Published online Dec 27, 2011. doi: 10.5496/wjmg.v1.i1.4
Revised: October 25, 2011
Accepted: December 17, 2011
Published online: December 27, 2011
Psychiatric disorders have traditionally been segregated from medical disorders in terms of drugs, treatment, insurance coverage and training of clinicians. This segregation is consistent with the long-standing observation that there are inherent differences between psychiatric disorders (diseases relating to thoughts, feelings and behavior) and medical disorders (diseases relating to physical processes). However, these differences are growing less distinct as we improve our understanding of the roles of epistasis and pleiotropy in medical genetics. Both psychiatric and medical disorders are predisposed in part by genetic variation, and psychiatric disorders tend to be comorbid with medical disorders. One hypothesis on this interaction posits that certain combinations of genetic variants (epistasis) influence psychiatric disorders due to their impact on the brain, but the associated genes are also expressed in other tissues so the same groups of variants influence medical disorders (pleiotropy). The observation that psychiatric and medical disorders may interact is not novel. Equally, both epistasis and pleiotropy are fundamental concepts in medical genetics. However, we are just beginning to understand how genetic variation can influence both psychiatric and medical disorders. In our recent work, we have discovered gene networks significantly associated with psychiatric and substance use disorders. Invariably, these networks are also significantly associated with medical disorders. Recognizing how genetic variation can influence both psychiatric and medical disorders will help us to understand the etiology of the individual and comorbid disease phenotypes, predict and minimize side effects in drug and other treatments, and help to reduce stigma associated with psychiatric disorders.
- Citation: McEachin RC, Cavalcoli JD. Overlap of genetic influences in phenotypes classically categorized as psychiatric vs medical disorders. World J Med Genet 2011; 1(1): 4-10
- URL: https://www.wjgnet.com/2220-3184/full/v1/i1/4.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v1.i1.4
Interaction between psychiatric and medical disorders has been observed for decades[1-3], although this interaction has not been well characterized with respect to the potential for common underlying genetic etiology. Disorders such as schizophrenia and diabetes are predisposed by multiple interacting genetic and environmental influences[4-6]. Identifying and understanding these interactions is critical to understanding the etiology of a range of common complex diseases[5,7-11], including psychiatric and medical disorders that have very significant public health consequences[4,12-14]. In assessing genetic influences on complex disease, we use the term “epistasis” to mean “interaction between genes”[15,16]. This definition is broad, including any mechanism by which the effect of one gene or genetic variant influences the effect of another gene or genetic variant, resulting in the observed phenotype (changes in protein-protein binding, regulation of expression, post-transcriptional processing, translation, post-translational processing, activation/deactivation, translocation, response to environmental stimuli, etc.). Complex diseases also tend to be comorbid[17-20], consistent with pleiotropy having an influence on them. We use the term “pleiotropy” to mean that multiple phenotypes are influenced by a single genetic variant or set of variants[21,22]. Again, we use a simple and broad definition, including cases where any two or more phenotypes are influenced by a single variant or set of variants. Based on our recent work[18,19,23], our observation is that epistasis and pleiotropy are both important in understanding the genetic etiology of complex diseases. Note that environmental factors are important in the etiology of complex diseases but here we focus on genetic etiology. Also, while epistasis and pleiotropy apply to any phenotypes, the focus of our work is on comorbid diseases, where two or more complex disease phenotypes are seen in a single individual.
We first highlight progress and challenges in assessing genetic influences on psychiatric disorders as well as interactions between psychiatric and medical disorders. We then illustrate the roles of epistasis and pleiotropy in a complex psychiatric/medical comorbidity, based on a set of candidate genes that are statistically enriched (over-represented) in both schizophrenia and breast cancer literature. Finally, we offer observations on the significance of the overlap of genetic influences on psychiatric and medical disorders.
Psychiatric disorders (schizophrenia, bipolar disorder, major depressive disorder, etc.) and medical disorders (diabetes, cancers, hypertension, etc.) are generally common complex diseases[24], predisposed by multiple interacting genetic and environmental influences. Substance use disorders (addiction to or dependence on cocaine, opium, alcohol, nicotine, etc.) are also complex diseases and they are often categorized as psychiatric disorders. Substance use disorders have characteristics of both psychiatric and medical disorders because they influence both behavior and physical condition, though they tend to have very clear environmental influences (i.e. the substances of abuse). Heritability estimates for psychiatric disorders range from 40% to 90%, depending on the disorder and population tested[24-26], strongly consistent with the hypothesis that genetic variation influences these disorders. However, efforts to find reproducible evidence of specific genetic influences have been frustrated due to locus heterogeneity, incomplete penetrance and interaction with environmental factors[24]. Efforts are currently under way to improve the success of these studies by the use of endophenotypes (sub-phenotypes), modeling the impact of environmental variation, identification of rare variants influencing the phenotype and the use of larger study populations or meta-analysis to improve power in hypothesis testing[27,28].
We have been pursuing a related approach for several years, leveraging epistasis and pleiotropy to improve the detection of sets of genetic variants associated with psychiatric disorders comorbid with substance use disorders[18,19,23]. We first noted that psychiatric and substance use disorders are often comorbid[29] and, in most populations tested, epidemiological evidence indicates that individuals diagnosed with psychiatric disorders are over-represented for substance use disorders and vice versa[29-37]. Data about genetic variants that influence psychiatric or substance use disorders is inherently noisy due to diverse populations and behaviorally based phenotype classifications. However, using the principles of epistasis and pleiotropy, we identify genetic variants at the intersection of a pair of comorbid diseases, effectively highlighting the association signal for the comorbidity and revealing biologically relevant gene sets which may be relevant to the molecular basis of the phenotype. Subsequently, we identify a network of genes that are candidates for influencing the comorbid phenotype. Interestingly, while we start out searching for candidate genes related to the psychiatric/substance use comorbidity, we invariably see medical disorders significantly over-represented in annotation for genes in these networks[18,19,23].
Consortia have been formed to improve the detection of genetic variation influencing medical disorders and these groups have seen some success[27,38,39], although the complexity of the genetic influences involved remain challenging[40-42]. Interestingly, interaction between psychiatric and medical disorders has been observed for many years[30,43-45] (e.g. colon cancer and breast cancer have been associated with schizophrenia[46], coronary disease has been associated with depression[47-49]), although the observed interactions do not necessarily point to genetic influences. For example, antipsychotics prescribed to schizophrenics (an environmental influence) may make them more vulnerable to hyperglycemia[50,51]. However, the observed interactions may well be due to some common element that predisposes both conditions[52] and common underlying genetic variation represents an important possible etiology. As a simple example, a variant in transcription factor TCF7L2 was recently found to increase risk for both diabetes and schizophrenia, although this single variant explains only a small amount of variation in either disease[53]. We hypothesize that explaining a greater portion of the genetic influence on a given comorbidity can be accomplished by leveraging epistasis and pleiotropy in modeling the combined phenotype.
We have previously reported modeling interactions among clusters of candidate genes for comorbid psychiatric and substance use disorders[18,19,23] to help us understand how the genetic influences impact the comorbidity and how the relevant substances interact with the genes involved. Summarizing the process, we firstly identify a set of candidate genes for the comorbidity via: genome wide expression or association studies, literature mining, Gene2MeSH[54] and/or the Genetic Association Database[55]; secondly, establish biological context for the set of genes by modeling their interactions via: Prioritizing Disease Genes by Analysis of Common Elements[56], Gene Relationships Among Implicated Loci[57] and/or GeneGo[58]; and thirdly, identify, report and interpret over-represented concepts in annotation for genes in these networks via: the Database for Annotation Visualization and Integrated Discovery[59], ConceptGen[60] and/or GeneGo[58].
As expected, the genes in the networks developed are significantly over-represented for annotation consistent with psychiatric and substance use disorders. However, we noted that each of these networks is also significantly over-represented by genes annotated for their roles in medical disorders. This observation in multiple studies leads us to hypothesize that, in a genetic sense, distinctions made between psychiatric and medical disorders are arbitrary.
To illustrate how psychiatric and medical disorders share constellations of genetic influences, we followed the observed interaction between schizophrenia and breast cancer[46]. To minimize the chance of pursuing a spurious association, we searched PubMed for [“schizophrenia”(MeSH Terms) AND “breast neoplasms”(MeSH Terms)] and found 43 papers annotated for this comorbidity. We then began a candidate gene search using Gene2MeSH[54], which identifies genes that are significantly over-represented (P-value < 10-5) in annotation for PubMed abstracts that are also annotated for given Medical Subject Headings (MeSH). Note that, while we recognize that a simple PubMed search may be subject to publication bias, Gene2MeSH overcomes this bias in calculating significance values. We searched for human genes over-represented for MeSH annotation “descriptor: Schizophrenia” (145 genes) and MeSH annotation “descriptor: Breast Neoplasms” (164 genes). We selected the 6 genes at the intersection of these sets for follow-up analysis (Table 1).
| Gene symbol | Entrez gene ID | MeSH descriptor | MeSH qualifier | PubMed citations | Citations expected | Fold change | χ2 | P value |
| AKT1 | 207 | Schizophrenia | Genetics | 26 | 8.97 | 2.9 | 31.4 | 2.97E-06 |
| Breast Neoplasms | Pathology | 91 | 36.40 | 2.5 | 78.2 | 1.63E-14 | ||
| COMT | 1312 | Schizophrenia | Genetics | 176 | 6.96 | 25.3 | 4107.4 | 5.06E-186 |
| Breast Neoplasms | Genetics | 68 | 28.33 | 2.4 | 55.3 | 8.43E-11 | ||
| CYP2D6 | 1565 | Schizophrenia | Drug therapy | 45 | 4.89 | 9.2 | 330.0 | 1.28E-28 |
| Breast Neoplasms | Drug therapy | 42 | 20.00 | 2.1 | 24.5 | 9.59E-06 | ||
| ERBB4 | 2066 | Schizophrenia | Genetics | 6 | 0.59 | 10.1 | 90.4 | 1.63E-08 |
| Breast Neoplasms | Pathology | 27 | 4.43 | 6.1 | 114.9 | 5.28E-14 | ||
| NRG1 | 3084 | Schizophrenia | Genetics | 98 | 1.85 | 53.1 | 5011.8 | 2.84E-140 |
| Breast Neoplasms | Pathology | 24 | 7.50 | 3.2 | 36.1 | 7.39E-07 | ||
| SOD2 | 6648 | Schizophrenia | Genetics | 16 | 3.64 | 4.4 | 42.8 | 1.07E-06 |
| Breast Neoplasms | Genetics | 50 | 14.71 | 3.4 | 85.1 | 9.33E-14 |
To place the 6 candidate genes identified in our Gene2MeSH search into context, we used GeneGo’s MetaCore database of protein-protein and protein-small molecule interactions to build a gene network model of the candidate genes, plus their closest interactors, using the following parameter settings: (1) shortest paths algorithm; (2) merged network; (3) no canonical pathways; (4) 2 maximum steps in the path; (5) show disconnected seed nodes; (6) show shortest path edges only; (7) discard low trust interactions; (8) use functional interactions; (9) use binding interactions; and (10) do not use compound-target interactions.
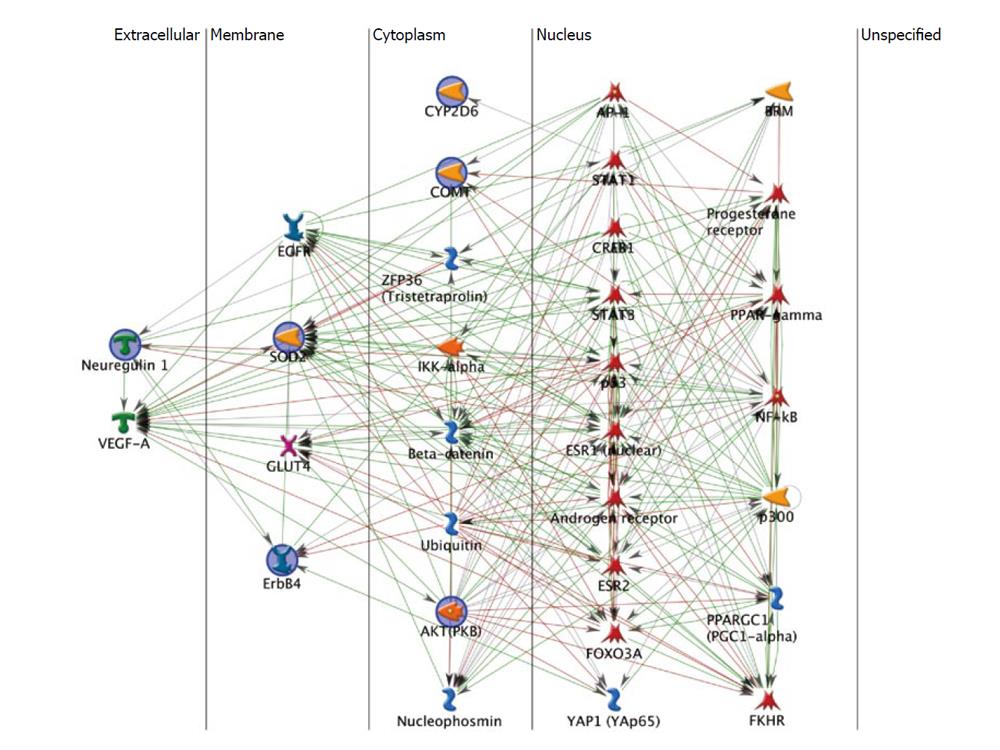
The resulting network (Figure 1) is organized by cellular compartment and shows the close and multi-layered interactions among these 6 genes and their interactors. Note that Figure 1 has 31 icons representing 43 genes because some of the icons represent dimerized proteins that act as a single unit. Based on MetaCore data, GeneGo calculates that the larger network is significantly enriched for genes annotated for brain diseases, cancers and hormone sensitive disorders (Table 2). This annotation is consistent with the hypothesis of shared genetic vulnerability to schizophrenia and breast cancer, and emphasizes that these disorders are genetically related to the broader phenotypes. The “hubs” for the network shown in Figure 1, the genes with the most interactions, are Androgen receptor (33 interactions) and Estrogen receptor 1 (32 interactions), consistent with hormone response. Other notable hubs include p53, strongly associated with cancers[61] (30 interactions), and Estrogen receptor 2, a homolog to Estrogen receptor 1 (22 interactions). Of the 43 genes in this network, 22 are transcription factors, consistent with an important role for regulation of gene expression.
| Disease | Percent of network genes annotated (%) | P value |
| Ovarian neoplasms | 76.74 | 5.80E-29 |
| Ovarian diseases | 76.74 | 3.24E-27 |
| Adnexal diseases | 76.74 | 3.89E-27 |
| Mesothelioma | 48.84 | 2.04E-26 |
| Neoplasms, mesothelial | 48.84 | 2.50E-26 |
| Gonadal disorders | 76.74 | 4.23E-26 |
| Genital neoplasms, female | 79.07 | 8.70E-26 |
| Hyperoxia | 30.23 | 2.10E-23 |
| Genital diseases, female | 83.72 | 4.75E-22 |
| Astrocytoma | 72.09 | 4.82E-22 |
| Neoplasms, connective and soft tissue | 62.79 | 1.64E-21 |
| Glioblastoma | 69.77 | 3.67E-21 |
“Adnexal Diseases” and “Hyperoxia” are over-represented phenotypes (Table 2) that we have not been able to place into the context of brain disorders or cancers. Equally, “positive regulation of nitrogen compound metabolic process” is the most significantly over-represented Gene Ontology biological process in the annotation for these genes (P-value: 3.49 × 10-37) and, again, we have not been able to place this process into context. These may represent spurious associations. However, based on our previous experience, we hesitate to dismiss the evidence for these associations. They could represent novel puzzle pieces that will be appreciated only when other pieces are discovered and put into context. This was the case when we first noticed that medical disorders were significantly enriched in annotation for networks based on psychiatric/substance use comorbidities.
Genetic variation’s influence on complex disease is agnostic to categorization of psychiatric vs medical disorders. We present a view on comorbidity research that opens the door to analysis of new combinations of related phenotypes, which could also shed light on single-disorder phenotypes. We propose that, based on epidemiological evidence, we can search for genetic influences on comorbidities that might otherwise seem unrelated[62-65]. Over-represented concepts found in annotation for genes in these networks serve as positive controls in model building and provide insight into the biological context of the genetic influences. These insights may also provide novel background on fundamental processes that would not be evident without the network model. These insights can then be applied to improve our understanding of the comorbidity and each of the single-disorder phenotypes. The methods we describe are not biased by disease terminology and are only biased based on the literature and gene set categorizations and annotations. By using multiple data sources we hope to minimize that bias but recognize that it still exists.
At least 16 genes in the network in Figure 1 are annotated in GeneGo’s MetaDrug database as being known targets for currently available therapeutic drugs. Arguably, any of these drugs has potential for therapeutic use in schizophrenia or breast cancer, as well as the related phenotypes seen in Table 2. The multiple positive and negative feedback loops evident in Figure 1 are also indicative of the complex nature of epistasis in this comorbidity. Development of drugs for therapeutic use can benefit from this work by incorporating the understanding that these genes have complex interactions that must be considered when targeting any one of them or any combination of them. In the larger sense, this network is not unique in modeling comorbid psychiatric and medical disorders; rather this is the pattern that we have seen in previous work and it is consistent with side effects seen in a range of treatment protocols[66-71]. By improving our understanding of the multiple interacting genetic and environmental influences on any disease phenotype, we should be able to better predict therapeutic interactions and side effects, and reduce their negative effects on the patient[72].
A challenging facet of complex disease research is the frequent failure to replicate significant findings in follow-on analyses[24]. The hypothesis that epistasis and pleiotropy are important in complex comorbidities is consistent with these observations where, for example, in one study the direction of association for a given comorbidity is positive[46] and in other study the direction is negative[73]. The alternate direction of correlation should actually be viewed as additional evidence of a relationship between common gene variants and these diseases. A model consistent with this phenomenon posits that a given set of genetic variants could be positively associated with both a psychiatric disorder and a medical disorder, while the same gene set with a slight change in variation, perhaps in a different population, could reverse the direction of effect for one of the disorders. Equally, minor changes in other interacting genes or environmental effects could yield population specific, gender specific or environment specific effects that hamper replication efforts.
A significant challenge for psychiatric patients is the stigma associated with diagnosis and treatment for their disorders, interfering with essentially every facet of their lives[12]. Part of this stigma comes from the preconception that there is something inherently different about psychiatric disorders when compared with medical disorders. This stigma is also the result of our lack of understanding of the etiology of psychiatric disorders and, in many cases, our inability to effectively treat these disorders. The growing recognition that psychiatric disorders are influenced by the same complex interacting genetic and environmental influences that predispose medical disorders, which are better accepted by society, may help to ease the stigma associated with psychiatric disorders. Overcoming this stigma may then lead individuals back to productive lifestyles and healthier relationships[12].
Psychiatric and medical disorders have traditionally been segregated, in part due to our understanding that psychiatric disorders are fundamentally different from medical disorders. In some ways this is true, although our improved understanding of both epistasis and pleiotropy in genetic predisposition to complex diseases makes the distinctions less clear. The methods we have been developing provide a way to identify common genetic influences on psychiatric and medical disorders that might otherwise seem distinct. These methods use existing annotation of genes and pathways, and the vast amount of biological and medical research literature currently available, to identify genes involved in comorbidities. The hope is that by identifying potential common genetic variants in comorbid diseases, we can improve medicine by better understanding basic molecular processes and gene or pathway interactions; leverage common therapeutic agents already developed for different diseases or disorders; and raise awareness of the potential for genetic etiology that is common across medical and psychiatric disorders.
Peer reviewer: Xiaoyi Gao, PhD, Assistant Professor, Department of Ophthalmology and Preventive Medicine, University of Southern California, 1450 San Pablo Street, Suite 4802, Los Angeles, CA 90033, United States
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
| 1. | Petty F. Depression and medical illness. Am J Med Sci. 1989;298:59-68. [PubMed] |
| 2. | Adler LE, Griffith JM. Concurrent medical illness in the schizophrenic patient. Epidemiology, diagnosis, and management. Schizophr Res. 1991;4:91-107. [PubMed] |
| 3. | Fiester SJ, Shefferman MM. Medical problems in hospitalized psychiatric patients. New Dir Ment Health Serv. 1994;59-70. [PubMed] |
| 4. | Risch N, Merikangas K. The future of genetic studies of complex human diseases. Science. 1996;273:1516-1517. [PubMed] |
| 5. | Franks PW. Gene × environment interactions in type 2 diabetes. Curr Diab Rep. 2011;11:552-561. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 6. | Pickard B. Progress in defining the biological causes of schizophrenia. Expert Rev Mol Med. 2011;13:e25. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 7. | Madsen AM, Ottman R, Hodge SE. Causal models for investigating complex genetic disease: II. what causal models can tell us about penetrance for additive, heterogeneity, and multiplicative two-locus models. Hum Hered. 2011;72:63-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 8. | Balistreri CR, Caruso C, Carruba G, Miceli V, Candore G. Genotyping of sex hormone-related pathways in benign and malignant human prostate tissues: data of a preliminary study. OMICS. 2011;15:369-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 9. | Basu S, Pan W, Oetting WS. A dimension reduction approach for modeling multi-locus interaction in case-control studies. Hum Hered. 2011;71:234-245. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 10. | Bell JT, Timpson NJ, Rayner NW, Zeggini E, Frayling TM, Hattersley AT, Morris AP, McCarthy MI. Genome-wide association scan allowing for epistasis in type 2 diabetes. Ann Hum Genet. 2011;75:10-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 26] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 11. | Hua L, Zhou P, Liu H, Li L, Yang Z, Liu ZC. Mining susceptibility gene modules and disease risk genes from SNP data by combining network topological properties with support vector regression. J Theor Biol. 2011;289:225-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 4] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 12. | Henderson M, Harvey SB, Overland S, Mykletun A, Hotopf M. Work and common psychiatric disorders. J R Soc Med. 2011;104:198-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 230] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 13. | Hart TC, Marazita ML, Wright JT. The impact of molecular genetics on oral health paradigms. Crit Rev Oral Biol Med. 2000;11:26-56. [PubMed] |
| 14. | Causey TN, Bodurtha JN, Ford N. A genetic perspective on infant mortality. South Med J. 2010;103:440-44; quiz 440-44;. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 15. | Cordell HJ. Epistasis: what it means, what it doesn't mean, and statistical methods to detect it in humans. Hum Mol Genet. 2002;11:2463-2468. [PubMed] |
| 16. | Zhang Y, Jiang B, Zhu J, Liu JS. Bayesian models for detecting epistatic interactions from genetic data. Ann Hum Genet. 2011;75:183-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 17. | Halaris A. Comorbidity between depression and cardiovascular disease. Int Angiol. 2009;28:92-99. [PubMed] |
| 18. | McEachin RC, Saccone NL, Saccone SF, Kleyman-Smith YD, Kar T, Kare RK, Ade AS, Sartor MA, Cavalcoli JD, McInnis MG. Modeling complex genetic and environmental influences on comorbid bipolar disorder with tobacco use disorder. BMC Med Genet. 2010;11:14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 20] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 19. | McEachin RC, Keller BJ, Saunders EF, McInnis MG. Modeling gene-by-environment interaction in comorbid depression with alcohol use disorders via an integrated bioinformatics approach. BioData Min. 2008;1:2. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 20. | Dick DM, Bierut LJ. The genetics of alcohol dependence. Curr Psychiatry Rep. 2006;8:151-157. [PubMed] |
| 21. | Wagner GP, Zhang J. The pleiotropic structure of the genotype-phenotype map: the evolvability of complex organisms. Nat Rev Genet. 2011;12:204-213. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 464] [Cited by in RCA: 452] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 22. | Ostman B, Hintze A, Adami C. Impact of epistasis and pleiotropy on evolutionary adaptation. Proc Biol Sci. 2012;279:247-256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 23. | McEachin RC, Chen H, Sartor MA, Saccone SF, Keller BJ, Prossin AR, Cavalcoli JD, McInnis MG. A genetic network model of cellular responses to lithium treatment and cocaine abuse in bipolar disorder. BMC Syst Biol. 2010;4:158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 18] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 24. | Burmeister M, McInnis MG, Zöllner S. Psychiatric genetics: progress amid controversy. Nat Rev Genet. 2008;9:527-540. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 354] [Cited by in RCA: 346] [Article Influence: 20.4] [Reference Citation Analysis (0)] |
| 25. | Dellava JE, Kendler KS, Neale MC. Generalized anxiety disorder and anorexia nervosa: evidence of shared genetic variation. Depress Anxiety. 2011;28:728-733. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 29] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 26. | Olvera RL, Bearden CE, Velligan DI, Almasy L, Carless MA, Curran JE, Williamson DE, Duggirala R, Blangero J, Glahn DC. Common genetic influences on depression, alcohol, and substance use disorders in Mexican-American families. Am J Med Genet B Neuropsychiatr Genet. 2011;156B:561-568. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 63] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 27. | Steinberg S, de Jong S, Andreassen OA, Werge T, Børglum AD, Mors O, Mortensen PB, Gustafsson O, Costas J, Pietiläinen OP. Common variants at VRK2 and TCF4 conferring risk of schizophrenia. Hum Mol Genet. 2011;20:4076-4081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 166] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 28. | Neale BM, Medland SE, Ripke S, Asherson P, Franke B, Lesch KP, Faraone SV, Nguyen TT, Schäfer H, Holmans P. Meta-analysis of genome-wide association studies of attention-deficit/hyperactivity disorder. J Am Acad Child Adolesc Psychiatry. 2010;49:884-897. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 387] [Cited by in RCA: 324] [Article Influence: 21.6] [Reference Citation Analysis (0)] |
| 29. | Grant BF, Stinson FS, Hasin DS, Dawson DA, Chou SP, Ruan WJ, Huang B. Prevalence, correlates, and comorbidity of bipolar I disorder and axis I and II disorders: results from the National Epidemiologic Survey on Alcohol and Related Conditions. J Clin Psychiatry. 2005;66:1205-1215. [PubMed] |
| 30. | Hughes JR, Hatsukami DK, Mitchell JE, Dahlgren LA. Prevalence of smoking among psychiatric outpatients. Am J Psychiatry. 1986;143:993-997. [PubMed] |
| 31. | Gonzalez-Pinto A, Gutierrez M, Ezcurra J, Aizpuru F, Mosquera F, Lopez P, de Leon J. Tobacco smoking and bipolar disorder. J Clin Psychiatry. 1998;59:225-228. [PubMed] |
| 32. | Lasser K, Boyd JW, Woolhandler S, Himmelstein DU, McCormick D, Bor DH. Smoking and mental illness: A population-based prevalence study. JAMA. 2000;284:2606-2610. [PubMed] |
| 33. | Uçok A, Polat A, Bozkurt O, Meteris H. Cigarette smoking among patients with schizophrenia and bipolar disorders. Psychiatry Clin Neurosci. 2004;58:434-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 48] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 34. | Wilens TE, Vitulano M, Upadhyaya H, Adamson J, Sawtelle R, Utzinger L, Biederman J. Cigarette smoking associated with attention deficit hyperactivity disorder. J Pediatr. 2008;153:414-419. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 73] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 35. | Wilens TE, Biederman J, Adamson JJ, Henin A, Sgambati S, Gignac M, Sawtelle R, Santry A, Monuteaux MC. Further evidence of an association between adolescent bipolar disorder with smoking and substance use disorders: a controlled study. Drug Alcohol Depend. 2008;95:188-198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 69] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 36. | Biederman J, Monuteaux MC, Spencer T, Wilens TE, Macpherson HA, Faraone SV. Stimulant therapy and risk for subsequent substance use disorders in male adults with ADHD: a naturalistic controlled 10-year follow-up study. Am J Psychiatry. 2008;165:597-603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 144] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 37. | Biederman J, Petty CR, Wilens TE, Fraire MG, Purcell CA, Mick E, Monuteaux MC, Faraone SV. Familial risk analyses of attention deficit hyperactivity disorder and substance use disorders. Am J Psychiatry. 2008;165:107-115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 38. | Plenge RM, Cotsapas C, Davies L, Price AL, de Bakker PI, Maller J, Pe'er I, Burtt NP, Blumenstiel B, DeFelice M. Two independent alleles at 6q23 associated with risk of rheumatoid arthritis. Nat Genet. 2007;39:1477-1482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 446] [Cited by in RCA: 436] [Article Influence: 24.2] [Reference Citation Analysis (0)] |
| 39. | Stančáková A, Paananen J, Soininen P, Kangas AJ, Bonnycastle LL, Morken MA, Collins FS, Jackson AU, Boehnke ML, Kuusisto J. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 2011;60:1608-1616. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 66] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 40. | Sanna S, Li B, Mulas A, Sidore C, Kang HM, Jackson AU, Piras MG, Usala G, Maninchedda G, Sassu A. Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet. 2011;7:e1002198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 116] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 41. | Lee W, Gusnanto A, Salim A, Magnusson P, Sim X, Tai ES, Pawitan Y. Estimating the number of true discoveries in genome-wide association studies. Stat Med. 2011;Epub ahead of print. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 42. | Cooper GM, Shendure J. Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat Rev Genet. 2011;12:628-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 424] [Cited by in RCA: 405] [Article Influence: 28.9] [Reference Citation Analysis (0)] |
| 43. | Carney CP, Jones L, Woolson RF. Medical comorbidity in women and men with schizophrenia: a population-based controlled study. J Gen Intern Med. 2006;21:1133-1137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 240] [Cited by in RCA: 250] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 44. | Carney CP, Jones LE. Medical comorbidity in women and men with bipolar disorders: a population-based controlled study. Psychosom Med. 2006;68:684-691. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 163] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 45. | Jones LE, Carney CP. Increased risk for metabolic syndrome in persons seeking care for mental disorders. Ann Clin Psychiatry. 2006;18:149-155. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 8] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 46. | Hippisley-Cox J, Vinogradova Y, Coupland C, Parker C. Risk of malignancy in patients with schizophrenia or bipolar disorder: nested case-control study. Arch Gen Psychiatry. 2007;64:1368-1376. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 47. | Burker EJ, Blumenthal JA, Feldman M, Burnett R, White W, Smith LR, Croughwell N, Schell R, Newman M, Reves JG. Depression in male and female patients undergoing cardiac surgery. Br J Clin Psychol. 1995;34:119-128. [PubMed] |
| 48. | Dwight MM, Stoudemire A. Effects of depressive disorders on coronary artery disease: a review. Harv Rev Psychiatry. 1997;5:115-122. [PubMed] |
| 49. | Thomas AJ, Kalaria RN, O'Brien JT. Depression and vascular disease: what is the relationship? J Affect Disord. 2004;79:81-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 184] [Cited by in RCA: 183] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 50. | Lindenmayer JP, Nathan AM, Smith RC. Hyperglycemia associated with the use of atypical antipsychotics. J Clin Psychiatry. 2001;62 Suppl 23:30-38. [PubMed] |
| 51. | Bai YM, Lin CC, Chen JY, Chen TT, Su TP, Chou P. Association of weight gain and metabolic syndrome in patients taking clozapine: an 8-year cohort study. J Clin Psychiatry. 2011;72:751-756. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 52. | Iacovides A, Siamouli M. Comorbid mental and somatic disorders: an epidemiological perspective. Curr Opin Psychiatry. 2008;21:417-421. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 47] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 53. | Hansen T, Ingason A, Djurovic S, Melle I, Fenger M, Gustafsson O, Jakobsen KD, Rasmussen HB, Tosato S, Rietschel M. At-risk variant in TCF7L2 for type II diabetes increases risk of schizophrenia. Biol Psychiatry. 2011;70:59-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 100] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 54. | Available from: http://gene2mesh.ncibi.org. |
| 55. | Zhang Y, De S, Garner JR, Smith K, Wang SA, Becker KG. Systematic analysis, comparison, and integration of disease based human genetic association data and mouse genetic phenotypic information. BMC Med Genomics. 2010;3:1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 73] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 56. | McEachin RC, Keller BJ, Zandi PP, Almani M, McInnis MG. Prioritizing Disease Genes by Analysis of Common Elements (PDG-ACE). AMIA Annu Symp Proc. 2007;1068. [PubMed] |
| 57. | Raychaudhuri S, Plenge RM, Rossin EJ, Ng AC, Purcell SM, Sklar P, Scolnick EM, Xavier RJ, Altshuler D, Daly MJ. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 349] [Cited by in RCA: 328] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 58. | Ekins S, Nikolsky Y, Bugrim A, Kirillov E, Nikolskaya T. Pathway mapping tools for analysis of high content data. Methods Mol Biol. 2007;356:319-350. [PubMed] |
| 59. | Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1-13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10350] [Cited by in RCA: 11158] [Article Influence: 656.4] [Reference Citation Analysis (0)] |
| 60. | Sartor MA, Mahavisno V, Keshamouni VG, Cavalcoli J, Wright Z, Karnovsky A, Kuick R, Jagadish HV, Mirel B, Weymouth T. ConceptGen: a gene set enrichment and gene set relation mapping tool. Bioinformatics. 2010;26:456-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 113] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 61. | Aylon Y, Oren M. New plays in the p53 theater. Curr Opin Genet Dev. 2011;21:86-92. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 62. | Ogunmakin KO, Rashid RM. Alopecia: the case for medical necessity. Skinmed. 2011;9:79-84. [PubMed] |
| 63. | Wolkowitz OM, Reus VI, Mellon SH. Of sound mind and body: depression, disease, and accelerated aging. Dialogues Clin Neurosci. 2011;13:25-39. [PubMed] |
| 64. | Ara R, Brazier J. Estimating health state utility values for comorbid health conditions using SF-6D data. Value Health. 2011;14:740-745. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 65. | Boyd CM, Leff B, Wolff JL, Yu Q, Zhou J, Rand C, Weiss CO. Informing clinical practice guideline development and implementation: prevalence of coexisting conditions among adults with coronary heart disease. J Am Geriatr Soc. 2011;59:797-805. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 66. | Praharaj SK, Jana AK, Goyal N, Sinha VK. Metformin for olanzapine-induced weight gain: a systematic review and meta-analysis. Br J Clin Pharmacol. 2011;71:377-382. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 67. | Pompili M, Serafini G, Innamorati M, Ambrosi E, Telesforo L, Venturini P, Giordano G, Battuello M, Lester D, Girardi P. Unmet treatment needs in schizophrenia patients: is asenapine a potential therapeutic option? Expert Rev Neurother. 2011;11:989-1006. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 68. | Sohn S, Kocher JP, Chute CG, Savova GK. Drug side effect extraction from clinical narratives of psychiatry and psychology patients. J Am Med Inform Assoc. 2011;18:i144-i149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 69. | Gonzalez JM, Thompson PM, Moore TA. Review of the safety, efficacy, and side effect profile of asenapine in the treatment of bipolar 1 disorder. Patient Prefer Adherence. 2011;5:333-341. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 70. | Brouwers L, Iskar M, Zeller G, van Noort V, Bork P. Network neighbors of drug targets contribute to drug side-effect similarity. PLoS One. 2011;6:e22187. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 78] [Cited by in RCA: 69] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 71. | Pauwels E, Stoven V, Yamanishi Y. Predicting drug side-effect profiles: a chemical fragment-based approach. BMC Bioinformatics. 2011;12:169. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 149] [Cited by in RCA: 160] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 72. | Buonaguro L, Wang E, Tornesello ML, Buonaguro FM, Marincola FM. Systems biology applied to vaccine and immunotherapy development. BMC Syst Biol. 2011;5:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 73. | Barak Y, Achiron A, Mandel M, Mirecki I, Aizenberg D. Reduced cancer incidence among patients with schizophrenia. Cancer. 2005;104:2817-2821. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |