Copyright
©2011 Baishideng Publishing Group Co.
World J Med Genet. Dec 27, 2011; 1(1): 14-22
Published online Dec 27, 2011. doi: 10.5496/wjmg.v1.i1.14
Published online Dec 27, 2011. doi: 10.5496/wjmg.v1.i1.14
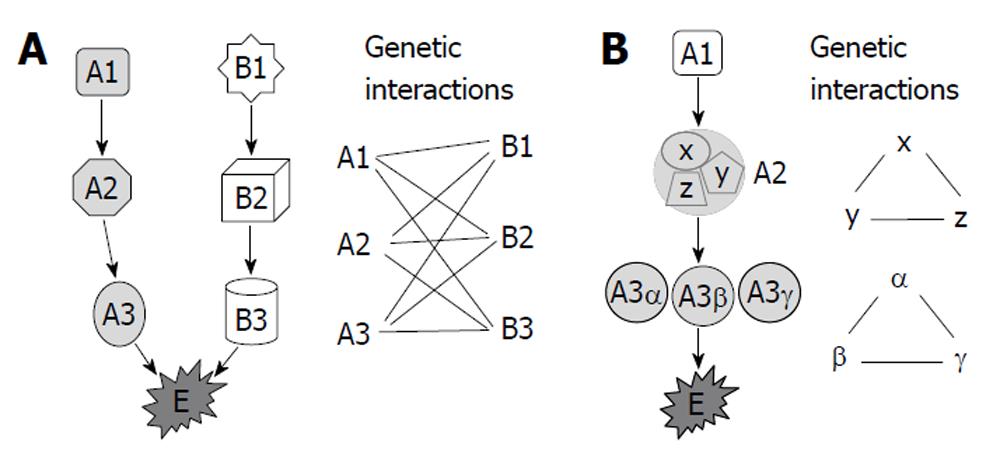
Figure 1 Diagram of genetic interactions.
A: The essential biological function E is regulated by pathways A and B. A functional change in either of these pathways, such as a mutation in A1 or B1, is insufficient to induce dysfunction of E. However, the simultaneous presence of a mutation in A1 and a mutation in any of B1, B2 or B3 induces dysfunction of E (or phenotype changes). Thus, A1 has genetic interaction with B1, B2 and B3, and vice versa; B: The essential biological function E is regulated by pathway A alone, in which A2 is a multiprotein complex composed of X, Y and Z, while A3 has homologues of α, β and γ. Genetic interaction may exist among X, Y and Z, and among A3α, β and γ.
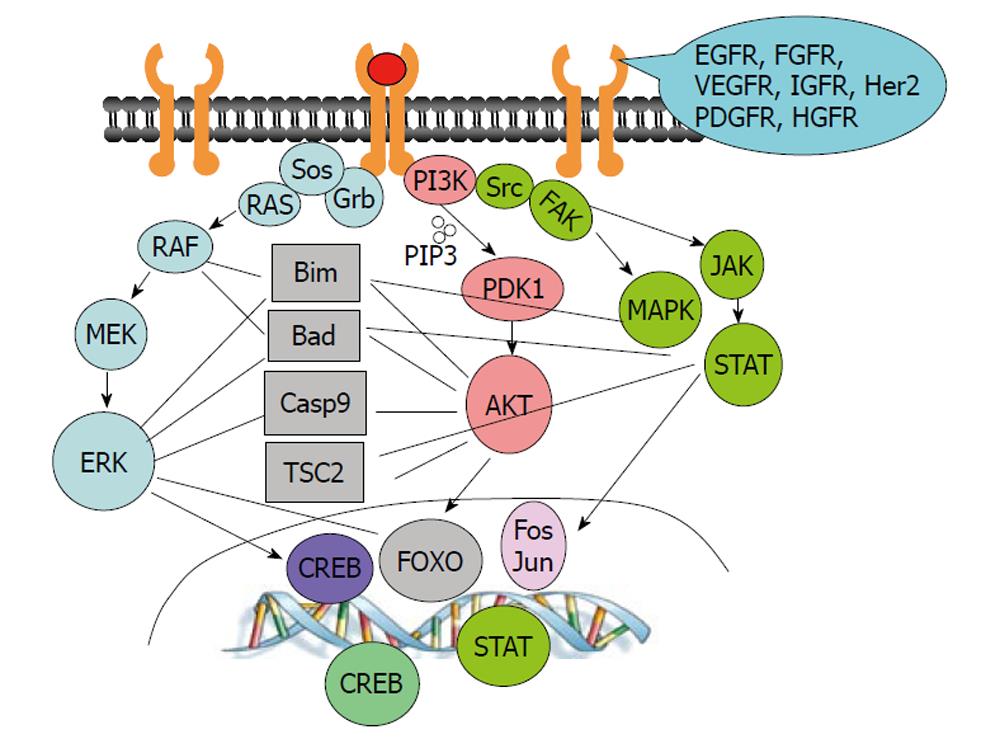
Figure 2 Growth factor activated pathways and their crosstalks.
Growth factors, such as epidermal growth factor (EGF), vascular endothelial grow factor (VEGF), fibroblast growth factor (FGF), hepatocyte growth factor (HGF), insulin-like growth factor (IGF) and platelet derived growth factors (PDGF), interact with their receptors and activate the common downstream pathways, such as PI3K/AKT, RAS/RAF/MAPK and SRC/JAK/STAT pathways. Moreover, there are crosstalks among those pathways in regulating transcription factors and apoptotic/survival proteins. Inhibiting a single target or blocking a single pathway is often not sufficient to induce apoptosis in cancer cells.
- Citation: Fang B. Genetic interactions in translational research on cancer. World J Med Genet 2011; 1(1): 14-22
- URL: https://www.wjgnet.com/2220-3184/full/v1/i1/14.htm
- DOI: https://dx.doi.org/10.5496/wjmg.v1.i1.14