Published online Dec 27, 2011. doi: 10.5496/wjmg.v1.i1.14
Revised: October 18, 2011
Accepted: December 17, 2011
Published online: December 27, 2011
Genetic interactions are functional crosstalk among different genetic loci that lead to phenotypic changes, such as health or viability alterations. A disease or lethal phenotype that results from the combined effects of gene mutations at different loci is termed a synthetic sickness or synthetic lethality, respectively. Studies of genetic interaction have provided insight on the relationships among biochemical processes or pathways. Cancer results from genetic interactions and is a major focus of current studies in genetic interactions. Various basic and translational cancer studies have explored the concept of genetic interactions, including studies of the mechanistic characterization of genes, drug discovery, biomarker identification and the rational design of combination therapies. This review discusses the implications of genetic interactions in the development of personalized cancer therapies, the identification of treatment-responsive genes, the delineation of mechanisms of chemoresistance and the rational design of combined therapeutic strategies to overcome drug resistance.
- Citation: Fang B. Genetic interactions in translational research on cancer. World J Med Genet 2011; 1(1): 14-22
- URL: https://www.wjgnet.com/2220-3184/full/v1/i1/14.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v1.i1.14
Genetic interactions are functional crosstalk among genes of different loci that regulate or compensate for one another in many signaling and/or metabolic pathways, leading to phenotypic changes, including disease status (sickness) or viability alterations (lethality or semilethality). Unlike the dominance caused by interactions between alleles of the same genetic locus, the interactions of different genetic loci may lead to unexpected phenotypic changes that are different from the effects of mutations in each individual gene. An example of a typical phenotypic change caused by genetic interaction is synthetic lethality or synthetic semilethality; in brief, homozygous mutations in two genes result in normal viability in living organisms when the mutations exist separately but become lethal or semilethal (viability reduced but not completely abolished) when they occur simultaneously[1].
Because a lethal phenotype can be easily identified, synthetic lethality has frequently been used as a research tool for identifying interactions among genes. Global gene knockout studies in yeast showed that about 20% of genes in Saccharomyces cerevisiae (S. cerevisiae) are essential for growth on a rich glucose-containing medium, whereas about 80% of the approximately 6200 predicted genes are nonessential, suggesting that the genome is buffered from the lethal effects of genetic disorders in more than 4700 genes that may have redundant functions associated with essential processes[2-4]. Thus far, global synthetic lethality analysis in yeast has generated substantial new information on genetic interactions that compensate for one another in biologically essential processes[5]. Information on genetic interactions has been used to predict the function of uncharacterized genes and decipher complex regulatory relationships among biochemical processes or pathways[5]. The principle of genetic interactions is also being exploited by various investigators to identify genes that are crucial to the survival of certain oncogene-transformed cells[6-9] or genes that sensitize cells to chemotherapy[10,11] or to find small molecules that selectively induce cell death in a subset of oncogene-transformed cells[12-14]. Thus, the principles of genetic interaction have become a research platform for characterizing gene functions, discovering novel anticancer agents, identifying molecular biomarkers for personalized therapy and designing effective combination therapies to overcome drug resistance. Applications of genetic interactions in anticancer drug discovery were recently reviewed in several articles[15-17]. This review will discuss potential applications of genetic interaction in personalized therapy and in the rational design of multimodality therapy.
The functional interactions among genes are more comprehensive than the physical interactions among proteins. Studies in yeast have shown that, on average, each gene may have more than 40 genetic interactions[18-20], whereas yeast proteins may have an average of 8 physical interactions per protein[21]. A study used 74 genes known to be involved in genomic integrity in S. cerevisiae to search for genetic interactions with those genes resulted in the identification of a network of 4956 unique pairs of genetic interactions involving 875 genes[19]. Within this network, several novel components and functional modules or minipathways were defined that are important for DNA integrity, including those involved in DNA replication, postreplication repair, homologous recombination and oxidative stress response[19]. More recently, several groups of researchers used a gene knockdown approach to search for genes that are synthetic lethals to the oncogenic KRAS gene and identified numerous synthetic lethal partners with mutant KRAS gene in various human cancer cells[6-9]. For example, a genome-wide RNAi screening in the isogenic human colon cancer cell line DLD-1 with and without oncogenic KRAS led to the identification of 368 lethal interaction candidate genes with a stringent cutoff and 1613 genes with relaxed statistical criteria[8]. Genes involved in the regulation of several biological processes or pathways, including nucleic acid metabolism, ribosome biogenesis, protein neddylation or sumoylation, RNA splicing, the cell cycle, mitosis and proteasome complexes, were found to be required as additional support to maintain the Ras oncogenic state[8]. Thus, genetic interactions are more complicated and comprehensive than physical interactions.
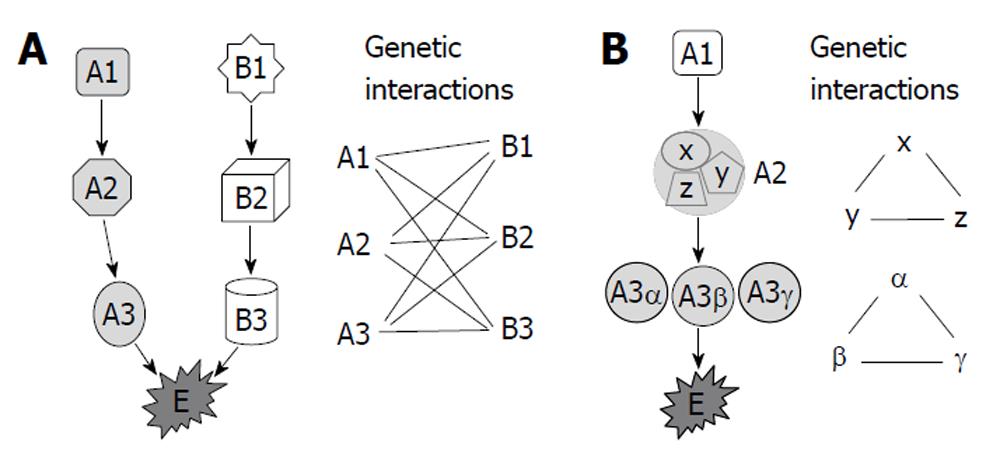
Several models have been proposed to account for genetic interactions[21-23], including the components of parallel pathways that together regulate an essential biological function, subunits of an essential multiprotein complex and components of a single linear essential pathway (Figure 1). Synthetic genetic array analysis and synthetic lethality analysis by microarray in yeast revealed that genetic interactions occurred the most frequently between genes with the same mutant phenotype, between genes encoding proteins with the same subcellular localization, and between genes involved in similar biological processes or bridging bioprocesses[5,18]. Although genetic interactions were more frequent than expected between genes encoding proteins within the same protein complex and among gene pairs encoding homologous proteins, relatively few synthetic lethal interactions (only 1%-2%) fall into these two categories[18]. Most of the genetic interactions were identified among functionally related genes or among genes that function in parallel or compensating pathways[2,5,18,24].
Activating mutations in oncogenes and growth factor receptors are known to play critical roles in tumorigenesis and in the malignant evolution of cancers[25,26]. Several oncogenes or growth factor receptors have been successfully targeted by small molecule inhibitors and/or monoclonal antibodies for cancer treatment. Genetic changes, such as gene amplifications or mutations in the corresponding genes, have been used as predictive biomarkers for identifying patients who would benefit from a particular treatment[27]. Cancers overexpressing HER2 were shown to respond favorably to the monoclonal antibody trastuzumab[28,29]. Similarly, the epidermal growth factor receptor (EGFR) inhibitors erlotinib and gefitinib were found to be more effective against EGFR mutant cancers[30], whereas imatinib was highly effective against cancer cells with BCR-Abl fusion protein[31].
Therapeutic benefits can also be obtained by targeting oncogenes and tumor suppressor genes indirectly through genetic lethal interactions. Functional alterations in some oncogenes or tumor suppressor genes may render the mutant cells more susceptible to a functional change in another gene. Therefore, the mutant cells can be eliminated through pharmaceutical intervention that leads to synthetic lethality. Selective cytotoxicity of poly (ADP-ribose) polymerase 1 (PARP1) inhibitors in BRCA1 and BRCA2 mutant cancer cells is mediated through genetic interaction between PARP1 and BRCAs. PARP1 is required for DNA single-strand break (SSB) repair because PARP1-/- mice have defective DNA SSB repair and increased homologous recombination, sister chromatid exchange and chromosome instability[32,33]. On the other hand, BRCA1 and BRCA2, whose loss-of-function mutations predispose carriers to breast, ovarian and other types of cancers[34,35], are required for homologous recombination of DNA double-strand break (DSB) repair[36,37]. PARP1 may not be directly involved in DSB repair and homologous recombination since PARP1-/- embryonic stem cells and embryonic fibroblasts exhibited normal repair of DNA DSBs[32]. Nevertheless, concurrent blockage of DNA DSB repair, resulting from a mutation in BRCA genes and DNA SSB repair due to PARP1 inhibition, is fatal to a cell[38,39]. As a result, BRCA mutant cells are 1000 times more sensitive to PARP1 than are BRCA wild-type cells[39]. Clinical trials also showed that cancer patients with BRCA1 or BRCA2 mutations responded favorably to an orally active PARP1 inhibitor, olaparib (AZD2281)[40-43].
Functional changes in several genes involved in DNA DSB repair pathways, such as ATM[44], RAD54[45] and BRIT1[46] genes, have been found to be highly associated with susceptibility to radiotherapy and the DNA cross-linking agent mitomycin C, suggesting that mutations in those genes may be used as biomarkers of susceptibility to radiotherapy or DNA-damaging chemotherapeutic agents. The ATM gene encodes the ataxia telangiectasia mutated (ATM) protein kinase that is rapidly activated when DNA DSBs occur in eukaryotic cells[47]. Activated ATM phosphorylates a variety of proteins involved in cell cycle checkpoint control, apoptosis and DNA repair pathways, including p53, CHK2, BRCA1, H2AX and FANCD2[47,48]. A recent study indicated that interactions of ATM and p53, two commonly mutated tumor suppressor genes, should be explored to determine their ability to predict clinical response to genotoxic chemotherapies[49]. In p53-deficient tumor cells, inactivation of ATM or of its downstream molecule CHK2 was sufficient to sensitize the cells to the genotoxic chemotherapeutic agents cisplatin and doxorubicin[49]. Interestingly, inhibition of ATM or of CHK2 resulted in a substantial survival benefit in p53 wild-type cells. Several clinical trials of CHK1/CHK2 inhibitors in combination with genotoxic agents for cancer treatment are currently under way[50]. The p53 inactivation that occurs in about 50% of human cancers because of genetic mutations[51] may serve as a biomarker for the efficacy of combination therapies containing cisplatin and doxorubicin plus inhibitors of ATM and CHK2.
Another indirect approach is targeting a downstream component in a single linear essential pathway. Evidence has shown that BRAF mutant cancer cells can be selectively killed by inhibitors of mitogen-activated protein (MAP) kinase (MEK), a substrate of Raf protein kinases[52]. The RAS/RAF/MEK/Erk pathway is one of the critical signal transduction cascades of most growth factor receptors and is pivotal in oncogenesis[53,54]. RAF kinases are activated by RAS upon the stimulation of extracellular ligands, such as growth factors, cytokines and hormones. Activated RAF phosphorylates and activates the dual-specificity protein kinase MEK, which in turn phosphorylates both tyrosine (Tyr185) and threonine (Thr183) residues of extracellular-signal-regulated kinase (ERK) proteins[55], leading to activation of ERK1/ERK2. Various constitutively active mutations of the BRAF gene have been identified in human cancers, including 60%-70% of malignant melanomas, 36%-50% of thyroid cancers, 5%-22% of colorectal cancers, 30% of serous ovarian cancers and lower percentages of other cancers[56]. The strong dependence of BRAF mutant tumors on MEK activity may provide a personalized therapeutic strategy for patients with this type of cancer[52].
Overexpression of the MYC oncogene was reported to upregulate the expression of the tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) death receptor DR5, thereby sensitizing tumor cells to TRAIL-induced apoptosis[57]. An analysis of the knockdown of 510 genes encoding known and predicted kinases, proteins with known functions in TRAIL-mediated signaling pathways, or proteins with unknown functions also revealed that siRNA against PAK1 and AKT1 strongly enhanced TRAIL activity, whereas siRNA against MYC or the WNT transducer TCF4 inhibited TRAIL-induced apoptosis, indicating that the MYC and WNT pathways are required for TRAIL-mediated apoptosis[58]. On the other hand, deficiency of the tumor suppressor gene adenomatous polyposis coli (APC) was found to cause accumulation of β-catenin in the nucleus, which interacts with TCF4 and promotes TCF4’s binding to c-MYC promoter and overexpression of c-MYC[59]. Deletion of the MYC gene rescued the phenotypes caused by deletion of the APC gene, despite the presence of high levels of nuclear β-catenin[60]. Thus, MYC overexpression is a critical component in the malignancy of APC-defective cancers. A recent study showed that the combination of TRAIL and all-trans-retinyl acetate, another death receptors inducer, significantly enhanced apoptosis induction in APC gene-defective tumor cells and premalignant cells[61], indicating that this combination can be useful for chemoprevention and personalized therapy in patients with APC-defective cancers.
Genetic interaction has been exploited as a research tool to identify genes or biomarkers associated with treatment responses. Studies of the Food and Drug Administration (FDA)-approved anticancer agents in a panel of yeast mutants revealed that the DNA cross-linking agent cisplatin displayed high specificity for mutants defective in postreplication repair, whereas the topoisomerase II inhibitor mitoxantrone was highly specific for defects in DNA DSB repair[62]. Because many human disease-related genes are conserved with their yeast counterparts[63,64], yeast has been exploited for mechanistic study of clinically relevant compounds[65,66]. A genome-wide screen of yeast heterozygotes with therapeutic compounds could reveal not only the possible targets but also synthetic lethal partners of the tested compounds[67]. For example, heterozygotes of TRX2, a nonessential gene involved in antioxidative stress, were found to be sensitive to camptothecin, whereas heterozygotes of genes involved in exosome rRNA processing were identified as possible lethal partners with 5-fluorouracil[67]. An analysis of more than 1000 structurally diverse compounds, including drugs approved by the FDA and the World Health Organization, in yeast whole-genome heterozygous and homozygous deletion collections showed that genes involved in endosomal transport, vacuolar degradation, aromatic amino acid biosynthesis or encoding of some transcription factors may function as multidrug-resistance genes because their deletion renders yeast sensitive to multiple drug treatments[68]. Nevertheless, information obtained from yeast studies needs to be validated in human cell systems before the results can be translated into clinical applications.
The advent of gene knockdown technology allows us to perform systematic analysis of genes associated with treatment response in human cancer cells. Whitehurst et al[10] used a library of more than 84 000 chemically synthesized siRNAs targeting 21 127 unique human genes to screen for gene targets that specifically reduce cell viability in the presence of an otherwise sublethal dose of paclitaxel in the human non-small cell lung cancer line NCI-H1155. Their study identified a set of 87 candidate genes whose knockdown sensitized cells to paclitaxel and some of the genes increased the sensitization of lung cancer cells to paclitaxel by more than 1000 times. Multiple genes encoding core components of the proteasome, proteins involved in the function of microtubules, posttranslational modification and cell adhesion, or cancer/testis antigens were found to be associated with the sensitivity of paclitaxel[10]. A similar approach has been used by Astsaturov et al[11] for identification of genes associated with response to EGFR inhibitors. Analysis of a siRNA library targeting 638 genes encoding proteins with evidence of functional interaction with the EGFR signaling network, including those transcriptionally responsive to inhibition or stimulation of EGFR, led Astsaturov et al[11] to identify 61 genes whose knockdown sensitized the A431 cervical adenocarcinoma cell line to the EGFR inhibitors erlotinib or cetuximab[11]. Most of those genes encode proteins connected in a physically interacting network, including kinases and phosphatases. Nevertheless, a further test in 7 other cell lines for sensitization to erlotinib or cetuximab showed that none of the tested genes sensitized all cell lines, although several of them sensitized 3-5 of the cell lines[11], suggesting that genetic interactions are highly dependent on cell context.
Genetic interactions could be the underlying mechanisms of resistance to targeted cancer therapies. The same concept may allow us to develop strategies to overcome this resistance. Mutation analyses of primary cancers for genes encoding kinases or genes with known associations with cancers have revealed that an individual tumor may harbor 50 or more mutations in such genes[25,69-71]. Several important signaling pathways might cooperatively be involved in the oncogenesis and malignant evolution of cancers[25,69-72]. Thus, cancer itself is a result of genetic interactions. Tumor cells, xenograft tumors and primary tumors may carry multiple concomitantly activated oncogenes or inactivated tumor suppressor genes. As a result, interrupting a single pathway is often insufficient to induce cell death in most cancer cells because redundant input from various pathways drives and maintains downstream signaling; thus, single-agent therapies have limited efficacy[73,74]. Consequently, combinations of targeted agents are frequently required for effective anticancer therapy or for overcoming drug resistance[73]. Numerous combination regimens of targeted agents are currently being investigated at either the preclinical or clinical level[74,75]. The information about networks of genetic interactions may facilitate the rational design of combinatorial therapy to enhance therapeutic efficacy.
The SRC oncogene encodes a nonreceptor tyrosine kinase that interacts with multiple receptor tyrosine kinases (RTKs), including EGFR, vascular endothelial grow factor receptor (VEGFR), platelet-derived growth factor receptor (PDGFR), fibroblast growth factor receptor, insulin-like growth factor 1 receptor, hepatocyte growth factor receptor and others[76,77]. Recruiting SRC to receptor tyrosine kinases activates SRC and triggers a cascade of downstream signaling promoting cell proliferation, survival and invasion, as well as angiogenesis. Moreover, SRC can interact synergistically with RTKs by phosphorylating RTKS and modulating their activities[78-80]. Increased SRC activity is associated with resistance to conventional anticancer agents, such as cisplatin[81] and gemcitabine[82], and targeted anticancer agents, such as gefinitib[83] and trastuzumab[84]. Simultaneous targeting of SRC and Her2 sensitizes multiple trastuzumab-resistant breast cancer cells to trastuzumab in vitro and in vivo[84]. Inhibiting SRC also sensitized KRAS mutant colorectal tumors to cetuximab[85]. Combined inhibition of SRC and EGFR sensitized pancreatic tumor cells to gemcitabine[86]. These results demonstrated that combination therapy consisting of SRC and RTKs inhibitors could be an effective strategy for overcoming resistances to a variety of anticancer agents.
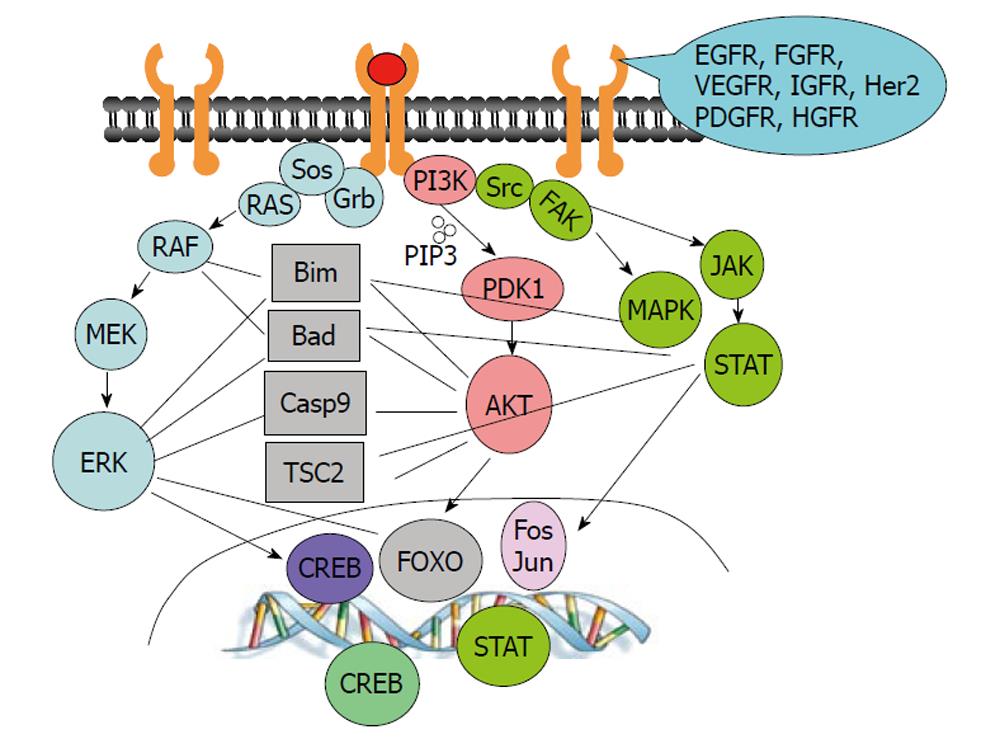
Both EGFR and hepatocyte growth factor receptor (MET)[87,88] play important roles in carcinogenesis[89,90]. Once activated by their ligands, EGF and hepatocyte growth factor, respectively, EGFR and MET activate common downstream pathways, including the PI3K/AKT, RAS/RAF/MAPK and SRC/JAK/STAT pathways (Figure 2). Therefore, elevated activity of MET may negate the effects of anti-EGFR therapeutic agents. Indeed, focal amplification of MET in EGFR-inhibitor-sensitive lung cancer cell lines rendered the cells resistant to anti-EGFR treatment by maintaining ERBB3/PI3K/AKT activity[91]. MET amplification was observed in lung cancer specimens that had developed resistance to gefitinib or erlotinib and in untreated tumors[91,92]. Treatment of resistant cells with a tyrosine kinase inhibitor for either MET or EGFR could not induce cytotoxicity in resistant cells, whereas combined targeting of MET and EGFR resulted in substantial growth inhibition of resistant cells and complete suppression of ERBB3/PI3K/AKT activity[91,92]. Such a therapeutic combination strategy overcame resistance to the EGFR inhibitor erlotinib in an EGFR mutant lung cancer tumor model, both in vitro and in vivo[93].
Crosstalk among downstream pathways of growth factors is also common. The RAS/RAF/MEK/ERK and PI3K/AKT pathways crosstalk and regulate many common downstream targets (Figure 2), such as forkhead transcription factors[94-96], the TSC2/mTOR complex[97-101], BAD[102-104] and caspase-9[105,106]. It is expected that high levels of PI3K/AKT activity can negate antitumor activity induced by MEK/ERK inhibition. Indeed, inhibition of MEK/ERK is sufficient to suppress cell growth or induce apoptosis in cells with low levels of AKT activity but is ineffective in cells with high levels of AKT activity[107]. Combination treatment with MEK and AKT inhibitors was more effective than either single agent alone in human non-small cell lung cancer models in vitro and in vivo[108].
Genetic interaction is likely to be involved in every biological process and has been used as a research platform in various areas of biomedical research. It will continue to be a powerful research tool for both basic and translational studies. Knowledge of the networks of genetic interactions is expected to be translated into clinical applications, in particular for the treatment of cancers.
Note that genetic interactions may be highly dependent on cell context. For a particular gene, genetic interaction may vary in different cell lines. Therefore, it is not unexpected that different candidate genes were obtained when the same oncogenic KRAS gene was used to query its genetic lethal interactions in various cell lines[6-9], or that a candidate gene identified in one cell line may not necessarily be applicable to another cell line[11]. Therefore, individualized therapeutic interventions will be required for patients with cancer, even though their cancers may harbor the same oncogene or tumor suppressor gene mutations. Nevertheless, it is possible that certain key nodes may exist in the networks of genetic interactions that will allow us to develop a common strategy to overcome resistance derived from different genetic interactions[84].
I thank Tamara Locke from the Department of Scientific Publications for her editorial review of this manuscript.
Peer reviewers: Enzo Lalli, MD, INSERM Research Director, Institut de Pharmacologie Moléculaire et Cellulaire CNRS, 660 route de Lucioles-Sophia Antipolis, 06560 Valbonne, France; Wei Li, MD, PhD, Project Staff, Department of Cell Biology/NC-10, Lerner Research Institute, Assistant Professor, Cleveland Clinic Lerner College of Medicine of Case Western Researve University, The Cleveland Clinic, 9500 Euclid Avenue, Cleveland, OH 44195, United States
S- Editor Wang JL L- Editor Roemmele A E- Editor Zheng XM
| 1. | Dobzhansky T. Genetics of Natural Populations. Xiii. Recombination and Variability in Populations of Drosophila Pseudoobscura. Genetics. 1946;31:269-290. [PubMed] |
| 2. | Tong AH, Evangelista M, Parsons AB, Xu H, Bader GD, Pagé N, Robinson M, Raghibizadeh S, Hogue CW, Bussey H. Systematic genetic analysis with ordered arrays of yeast deletion mutants. Science. 2001;294:2364-2368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1635] [Cited by in RCA: 1598] [Article Influence: 66.6] [Reference Citation Analysis (0)] |
| 3. | Goebl MG, Petes TD. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986;46:983-992. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 117] [Cited by in RCA: 130] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 4. | Giaever G, Chu AM, Ni L, Connelly C, Riles L, Véronneau S, Dow S, Lucau-Danila A, Anderson K, André B. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387-391. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3306] [Cited by in RCA: 3182] [Article Influence: 138.3] [Reference Citation Analysis (0)] |
| 5. | Costanzo M, Baryshnikova A, Bellay J, Kim Y, Spear ED, Sevier CS, Ding H, Koh JL, Toufighi K, Mostafavi S. The genetic landscape of a cell. Science. 2010;327:425-431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1783] [Cited by in RCA: 1624] [Article Influence: 108.3] [Reference Citation Analysis (0)] |
| 6. | Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, Schinzel AC, Sandy P, Meylan E, Scholl C. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462:108-112. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1822] [Cited by in RCA: 2683] [Article Influence: 167.7] [Reference Citation Analysis (0)] |
| 7. | Licciulli S, Kissil JL. WT1: a weak spot in KRAS-induced transformation. J Clin Invest. 2010;120:3804-3807. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 6] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 8. | Luo J, Emanuele MJ, Li D, Creighton CJ, Schlabach MR, Westbrook TF, Wong KK, Elledge SJ. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137:835-848. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 859] [Cited by in RCA: 816] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 9. | Scholl C, Fröhling S, Dunn IF, Schinzel AC, Barbie DA, Kim SY, Silver SJ, Tamayo P, Wadlow RC, Ramaswamy S. Synthetic lethal interaction between oncogenic KRAS dependency and STK33 suppression in human cancer cells. Cell. 2009;137:821-834. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 421] [Cited by in RCA: 430] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 10. | Whitehurst AW, Bodemann BO, Cardenas J, Ferguson D, Girard L, Peyton M, Minna JD, Michnoff C, Hao W, Roth MG. Synthetic lethal screen identification of chemosensitizer loci in cancer cells. Nature. 2007;446:815-819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 384] [Cited by in RCA: 371] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 11. | Astsaturov I, Ratushny V, Sukhanova A, Einarson MB, Bagnyukova T, Zhou Y, Devarajan K, Silverman JS, Tikhmyanova N, Skobeleva N. Synthetic lethal screen of an EGFR-centered network to improve targeted therapies. Sci Signal. 2010;3:ra67. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 115] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 12. | Torrance CJ, Agrawal V, Vogelstein B, Kinzler KW. Use of isogenic human cancer cells for high-throughput screening and drug discovery. Nat Biotechnol. 2001;19:940-945. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 200] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 13. | Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 709] [Cited by in RCA: 1047] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 14. | Guo W, Wu S, Liu J, Fang B. Identification of a small molecule with synthetic lethality for K-ras and protein kinase C iota. Cancer Res. 2008;68:7403-7408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 68] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 15. | Kaelin WG. Synthetic lethality: a framework for the development of wiser cancer therapeutics. Genome Med. 2009;1:99. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 63] [Cited by in RCA: 69] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 16. | Reinhardt HC, Jiang H, Hemann MT, Yaffe MB. Exploiting synthetic lethal interactions for targeted cancer therapy. Cell Cycle. 2009;8:3112-3119. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 90] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 17. | Chan DA, Giaccia AJ. Harnessing synthetic lethal interactions in anticancer drug discovery. Nat Rev Drug Discov. 2011;10:351-364. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 201] [Cited by in RCA: 206] [Article Influence: 14.7] [Reference Citation Analysis (0)] |
| 18. | Tong AH, Lesage G, Bader GD, Ding H, Xu H, Xin X, Young J, Berriz GF, Brost RL, Chang M. Global mapping of the yeast genetic interaction network. Science. 2004;303:808-813. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1560] [Cited by in RCA: 1495] [Article Influence: 71.2] [Reference Citation Analysis (0)] |
| 19. | Pan X, Ye P, Yuan DS, Wang X, Bader JS, Boeke JD. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell. 2006;124:1069-1081. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 420] [Cited by in RCA: 435] [Article Influence: 22.9] [Reference Citation Analysis (0)] |
| 20. | Lin YY, Qi Y, Lu JY, Pan X, Yuan DS, Zhao Y, Bader JS, Boeke JD. A comprehensive synthetic genetic interaction network governing yeast histone acetylation and deacetylation. Genes Dev. 2008;22:2062-2074. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 129] [Cited by in RCA: 120] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 21. | Ooi SL, Pan X, Peyser BD, Ye P, Meluh PB, Yuan DS, Irizarry RA, Bader JS, Spencer FA, Boeke JD. Global synthetic-lethality analysis and yeast functional profiling. Trends Genet. 2006;22:56-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 124] [Cited by in RCA: 113] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 22. | Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5:689-698. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1064] [Cited by in RCA: 1047] [Article Influence: 52.4] [Reference Citation Analysis (0)] |
| 23. | Le Meur N, Gentleman R. Modeling synthetic lethality. Genome Biol. 2008;9:R135. [PubMed] |
| 24. | Ye P, Peyser BD, Pan X, Boeke JD, Spencer FA, Bader JS. Gene function prediction from congruent synthetic lethal interactions in yeast. Mol Syst Biol. 2005;1:2005.0026. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 99] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 25. | Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069-1075. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2199] [Cited by in RCA: 2082] [Article Influence: 122.5] [Reference Citation Analysis (0)] |
| 26. | Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, Shen D, Boca SM, Barber T, Ptak J. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2283] [Cited by in RCA: 2263] [Article Influence: 125.7] [Reference Citation Analysis (0)] |
| 27. | Sawyers CL. The cancer biomarker problem. Nature. 2008;452:548-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 673] [Cited by in RCA: 678] [Article Influence: 39.9] [Reference Citation Analysis (0)] |
| 28. | Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783-792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8204] [Cited by in RCA: 8128] [Article Influence: 338.7] [Reference Citation Analysis (0)] |
| 29. | Vogel CL, Cobleigh MA, Tripathy D, Gutheil JC, Harris LN, Fehrenbacher L, Slamon DJ, Murphy M, Novotny WF, Burchmore M. Efficacy and safety of trastuzumab as a single agent in first-line treatment of HER2-overexpressing metastatic breast cancer. J Clin Oncol. 2002;20:719-726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1128] [Cited by in RCA: 905] [Article Influence: 39.3] [Reference Citation Analysis (0)] |
| 30. | Lynch TJ, Bell DW, Sordella R, Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat SM, Supko JG, Haluska FG. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350:2129-2139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8739] [Cited by in RCA: 8778] [Article Influence: 418.0] [Reference Citation Analysis (0)] |
| 31. | Druker BJ, Tamura S, Buchdunger E, Ohno S, Segal GM, Fanning S, Zimmermann J, Lydon NB. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2:561-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2692] [Cited by in RCA: 2581] [Article Influence: 89.0] [Reference Citation Analysis (0)] |
| 32. | Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872-3882. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 195] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 33. | de Murcia JM, Niedergang C, Trucco C, Ricoul M, Dutrillaux B, Mark M, Oliver FJ, Masson M, Dierich A, LeMeur M. Requirement of poly(ADP-ribose) polymerase in recovery from DNA damage in mice and in cells. Proc Natl Acad Sci U S A. 1997;94:7303-7307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 778] [Cited by in RCA: 802] [Article Influence: 28.6] [Reference Citation Analysis (0)] |
| 34. | Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A. Average risks of breast and ovarian cancer associated with BRCA1 or BRCA2 mutations detected in case Series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117-1130. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2571] [Cited by in RCA: 2554] [Article Influence: 116.1] [Reference Citation Analysis (0)] |
| 35. | Thompson D, Easton DF. Cancer Incidence in BRCA1 mutation carriers. J Natl Cancer Inst. 2002;94:1358-1365. [PubMed] |
| 36. | Chen J, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM. Stable interaction between the products of the BRCA1 and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol Cell. 1998;2:317-328. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 449] [Cited by in RCA: 445] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 37. | Patel KJ, Yu VP, Lee H, Corcoran A, Thistlethwaite FC, Evans MJ, Colledge WH, Friedman LS, Ponder BA, Venkitaraman AR. Involvement of Brca2 in DNA repair. Mol Cell. 1998;1:347-357. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 456] [Cited by in RCA: 455] [Article Influence: 16.9] [Reference Citation Analysis (0)] |
| 38. | Bryant HE, Schultz N, Thomas HD, Parker KM, Flower D, Lopez E, Kyle S, Meuth M, Curtin NJ, Helleday T. Specific killing of BRCA2-deficient tumours with inhibitors of poly(ADP-ribose) polymerase. Nature. 2005;434:913-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3368] [Cited by in RCA: 3829] [Article Influence: 191.5] [Reference Citation Analysis (0)] |
| 39. | Farmer H, McCabe N, Lord CJ, Tutt AN, Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I, Knights C. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917-921. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4316] [Cited by in RCA: 4874] [Article Influence: 243.7] [Reference Citation Analysis (0)] |
| 40. | Fong PC, Boss DS, Yap TA, Tutt A, Wu P, Mergui-Roelvink M, Mortimer P, Swaisland H, Lau A, O'Connor MJ. Inhibition of poly(ADP-ribose) polymerase in tumors from BRCA mutation carriers. N Engl J Med. 2009;361:123-134. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3070] [Cited by in RCA: 2854] [Article Influence: 178.4] [Reference Citation Analysis (0)] |
| 41. | Fong PC, Yap TA, Boss DS, Carden CP, Mergui-Roelvink M, Gourley C, De Greve J, Lubinski J, Shanley S, Messiou C. Poly(ADP)-ribose polymerase inhibition: frequent durable responses in BRCA carrier ovarian cancer correlating with platinum-free interval. J Clin Oncol. 2010;28:2512-2519. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 762] [Article Influence: 50.8] [Reference Citation Analysis (0)] |
| 42. | Audeh MW, Carmichael J, Penson RT, Friedlander M, Powell B, Bell-McGuinn KM, Scott C, Weitzel JN, Oaknin A, Loman N. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and recurrent ovarian cancer: a proof-of-concept trial. Lancet. 2010;376:245-251. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1055] [Cited by in RCA: 1077] [Article Influence: 71.8] [Reference Citation Analysis (0)] |
| 43. | Tutt A, Robson M, Garber JE, Domchek SM, Audeh MW, Weitzel JN, Friedlander M, Arun B, Loman N, Schmutzler RK. Oral poly(ADP-ribose) polymerase inhibitor olaparib in patients with BRCA1 or BRCA2 mutations and advanced breast cancer: a proof-of-concept trial. Lancet. 2010;376:235-244. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1352] [Cited by in RCA: 1364] [Article Influence: 90.9] [Reference Citation Analysis (0)] |
| 44. | Barlow C, Hirotsune S, Paylor R, Liyanage M, Eckhaus M, Collins F, Shiloh Y, Crawley JN, Ried T, Tagle D. Atm-deficient mice: a paradigm of ataxia telangiectasia. Cell. 1996;86:159-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1132] [Cited by in RCA: 1120] [Article Influence: 38.6] [Reference Citation Analysis (0)] |
| 45. | Essers J, Hendriks RW, Swagemakers SM, Troelstra C, de Wit J, Bootsma D, Hoeijmakers JH, Kanaar R. Disruption of mouse RAD54 reduces ionizing radiation resistance and homologous recombination. Cell. 1997;89:195-204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 316] [Cited by in RCA: 324] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 46. | Liang Y, Gao H, Lin SY, Peng G, Huang X, Zhang P, Goss JA, Brunicardi FC, Multani AS, Chang S. BRIT1/MCPH1 is essential for mitotic and meiotic recombination DNA repair and maintaining genomic stability in mice. PLoS Genet. 2010;6:e1000826. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 85] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 47. | Shiloh Y. The ATM-mediated DNA-damage response: taking shape. Trends Biochem Sci. 2006;31:402-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 438] [Cited by in RCA: 429] [Article Influence: 22.6] [Reference Citation Analysis (0)] |
| 48. | Matsuoka S, Ballif BA, Smogorzewska A, McDonald ER, Hurov KE, Luo J, Bakalarski CE, Zhao Z, Solimini N, Lerenthal Y. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science. 2007;316:1160-1166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2234] [Cited by in RCA: 2399] [Article Influence: 133.3] [Reference Citation Analysis (0)] |
| 49. | Jiang H, Reinhardt HC, Bartkova J, Tommiska J, Blomqvist C, Nevanlinna H, Bartek J, Yaffe MB, Hemann MT. The combined status of ATM and p53 link tumor development with therapeutic response. Genes Dev. 2009;23:1895-1909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 239] [Cited by in RCA: 243] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 50. | Bolderson E, Richard DJ, Zhou BB, Khanna KK. Recent advances in cancer therapy targeting proteins involved in DNA double-strand break repair. Clin Cancer Res. 2009;15:6314-6320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 144] [Cited by in RCA: 141] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 51. | Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307-310. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5100] [Cited by in RCA: 5113] [Article Influence: 204.5] [Reference Citation Analysis (0)] |
| 52. | Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, Ye Q, Lobo JM, She Y, Osman I. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358-362. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1117] [Cited by in RCA: 1052] [Article Influence: 55.4] [Reference Citation Analysis (0)] |
| 53. | Chang F, Steelman LS, Lee JT, Shelton JG, Navolanic PM, Blalock WL, Franklin RA, McCubrey JA. Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from cytokine receptors to transcription factors: potential targeting for therapeutic intervention. Leukemia. 2003;17:1263-1293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 513] [Cited by in RCA: 537] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 54. | Colicelli J. Human RAS superfamily proteins and related GTPases. Sci STKE. 2004;2004:RE13. [PubMed] |
| 55. | Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4:937-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 884] [Article Influence: 42.1] [Reference Citation Analysis (0)] |
| 56. | Davies H, Bignell GR, Cox C, Stephens P, Edkins S, Clegg S, Teague J, Woffendin H, Garnett MJ, Bottomley W. Mutations of the BRAF gene in human cancer. Nature. 2002;417:949-954. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7459] [Cited by in RCA: 7631] [Article Influence: 331.8] [Reference Citation Analysis (0)] |
| 57. | Wang Y, Engels IH, Knee DA, Nasoff M, Deveraux QL, Quon KC. Synthetic lethal targeting of MYC by activation of the DR5 death receptor pathway. Cancer Cell. 2004;5:501-512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 125] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 58. | Aza-Blanc P, Cooper CL, Wagner K, Batalov S, Deveraux QL, Cooke MP. Identification of modulators of TRAIL-induced apoptosis via RNAi-based phenotypic screening. Mol Cell. 2003;12:627-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 248] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 59. | He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509-1512. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3533] [Cited by in RCA: 3601] [Article Influence: 133.4] [Reference Citation Analysis (0)] |
| 60. | Sansom OJ, Meniel VS, Muncan V, Phesse TJ, Wilkins JA, Reed KR, Vass JK, Athineos D, Clevers H, Clarke AR. Myc deletion rescues Apc deficiency in the small intestine. Nature. 2007;446:676-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 496] [Cited by in RCA: 484] [Article Influence: 26.9] [Reference Citation Analysis (0)] |
| 61. | Zhang L, Ren X, Alt E, Bai X, Huang S, Xu Z, Lynch PM, Moyer MP, Wen XF, Wu X. Chemoprevention of colorectal cancer by targeting APC-deficient cells for apoptosis. Nature. 2010;464:1058-1061. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 113] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 62. | Hartwell LH, Szankasi P, Roberts CJ, Murray AW, Friend SH. Integrating genetic approaches into the discovery of anticancer drugs. Science. 1997;278:1064-1068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 590] [Cited by in RCA: 563] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 63. | Steinmetz LM, Scharfe C, Deutschbauer AM, Mokranjac D, Herman ZS, Jones T, Chu AM, Giaever G, Prokisch H, Oefner PJ. Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400-404. [PubMed] |
| 64. | Yu L, Lopez A, Anaflous A, El Bali B, Hamal A, Ericson E, Heisler LE, McQuibban A, Giaever G, Nislow C. Chemical-genetic profiling of imidazo[1,2-a]pyridines and -pyrimidines reveals target pathways conserved between yeast and human cells. PLoS Genet. 2008;4:e1000284. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 29] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 65. | Heitman J, Movva NR, Hall MN. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science. 1991;253:905-909. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1427] [Cited by in RCA: 1463] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 66. | Heitman J, Movva NR, Hiestand PC, Hall MN. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1991;88:1948-1952. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 225] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 67. | Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 380] [Cited by in RCA: 367] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 68. | Hillenmeyer ME, Fung E, Wildenhain J, Pierce SE, Hoon S, Lee W, Proctor M, St Onge RP, Tyers M, Koller D. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science. 2008;320:362-365. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 832] [Cited by in RCA: 750] [Article Influence: 44.1] [Reference Citation Analysis (0)] |
| 69. | Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2512] [Cited by in RCA: 2295] [Article Influence: 127.5] [Reference Citation Analysis (0)] |
| 70. | Kan Z, Jaiswal BS, Stinson J, Janakiraman V, Bhatt D, Stern HM, Yue P, Haverty PM, Bourgon R, Zheng J. Diverse somatic mutation patterns and pathway alterations in human cancers. Nature. 2010;466:869-873. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 767] [Cited by in RCA: 826] [Article Influence: 55.1] [Reference Citation Analysis (0)] |
| 71. | Stephens P, Edkins S, Davies H, Greenman C, Cox C, Hunter C, Bignell G, Teague J, Smith R, Stevens C. A screen of the complete protein kinase gene family identifies diverse patterns of somatic mutations in human breast cancer. Nat Genet. 2005;37:590-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 257] [Article Influence: 12.9] [Reference Citation Analysis (0)] |
| 72. | Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2010;463:191-196. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1350] [Cited by in RCA: 1255] [Article Influence: 83.7] [Reference Citation Analysis (0)] |
| 73. | Stommel JM, Kimmelman AC, Ying H, Nabioullin R, Ponugoti AH, Wiedemeyer R, Stegh AH, Bradner JE, Ligon KL, Brennan C. Coactivation of receptor tyrosine kinases affects the response of tumor cells to targeted therapies. Science. 2007;318:287-290. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 682] [Cited by in RCA: 713] [Article Influence: 39.6] [Reference Citation Analysis (0)] |
| 74. | Grant S. Is the focus moving toward a combination of targeted drugs? Best Pract Res Clin Haematol. 2008;21:629-637. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 19] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 75. | Faivre S, Djelloul S, Raymond E. New paradigms in anticancer therapy: targeting multiple signaling pathways with kinase inhibitors. Semin Oncol. 2006;33:407-420. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 195] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 76. | Bromann PA, Korkaya H, Courtneidge SA. The interplay between Src family kinases and receptor tyrosine kinases. Oncogene. 2004;23:7957-7968. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 337] [Cited by in RCA: 362] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 77. | Kim LC, Song L, Haura EB. Src kinases as therapeutic targets for cancer. Nat Rev Clin Oncol. 2009;6:587-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 494] [Cited by in RCA: 562] [Article Influence: 35.1] [Reference Citation Analysis (0)] |
| 78. | Huang YZ, McNamara JO. Mutual regulation of Src family kinases and the neurotrophin receptor TrkB. J Biol Chem. 2010;285:8207-8217. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 79. | Emaduddin M, Bicknell DC, Bodmer WF, Feller SM. Cell growth, global phosphotyrosine elevation, and c-Met phosphorylation through Src family kinases in colorectal cancer cells. Proc Natl Acad Sci U S A. 2008;105:2358-2362. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 80. | Peterson JE, Jelinek T, Kaleko M, Siddle K, Weber MJ. c phosphorylation and activation of the IGF-I receptor in src-transformed cells. J Biol Chem. 1994;269:27315-27321. [PubMed] |
| 81. | Peterson-Roth E, Brdlik CM, Glazer PM. Src-Induced cisplatin resistance mediated by cell-to-cell communication. Cancer Res. 2009;69:3619-3624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 82. | Duxbury MS, Ito H, Zinner MJ, Ashley SW, Whang EE. Inhibition of SRC tyrosine kinase impairs inherent and acquired gemcitabine resistance in human pancreatic adenocarcinoma cells. Clin Cancer Res. 2004;10:2307-2318. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 142] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 83. | Qin B, Ariyama H, Baba E, Tanaka R, Kusaba H, Harada M, Nakano S. Activated Src and Ras induce gefitinib resistance by activation of signaling pathways downstream of epidermal growth factor receptor in human gallbladder adenocarcinoma cells. Cancer Chemother Pharmacol. 2006;58:577-584. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 31] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 84. | Zhang S, Huang WC, Li P, Guo H, Poh SB, Brady SW, Xiong Y, Tseng LM, Li SH, Ding Z. Combating trastuzumab resistance by targeting SRC, a common node downstream of multiple resistance pathways. Nat Med. 2011;17:461-469. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 378] [Cited by in RCA: 425] [Article Influence: 30.4] [Reference Citation Analysis (0)] |
| 85. | Dunn EF, Iida M, Myers RA, Campbell DA, Hintz KA, Armstrong EA, Li C, Wheeler DL. Dasatinib sensitizes KRAS mutant colorectal tumors to cetuximab. Oncogene. 2011;30:561-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 110] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 86. | Nagaraj NS, Washington MK, Merchant NB. Combined blockade of Src kinase and epidermal growth factor receptor with gemcitabine overcomes STAT3-mediated resistance of inhibition of pancreatic tumor growth. Clin Cancer Res. 2011;17:483-493. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 82] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 87. | Giordano S, Ponzetto C, Di Renzo MF, Cooper CS, Comoglio PM. Tyrosine kinase receptor indistinguishable from the c-met protein. Nature. 1989;339:155-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 349] [Cited by in RCA: 366] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 88. | Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2000;103:211-225. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3110] [Cited by in RCA: 3089] [Article Influence: 123.6] [Reference Citation Analysis (0)] |
| 89. | Velu TJ, Beguinot L, Vass WC, Willingham MC, Merlino GT, Pastan I, Lowy DR. Epidermal-growth-factor-dependent transformation by a human EGF receptor proto-oncogene. Science. 1987;238:1408-1410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 354] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 90. | Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. Targeting MET as a strategy to overcome crosstalk-related resistance to EGFR inhibitors. Lancet Oncol. 2009;10:709-717. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 91. | Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, Lindeman N, Gale CM, Zhao X, Christensen J. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3442] [Cited by in RCA: 3664] [Article Influence: 203.6] [Reference Citation Analysis (0)] |
| 92. | Turke AB, Zejnullahu K, Wu YL, Song Y, Dias-Santagata D, Lifshits E, Toschi L, Rogers A, Mok T, Sequist L. Preexistence and clonal selection of MET amplification in EGFR mutant NSCLC. Cancer Cell. 2010;17:77-88. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 842] [Cited by in RCA: 854] [Article Influence: 56.9] [Reference Citation Analysis (0)] |
| 93. | Tang Z, Du R, Jiang S, Wu C, Barkauskas DS, Richey J, Molter J, Lam M, Flask C, Gerson S. Dual MET-EGFR combinatorial inhibition against T790M-EGFR-mediated erlotinib-resistant lung cancer. Br J Cancer. 2008;99:911-922. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 127] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 94. | Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell. 1999;96:857-868. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4896] [Cited by in RCA: 5073] [Article Influence: 195.1] [Reference Citation Analysis (0)] |
| 95. | Medema RH, Kops GJ, Bos JL, Burgering BM. AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature. 2000;404:782-787. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1071] [Cited by in RCA: 1119] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 96. | Yang JY, Zong CS, Xia W, Yamaguchi H, Ding Q, Xie X, Lang JY, Lai CC, Chang CJ, Huang WC. ERK promotes tumorigenesis by inhibiting FOXO3a via MDM2-mediated degradation. Nat Cell Biol. 2008;10:138-148. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 553] [Cited by in RCA: 570] [Article Influence: 33.5] [Reference Citation Analysis (0)] |
| 97. | Liu H, Radisky DC, Nelson CM, Zhang H, Fata JE, Roth RA, Bissell MJ. Mechanism of Akt1 inhibition of breast cancer cell invasion reveals a protumorigenic role for TSC2. Proc Natl Acad Sci U S A. 2006;103:4134-4139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 150] [Cited by in RCA: 157] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 98. | Potter CJ, Pedraza LG, Xu T. Akt regulates growth by directly phosphorylating Tsc2. Nat Cell Biol. 2002;4:658-665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 714] [Cited by in RCA: 732] [Article Influence: 31.8] [Reference Citation Analysis (0)] |
| 99. | Inoki K, Li Y, Zhu T, Wu J, Guan KL. TSC2 is phosphorylated and inhibited by Akt and suppresses mTOR signalling. Nat Cell Biol. 2002;4:648-657. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2234] [Cited by in RCA: 2350] [Article Influence: 102.2] [Reference Citation Analysis (0)] |
| 100. | Ma L, Teruya-Feldstein J, Bonner P, Bernardi R, Franz DN, Witte D, Cordon-Cardo C, Pandolfi PP. Identification of S664 TSC2 phosphorylation as a marker for extracellular signal-regulated kinase mediated mTOR activation in tuberous sclerosis and human cancer. Cancer Res. 2007;67:7106-7112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 110] [Cited by in RCA: 118] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 101. | Ma L, Chen Z, Erdjument-Bromage H, Tempst P, Pandolfi PP. Phosphorylation and functional inactivation of TSC2 by Erk implications for tuberous sclerosis and cancer pathogenesis. Cell. 2005;121:179-193. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 934] [Cited by in RCA: 1017] [Article Influence: 50.9] [Reference Citation Analysis (0)] |
| 102. | Scheid MP, Duronio V. Dissociation of cytokine-induced phosphorylation of Bad and activation of PKB/akt: involvement of MEK upstream of Bad phosphorylation. Proc Natl Acad Sci U S A. 1998;95:7439-7444. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 244] [Cited by in RCA: 246] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 103. | Scheid MP, Schubert KM, Duronio V. Regulation of bad phosphorylation and association with Bcl-x(L) by the MAPK/Erk kinase. J Biol Chem. 1999;274:31108-31113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 304] [Cited by in RCA: 311] [Article Influence: 12.0] [Reference Citation Analysis (0)] |
| 104. | Datta SR, Dudek H, Tao X, Masters S, Fu H, Gotoh Y, Greenberg ME. Akt phosphorylation of BAD couples survival signals to the cell-intrinsic death machinery. Cell. 1997;91:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4235] [Cited by in RCA: 4318] [Article Influence: 154.2] [Reference Citation Analysis (0)] |
| 105. | Cardone MH, Roy N, Stennicke HR, Salvesen GS, Franke TF, Stanbridge E, Frisch S, Reed JC. Regulation of cell death protease caspase-9 by phosphorylation. Science. 1998;282:1318-1321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2297] [Cited by in RCA: 2291] [Article Influence: 84.9] [Reference Citation Analysis (0)] |
| 106. | Allan LA, Morrice N, Brady S, Magee G, Pathak S, Clarke PR. Inhibition of caspase-9 through phosphorylation at Thr 125 by ERK MAPK. Nat Cell Biol. 2003;5:647-654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 363] [Cited by in RCA: 363] [Article Influence: 16.5] [Reference Citation Analysis (0)] |
| 107. | Meng J, Peng H, Dai B, Guo W, Wang L, Ji L, Minna JD, Chresta CM, Smith PD, Fang B. High level of AKT activity is associated with resistance to MEK inhibitor AZD6244 (ARRY-142886). Cancer Biol Ther. 2009;8:2073-2080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 108. | Meng J, Dai B, Fang B, Bekele BN, Bornmann WG, Sun D, Peng Z, Herbst RS, Papadimitrakopoulou V, Minna JD. Combination treatment with MEK and AKT inhibitors is more effective than each drug alone in human non-small cell lung cancer in vitro and in vivo. PLoS One. 2010;5:e14124. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |