array) genotype data were obtained from the CARDIA study (dbGAP accession numbers phg000092.v2 and phg000098.v2). Genetic data were pre-processed to ensure uniformity from the original plink data. Briefly, the binary plink file sets were merged and filtered for autosomal genotypes with less than 10% missing genotype calls (sample or locus), a minor allele frequencies threshold was set at 1%, and a Hardy-Weinberg equilibrium threshold of 0.0001 was used for filtering out spurious variants. After pre-processing, they were split into the training and validation sets using the sample identifiers defined in the phenotype data splitting.
We conducted GWAS using plink 1.9 evaluating only the total scores (not scores for the questionnaire responses) and high/Low status for each trait, totaling 6 phenotypes. We used the following command for the association: plink --bfile <training dataset prefix> --memory 15000 --pheno <phenotype filename>.csv --all-pheno --assoc --pfilter 0.001 --adjust qq-plot --out <output filename>. For assessing the replication of associations in continuous variables traits, we repeated the GWAS in plink restricting the input variants to only those candidates identified in the discovery phase. For dichotomous traits, the set of candidate single nucleotide polymorphisms (SNPs) identified as significantly associated with the trait (P < 5 × 10-8) or suggestively associated (P < 5 × 10-6) were then evaluated by chi-square test using the test dataset. Additionally, each analysis was repeated conditioning upon the highest associated SNP.
Our method of PRS development was using the machine learning package scikit-learn[30]. We used four supervised classifiers [Ridge, multi-layer perceptron, random forest, and k-nearest neighbors (KNN)] and iterated through the relevant parameter space for each classifier (e.g., number of neighbors for KNN). The classifiers were trained on the training set (n = 1852) and validated using the test dataset (n = 465). Each classifier was evaluated using the area under the receiver operator characteristics (ROC) curve (AUC) and the model with the best AUC was saved for each classifier. Finally, a ROC curve comparing the best models of each classifier was created.
We performed two-sample Mendelian randomization (2SMR) estimates using the TwoSampleMR R package[24]. We used the MR Egger regression[31], inverse variance weighted estimator[32], weighted median estimator[33], and Wald ratio estimator[34] algorithms. Instrumental variables (IV, SNPs associated with the exposure) were extracted by P value thresholds 5 × 10-6 and 5 × 10-8. We excluded SNPs in strong linkage disequilibrium (LD) to reduce bias and used a clumping process with European samples from the 1000 Genomes Project (r2 < 0.001, window size = 10000). If SNPs identified in the exposure dataset were not in the outcome dataset, proxy SNPs in LD (r2 > 0.9) were used as instrumental variables. For the sensitivity analysis, we performed heterogeneity testing using Cochran’s Q and I2 analyses[35] and tested pleiotropy on the weighted median estimation results[33].
Statistical analyses for assessing variant associations were performed in R. To confirm the association of the variants identified for the high cynicism and high CD variables, we created a matrix containing the allele counts for each candidate variant. We then performed a chi-square analysis using the chisq.test function within base R using the allele count matrix as the input argument. The statistical analyses of the PRS estimators were performed in the scikit-learn package. As the class allocation displayed only slight imbalance (control/case group sample size ratios of 1:2.1, 1:2.5, and 1:2.7 for JH, cynicism, and CD, respectively), we used the ROC AUC for evaluating performance of the PRS classification models.
The initial phenotype dataset consisted of 3111 samples (Figure 1). The median scores for JH, cynicism, and CD were 49, 6, and 3, respectively. Score ranges were 26 to 60 for JH, 0 to 13 for cynicism, and 0 to 8 for CD. There were 1497 (48.1%) samples with high JH scores (JH > 49), 1228 (39.5%) with high cynicism (> 6), and 1163 (37.4%) with high CD scores (> 3). We found minimal correlation between JH and cynicism or CD (Pearson r = 0.159 and 0.118, respectively). After splitting the data into training (80%) and validation (20%) sets, the median scores were 49, 6, and 3, respectively, for the training set and 50, 5, and 3, respectively, for the validation set. The percent of samples above the population median scores was 47.1%, 40.0%, and 37.8%, respectively, for the training set and 52.3%, 37.4%, and 35.6%, respectively, for the validation set. T-tests showed that the distribution of samples in the subsets was not statistically different from the original distribution (JH training vs original P = 0.52; JH validation vs original P = 0.12; cynicism training vs original P = 0.48; cynicism validation vs original P = 0.08; CD training vs original P = 0.62; and CD validation vs original P = 0.22). Evaluating the Shapiro-Wilk normality test in R reveals that none of the six variables are normally distributed (all have P < 2.2 × 10-16). The Shapiro-Wilk W constants for the distributions of quantitative scores were 0.980, 0.935, and 0.970 for JH, CD, and cynicism, respectively, while the W constants for the dichotomous traits were 0.635, 0.615, and 0.622, respectively. The skewness of the three continuous traits was -0.511, 0.118, and 0.373, respectively, while the kurtosis values were 3.11, 1.01, and 2.18, respectively. From these results, we see that the data are generally positively skewed and primarily platykurtic.
The merged genetic dataset contained 2466 samples (1162 from accession phg000092 and 1441 from accession phg000098, with 137 overlapping between the two studies), of which 978 (42.1%) were male. The two datasets contained 909662 and 720622 markers, respectively. Following SNP filtering, there were 561045 variants remaining (153333 for missingness, 13410 below 1% MAF, and 144454 not passing the Hardy-Weinberg filter). 136 samples were removed for having greater than 10% missing genotype calls, creating a final dataset of 2330 samples. The total genotyping rate for the dataset was 99.4%. After splitting samples using the training and validation sets defined in the phenotype stage, there were 1852 samples in the training set, 465 samples in the validation set, and 13 (0.6%) of the samples with genetic data did not have phenotype data (Table 1).
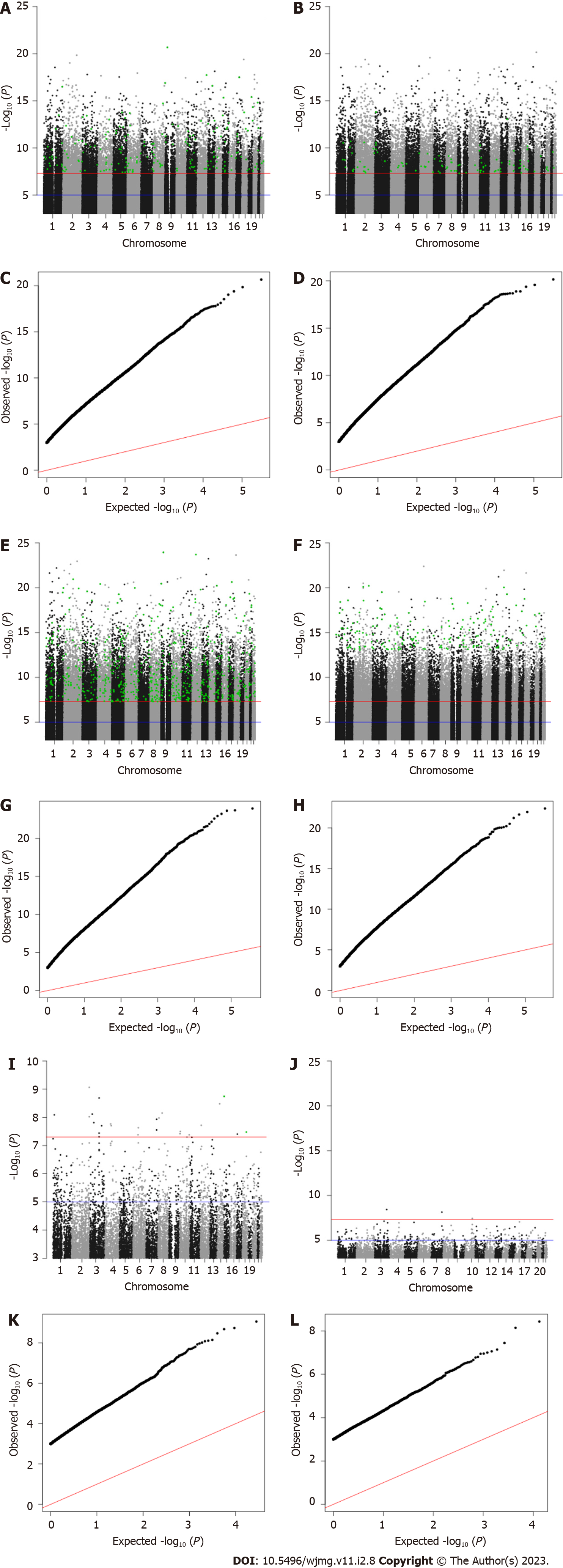
Our discovery phase GWAS analysis of 1852 samples revealed 25 candidate variants with Bonferroni-corrected P values below 5 × 10-8 associated with JH, 28926 candidates associated with cynicism, and 14134 candidate associations with CD (Figure 2). For each of the candidate variants, we performed a second GWAS on the test sample set (n = 465) using only the candidate SNPs identified in the first analysis. We found that 2 SNPs replicated as associated with the quantitative JH trait (P = 0.002), 727 replicated for association with cynicism (P = 1.73 × 10-6), and 204 replicated for association with CD (P = 3.54 × 10-6).
Similar to how we analyzed the continuous variables, we identified in the dichotomous variable analysis 3 candidate variants associated with high JH, 708 associations with high cynicism, and 17507 associations with high CD. After evaluating for replication of significance (P < 0.05) in the validation set of 465 samples, we found 0, 173, and 109 significantly associated variants that replicated in our test set for JH, cynicism, and CD, respectively. Of the 173 replicated variants associated with cynicism, 19 were located across 9 distinct loci (defined as within a 250000-base pair window on the same chromosome) and the other 154 were distinct variants. We also observed that 79 of the 173 high cynicism-associated variants were also associated with the quantitative trait, while none of the high CD-associated variants were present in the quantitative trait CD list. Lists of all candidate SNPs and replicated SNPs are available from the corresponding author upon reasonable request.
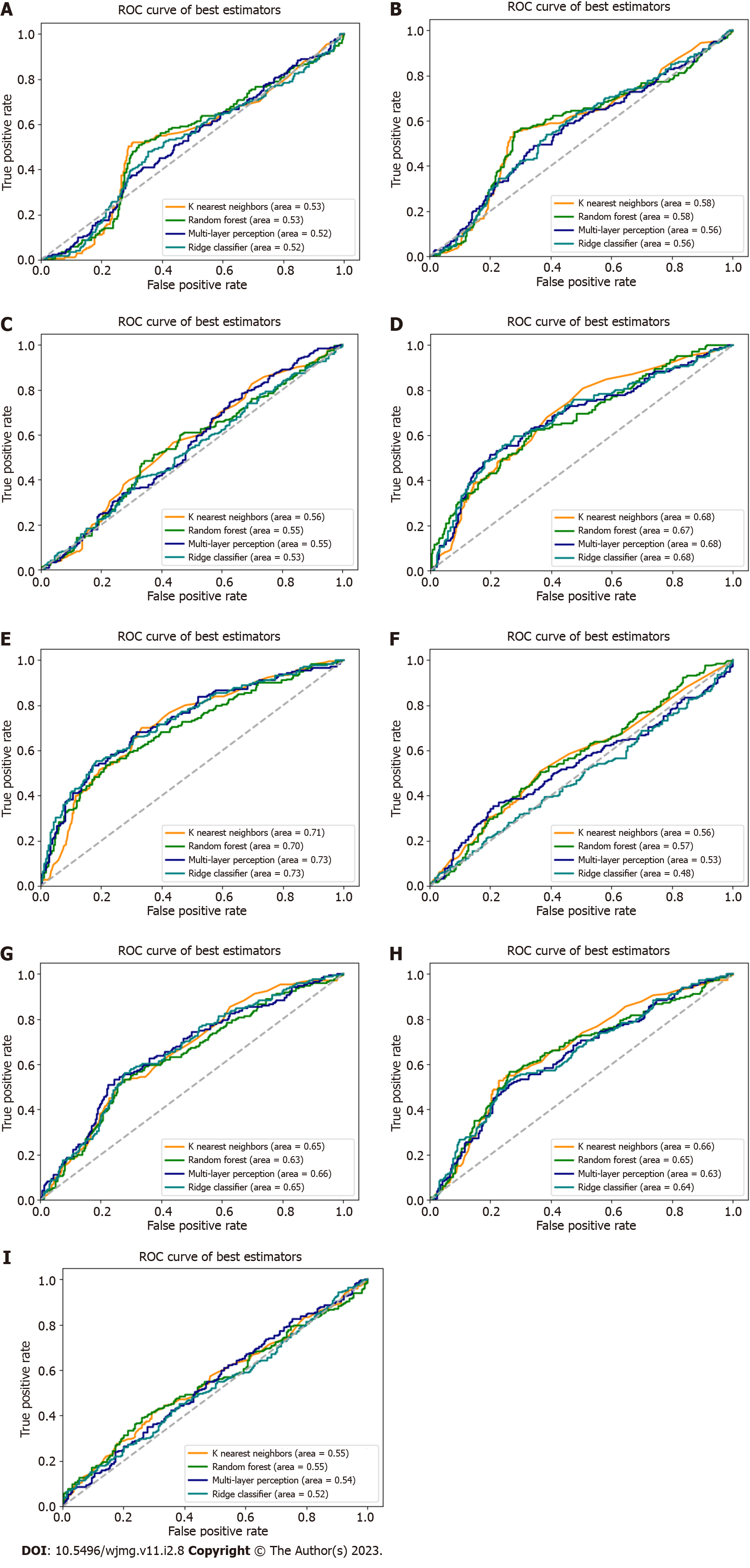
We used the replicated SNPs for each condition cynicism and CD to evaluate the predictive capability for each, addressing the question of whether genetic variants associated with high JH, for example, were predictive of high cynicism. As no SNPs replicated for high JH, we used the candidate SNPs. Using the scikit-learn software package, we performed the nine tests for each input/output combination (e.g., cynicism vs CD, cynicism, and JH) with the four classification methods. Our results (Figure 3, Table 2) reveal that PRS algorithms based on genetic variants associated with high cynicism are predictive of high cynicism (AUC range = 0.696-0.732) and high CD (AUC range = 0.652-0.684), whereas algorithms trained on genetic markers of high JH or high CD are not predictive for any trait. These classifiers would be considered to be acceptable predictors[36] with AUC values near 0.7, these results show that cynicism and CD are genetically related, reinforcing the psychological relationship, and that JH is a distinct trait deriving from different genetic contributions.
| Input vs output pair | Ridge | MLP | RFC | KNN |
| JH vs JH | 0.524 | 0.542 | 0.545 | 0.550 |
| JH vs cynicism | 0.638 | 0.634 | 0.649 | 0.660 |
| JH vs CD | 0.653 | 0.656 | 0.651 | 0.630 |
| Cynicism vs JH | 0.482 | 0.533 | 0.565 | 0.563 |
| Cynicism vs cynicism | 0.732 | 0.728 | 0.696 | 0.712 |
| Cynicism vs CD | 0.681 | 0.676 | 0.671 | 0.684 |
| CD vs JH | 0.526 | 0.545 | 0.546 | 0.563 |
| CD vs cynicism | 0.563 | 0.556 | 0.577 | 0.580 |
| CD vs CD | 0.516 | 0.516 | 0.526 | 0.533 |
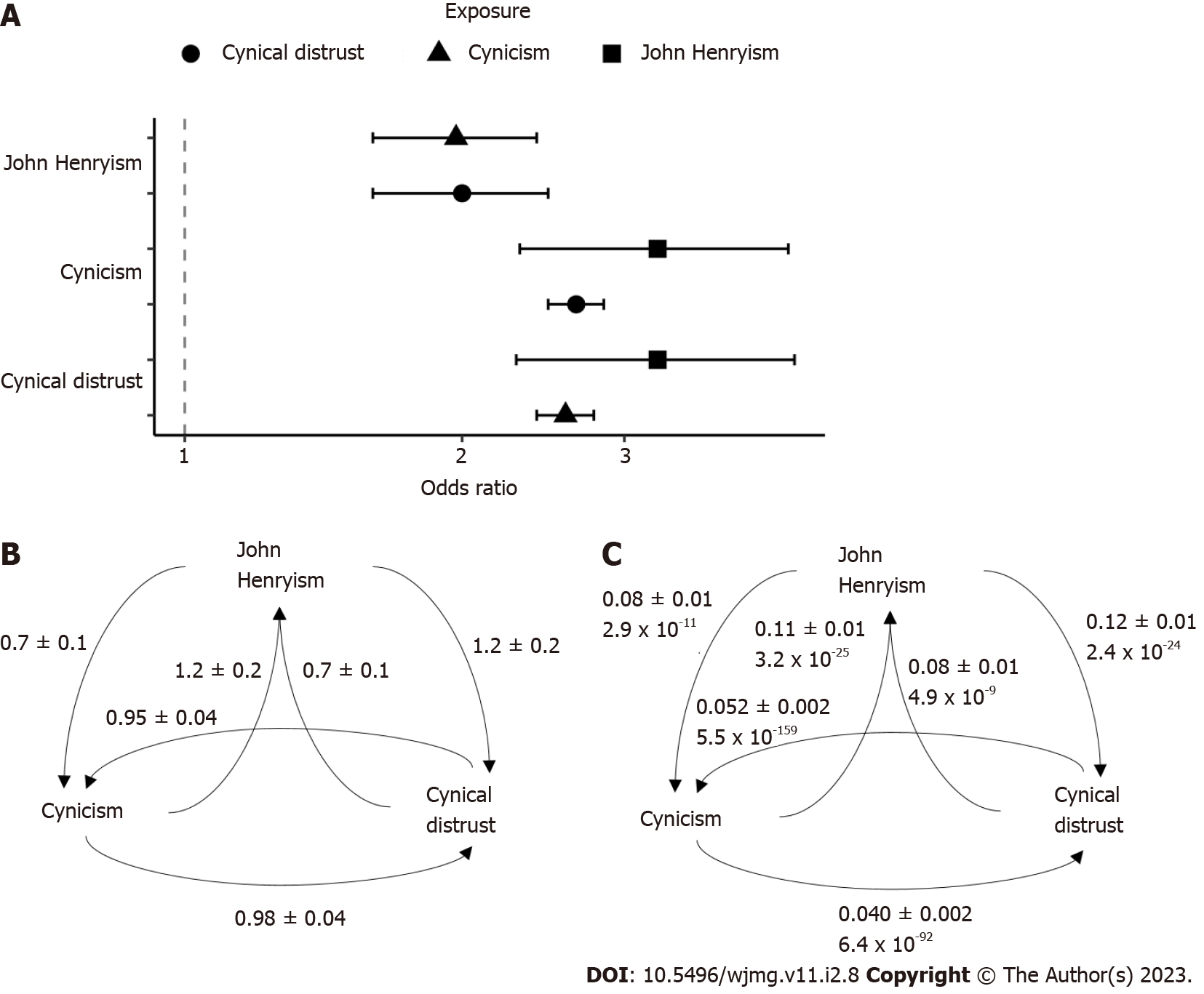
Our two-sample Mendelian randomization analyses using the GWAS summary statistics previously described revealed significant causal relationships between all three traits (Figure 4). The odds ratios (Figure 4A) and beta coefficients (Figure 4B) for the relationships were highly similar, with odds ratios ranging from 2.0 (for the causal effect of JH on cynicism and the causal effect of CD on JH) to 3.3 (for the inverse effects), with the CD/cynicism relationship near 2.6. The mendelian randomization (MR) comparisons involved 4680 variants for the JH/cynicism relationship, 4262 variants for the JH/CD relationship, and 131185 variants for the CD/cynicism relationship. We performed pleiotropy and heterogeneity tests to assess the sensitivity of the MR estimates. We found significant pleiotropy in all cases, with the Egger intercepts significantly different from zero (Figure 4C)[37].
Our results from genetic analyses suggest that there is not a significant relationship between JH and cynicism. These observations are consistent with previously published literature using only psychometric approaches. Adams and co-workers[38] demonstrated that JH scores were not significantly correlated with the Cook Medley Hostility Scale, of which we used the cynicism subscale herein. In contrast, a large study investigating the effects of various psychosocial factors on chronic kidney disease found that JH and hostility exhibited inverse risk factor loading[39]. Finally, initial reports from the CARDIA study suggest a weak but significant correlation between hostility and effortful coping (r = 0.14, P < 0.05), especially among younger, less educated individuals that were more likely to consume alcohol and be smokers[40].
Although our findings do not substantiate a relationship between JH and cynicism, and subsequently burnout syndrome, they do suggest that there is a genetic component to both outcomes. Research among the general population has shown that hostility is an independent risk factor for cardiovascular disease[41], which has also been reinforced with evidence in population-specific work on African Americans[42]. Our results suggest that a significant predisposition to hostility and cynicism, when coupled with chronic occupational stress and hostility, could lead to elevated levels of burnout and significant long-term health decline.
Our assessment of causal effects revealed significant pleiotropy in the relationships, suggesting that any interaction which may exist is indirect. Our results do not support a causal effect, but neither do they provide sufficient evidence to refute any such relationship. Indeed, our assessment using multiple MR estimators provides limited support that there is a relationship between these traits, but that such an interaction may not be mediated by genetics. We used the MR Egger estimate to evaluate violations of the instrumental variable assumptions. We found that these assumptions are not valid in our analysis (the Egger intercepts were significantly non-zero). However, the magnitude of the impact of any deviations must be small as none of the six beta coefficients determined by the MR Egger method were outliers from the other four estimates (i.e., all MR Egger estimates were within the 95% confidence interval of the mean observed beta coefficient). Therefore, it appears that using the MR Egger estimator did not introduce additional bias or increase the Type I error rate[32].
There are three major limitations of our study. First, the pleiotropic analysis suggests that there are confounders involved in the interaction and, therefore, this pleiotropy highlights one limitation of our study in that we did not consider covariates in the initial analysis. The CARDIA study included many demographic and phenotypic datasets from which covariates could be identified. As the outcomes we measured tend to change with age[40], at least age at the time the questionnaire could be used as a future covariate. Indeed, the weak correlations reported by Albanese were largely derived from stratified data. While it was beyond the scope of our initial view of the interactions between genetics, JH, and cynicism to assess covariates, additional covariates could include gender, race, or personality, among others.
A second limitation is that the sample size is relatively small for a large-scale genomics study. Our discovery phase was comprised of only 1852 samples and our replication phase consisted of another 465 samples. While an a priori power analysis revealed that these sample sizes should have been powered to identify significant associations, it would be prudent to repeat these observations in larger populations to confirm which, if any, variants remain significantly associated with JH, cynicism, and/or CD. Due to this sample size limitation, the generalizability of our results should be considered alongside larger-scale studies, especially with regard to burnout syndrome, for which not much is published in the genetic literature.
Finally, there is a significant deviation from normality in all of the datasets, as determined by analyzing qq-plots (Figure 2). Considering the non-normal distribution of the data and the large sample distribution within the dataset, this deviation from normality was expected (both Cook-Medley and JH scores were previously acknowledged to have skewed distributions)[40]. What was unexpected was the relatively large number of significantly associated variants for cynicism and CD found in both the discovery and replication phases. Viewed alone, the large number of associated variants is cause for skepticism in the results. However, when taken into consideration along with the predictive ability of ML algorithms (Figure 3) developed based upon these replicated variants, the evidence for a significant genetic contribution to cynicism and CD becomes stronger.
In conclusion, our results suggest that high levels of cynicism and CD have a significant genetic component and there may some genetic component to levels of JH. This genetic component of cynicism and CD appears to have some common effects as polygenic risk scores developed to classify individuals with high scores in one trait are reasonably effective at classifying individuals with the other trait (AUC > 0.6). Our results are insufficient to determine if this correlation is based upon causation, however, as there is a significant amount of observed pleiotropy in the MR analysis, suggesting the existence of confounding variables.
Our hypothesis was that cynicism, a distrustful attitude that assumes people are mainly driven by self-interest, would result from John Henryism (JH), a coping strategy that involves working hard and exerting high levels of effort to deal with chronic stress. We reasoned that as JH is a way of handling discrimination, and that cynicism involves skepticism and negativity towards others, the two traits would be related. Furthermore, both JH and cynicism are linked to a higher risk of cardiovascular disease, while cynicism also often accompanies burnout syndrome, another cardiovascular risk factor.
Rates of burnout are increasing broadly across the globe. Burnout can cause physiological and emotional distress. Understanding the role of stress coping skills such as JH may help clarify the role of occupational stress in overall health and well-being.
The present study aimed to determine if JH is correlated with the cynicism component of burnout by using approaches from statistical genetics.
Genotype and phenotype data from the “CARDIA Cohort” study were obtained from the Database of Genotypes and Phenotypes. Genome-wide association studies (GWAS) were performed in plink version 1.9 on the log-normalized JH, cynicism, and cynical distrust (CD) phenotypes as well as a binary “high/Low” trait for each phenotype using a two-stage discovery and replication approach. The GWAS results were then used to develop polygenic risk score (PRS) algorithms using supervised machine learning in scikit-learn. Significant variants identified in the discovery stage were tested for replication by performing GWAS in a second, independent set of data restricting variants only to those candidates identified in discovery (for continuous variables) or through chi-square testing for the binary variable. The performance of the PRS algorithms at classifying individuals as “high” or “low” for the phenotype was evaluated in scikit-learn using the area under the receiver operator characteristics curve.
The GWAS identified significant variant associations with JH (2), cynicism (727), or CD (204) scores and with high cynicism (173) or CD (109). PRS classifiers were successfully developed for cynicism and CD (AUC > 0.65), but not for JH. Neither of the classifiers for cynicism or CD could predict JH traits, nor could the JH classifier predict cynicism or CD.
There are genetic variants associated with each trait, however JH active coping does not appear to be correlated with cynicism or CD levels.
The genetic associations with these phenotypes suggest that further research could provide insight into how each trait results in health impacts such as cardiovascular disease.
The authors wish to thank Mr. Billy Thompson, Ms. CharLee Martin, Mr. Eric Rigby, and Mr. Warren Fridy for their assistance in coordinating data access and security. The authors also wish to thank the participants and the original depositors of the CARDIA study.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Genetics and heredity
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Arumugam VA, India S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Freudenberger HJ. Staff burnout. J Soc Issues. 1974;30:159-165. [DOI] [Full Text] |
| 2. | Maslach C, Jackson SE. The measurement of experienced burnout. J Occupational Behaviour. 1981;2:99-113. [DOI] [Full Text] |
| 3. | Güler Y, Şengül S, Çaliş H, Karabulut Z. Burnout syndrome should not be underestimated. Rev Assoc Med Bras (1992). 2019;65:1356-1360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 9] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 4. | Jackson SE, Schwab RL, Schuler RS. Toward an understanding of the burnout phenomenon. J Appl Psychol. 1986;71:630-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 454] [Cited by in RCA: 450] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 5. | Honkonen T, Ahola K, Pertovaara M, Isometsä E, Kalimo R, Nykyri E, Aromaa A, Lönnqvist J. The association between burnout and physical illness in the general population--results from the Finnish Health 2000 Study. J Psychosom Res. 2006;61:59-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 169] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 6. | Melamed S, Kushnir T, Shirom A. Burnout and risk factors for cardiovascular diseases. Behav Med. 1992;18:53-60. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 277] [Cited by in RCA: 281] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 7. | Koutsimani P, Montgomery A, Masoura E, Panagopoulou E. Burnout and Cognitive Performance. Int J Environ Res Public Health. 2021;18. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 8. | Amaral KV, Galdino MJQ, Martins JT. Burnout, daytime sleepiness and sleep quality among technical-level Nursing students. Rev Lat Am Enfermagem. 2021;29:e3487. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 9. | Stewart NH, Arora VM. The Impact of Sleep and Circadian Disorders on Physician Burnout. Chest. 2019;156:1022-1030. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 121] [Article Influence: 20.2] [Reference Citation Analysis (0)] |
| 10. | Melamed S, Shirom A, Toker S, Shapira I. Burnout and risk of type 2 diabetes: a prospective study of apparently healthy employed persons. Psychosom Med. 2006;68:863-869. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 125] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Shirom A, Westman M, Shamai O, Carel RS. Effects of work overload and burnout on cholesterol and triglycerides levels: the moderating effects of emotional reactivity among male and female employees. J Occup Health Psychol. 1997;2:275-288. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 61] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 12. | Rolle T, Vue Z, Murray SA, Shareef SA, Shuler HD, Beasley HK, Marshall AG, Hinton A. Toxic stress and burnout: John Henryism and social dominance in the laboratory and STEM workforce. Pathog Dis. 2021;79. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 28] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 13. | James SA, Hartnett SA, Kalsbeek WD. John Henryism and blood pressure differences among black men. J Behav Med. 1983;6:259-278. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 290] [Cited by in RCA: 293] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 14. | James SA. John Henryism and the health of African-Americans. Cult Med Psychiatry. 1994;18:163-182. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 379] [Cited by in RCA: 355] [Article Influence: 11.5] [Reference Citation Analysis (0)] |
| 15. | LeBrón AM, Schulz AJ, Mentz G, White Perkins D. John Henryism, socioeconomic position, and blood pressure in a multi-ethnic urban community. Ethn Dis. 2015;25:24-30. [PubMed] |
| 16. | Vargas EA, Li Y, Mahalingham R, Hui P, Liu G, Lapedis M, Liu JR. The double edge sword of John Henryism: Impact on patients' health in the People's Republic of China. J Health Psychol. 2020;25:2374-2387. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 17. | Robinson MN, Thomas Tobin CS. Is John Henryism a Health Risk or Resource?: Exploring the Role of Culturally Relevant Coping for Physical and Mental Health among Black Americans. J Health Soc Behav. 2021;62:136-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 36] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 18. | Cates DS, Houston BK, Vavak CR, Crawford MH, Uttley M. Heritability of hostility-related emotions, attitudes, and behaviors. J Behav Med. 1993;16:237-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 39] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 19. | Carmelli D, Rosenman RH, Swan GE. The Cook and Medley HO scale: a heritability analysis in adult male twins. Psychosom Med. 1988;50:165-174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 21] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 20. | Rose RJ. Genetic and environmental variance in content dimensions of the MMPI. J Pers Soc Psychol. 1988;55:302-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 6] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 21. | Smith TW, McGonigle M, Turner CW, Ford MH, Slattery ML. Cynical hostility in adult male twins. Psychosom Med. 1991;53:684-692. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 21] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 22. | Wang X, Trivedi R, Treiber F, Snieder H. Genetic and environmental influences on anger expression, John Henryism, and stressful life events: the Georgia Cardiovascular Twin Study. Psychosom Med. 2005;67:16-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 28] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Whitfield KE, Brandon DT, Robinson E, Bennett G, Merritt M, Edwards C. Sources of variability in John Henryism. J Natl Med Assoc. 2006;98:641-647. [PubMed] |
| 24. | Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R, Tan VY, Yarmolinsky J, Shihab HA, Timpson NJ, Evans DM, Relton C, Martin RM, Davey Smith G, Gaunt TR, Haycock PC. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4747] [Cited by in RCA: 4852] [Article Influence: 693.1] [Reference Citation Analysis (0)] |
| 25. | Friedman GD, Cutter GR, Donahue RP, Hughes GH, Hulley SB, Jacobs DR Jr, Liu K, Savage PJ. CARDIA: study design, recruitment, and some characteristics of the examined subjects. J Clin Epidemiol. 1988;41:1105-1116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1112] [Cited by in RCA: 1358] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 26. | Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, Khoury MJ, Cohen B, Davey-Smith G, Grimshaw J, Scheet P, Gwinn M, Williamson RE, Zou GY, Hutchings K, Johnson CY, Tait V, Wiens M, Golding J, van Duijn C, McLaughlin J, Paterson A, Wells G, Fortier I, Freedman M, Zecevic M, King R, Infante-Rivard C, Stewart A, Birkett N; STrengthening the REporting of Genetic Association Studies. STrengthening the REporting of Genetic Association Studies (STREGA): an extension of the STROBE statement. PLoS Med. 2009;6:e22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 361] [Cited by in RCA: 341] [Article Influence: 21.3] [Reference Citation Analysis (0)] |
| 27. | Barefoot JC, Peterson BL, Dahlstrom WG, Siegler IC, Anderson NB, Williams RB Jr. Hostility patterns and health implications: correlates of Cook-Medley Hostility Scale scores in a national survey. Health Psychol. 1991;10:18-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 51] [Reference Citation Analysis (0)] |
| 28. | Greenglass ER, Julkunen J. Cook-Medley hostility, anger, and the Type A behavior pattern in Finland. Psychol Rep. 1991;68:1059-1066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 34] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Wong JM, Na B, Regan MC, Whooley MA. Hostility, health behaviors, and risk of recurrent events in patients with stable coronary heart disease: findings from the Heart and Soul Study. J Am Heart Assoc. 2013;2:e000052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 30. | Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J. Scikit-learn: machine learning in Python. J Mach Learn Res 2011; 12: 2825–2830. Available from: https://jmlr.csail.mit.edu/papers/v12/pedregosa11a. html. |
| 31. | Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512-525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2275] [Cited by in RCA: 6281] [Article Influence: 628.1] [Reference Citation Analysis (0)] |
| 32. | Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37:658-665. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1330] [Cited by in RCA: 3830] [Article Influence: 319.2] [Reference Citation Analysis (1)] |
| 33. | Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40:304-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4015] [Cited by in RCA: 5754] [Article Influence: 639.3] [Reference Citation Analysis (0)] |
| 34. | Teumer A. Common Methods for Performing Mendelian Randomization. Front Cardiovasc Med. 2018;5:51. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 108] [Article Influence: 15.4] [Reference Citation Analysis (0)] |
| 35. | Greco M FD, Minelli C, Sheehan NA, Thompson JR. Detecting pleiotropy in Mendelian randomisation studies with summary data and a continuous outcome. Stat Med. 2015;34:2926-2940. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 286] [Cited by in RCA: 992] [Article Influence: 99.2] [Reference Citation Analysis (0)] |
| 36. | Mandrekar JN. Receiver operating characteristic curve in diagnostic test assessment. J Thorac Oncol. 2010;5:1315-1316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1283] [Cited by in RCA: 2558] [Article Influence: 170.5] [Reference Citation Analysis (0)] |
| 37. | Burgess S, Thompson SG. Interpreting findings from Mendelian randomization using the MR-Egger method. Eur J Epidemiol. 2017;32:377-389. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1644] [Cited by in RCA: 2903] [Article Influence: 362.9] [Reference Citation Analysis (0)] |
| 38. | Adams JH, Aubert RE, Clark VR. The relationship among John Henryism, hostility, perceived stress, social support, and blood pressure in African-American college students. Ethn Dis. 1999;9:359-368. [PubMed] |
| 39. | Lunyera J, Davenport CA, Bhavsar NA, Sims M, Scialla J, Pendergast J, Hall R, Tyson CC, Russell JSC, Wang W, Correa A, Boulware LE, Diamantidis CJ. Nondepressive Psychosocial Factors and CKD Outcomes in Black Americans. Clin J Am Soc Nephrol. 2018;13:213-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 40. | Albanese E, Matthews KA, Zhang J, Jacobs DR Jr, Whitmer RA, Wadley VG, Yaffe K, Sidney S, Launer LJ. Hostile attitudes and effortful coping in young adulthood predict cognition 25 years later. Neurology. 2016;86:1227-1234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 8] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 41. | Matthews KA, Gump BB, Harris KF, Haney TL, Barefoot JC. Hostile behaviors predict cardiovascular mortality among men enrolled in the Multiple Risk Factor Intervention Trial. Circulation. 2004;109:66-70. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 74] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 42. | Bhavsar NA, Davenport CA, Yang LZ, Peskoe S, Scialla JJ, Hall RK, Tyson CC, Strigo T, Sims M, Pendergast J, Curtis LH, Boulware LE, Diamantidis CJ. Psychosocial determinants of cardiovascular events among black Americans with chronic kidney disease or associated risk factors in the Jackson heart study. BMC Nephrol. 2021;22:375. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |