Published online Nov 25, 2015. doi: 10.5495/wjcid.v5.i4.86
Peer-review started: February 10, 2015
First decision: March 6, 2015
Revised: May 11, 2015
Accepted: June 9, 2015
Article in press: June 11, 2015
Published online: November 25, 2015
Processing time: 291 Days and 0.1 Hours
AIM: To study the presence of various nucleic acids targets of Staphylococcus aureus (S. aureus) during bacterial growth and antibiotic induced killing in relation to viability.
METHODS: S. aureus was cultured to log phase and spiked in Todd Hewitt (TH) broth and whole blood of healthy human volunteers. Viability of S. aureus after flucloxacillin treatment (0, 1, 3 and 6 d) was assessed by culture on bloodagar plates. DNA and RNA were isolated from 200 μL. cDNA synthesis was performed by using random primers. The presence of S. aureus DNA, rRNA, and mRNA were determined by real-time polymerase chain reaction of the 16S rDNA and tuf gene (elongation factor Tu).
RESULTS: S. aureus spiked in TH broth without antibiotics grew from day 0-6 and DNA (tuf and 16S), and 16S rRNA remained detectable during this whole period. During flucloxacillin treatment S. aureus lost viability from day 3 onwards, while the 16S rRNA-gene and its RNA transcripts remained detectable. DNA and rRNA can be detected in flucloxacillin treated S. aureus cultures that do not further contain culturable bacteria. However, tuf mRNA became undetectable from day 3 onwards. Tuf mRNA can only be detected from samples with culturable bacteria. When spiking S. aureus in whole blood instead of broth no bacterial growth was seen, neither in the absence nor in the presence of flucloxacillin. Accordingly, no increase in DNA and RNA levels of both 16S rDNA and the tuf gene were detected.
CONCLUSION: Tuf mRNA expression is associated with culturable S. aureus and might be used to monitor antibiotic effects.
Core tip: We report our first results from a proof-of-principle study where we show that tuf mRNA expression seems to correlate with active Staphylococcus aureus (S. aureus) infection. The commonly used target, 16S rRNA, seems unsuitable for viability measurements as it can be detected from samples containing unculturable bacteria. This study indicates that tuf mRNA expression is associated with viable S. aureus, as determined by culture.
-
Citation: Loonen AJ, Wolffs PF, de Bresser M, Habraken M, Bruggeman CA, Hermans MH, van den Brule AJ.
Tuf mRNA rather than 16S rRNA is associated with culturableStaphylococcus aureus . World J Clin Infect Dis 2015; 5(4): 86-93 - URL: https://www.wjgnet.com/2220-3176/full/v5/i4/86.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v5.i4.86
Bacteremia is defined as the presence of viable bacteria in the bloodstream[1]. The current gold standard method for the detection of microorganisms in the bloodstream is blood culture and subsequent identification of the bacteria by conventional (sometimes automated) biochemical techniques or MALDI-TOF MS[2-4]. An important advantage of this method is that only viable microorganisms are detected. A major disadvantage of the method is that the time-to-results is long (24-72 h) due to the involvement of culturing steps. Because fast and accurate diagnosis is of crucial importance for patients suffering from bloodstream infection (BSI), molecular (real-time) polymerase chain reaction (PCR) applications are increasingly being applied to decrease time to pathogen identification, thereby improving patient outcome[5-8]. However, all commercially available sepsis tests [SeptiFAST (Roche), SepsiTest (Molzym), and MagicPlex Sepsis Test (Seegene)] are based on DNA detection. DNA is a stable molecule and the presence of DNA of a certain pathogen does not provide information about the viability status of that pathogen as the DNA can originate from either living or dead pathogens[9-12]. In contrast to DNA, bacterial messenger RNA molecules have a half-live of only minutes[13]. For that reason, several studies have evaluated the detection of mRNA as a marker for the presence of actively growing bacteria[9,11,14-18]. Some of these studies have focused on detection of viable pathogens from food and environmental samples[14], while other studies focused on human disease and viability of pathogens from spiked culture broths (i.e., Borrelia burgdorferi, Escherichia coli, Salmonella typhi, Shigella sonnei, Mycobacterium smegmatis)[10,11,17,19]. Few studies used clinical specimens (Mycobacterium tuberculosis from sputum samples, Aspergillus spp. from blood samples, Chlamydia trachomatis from cervical smears and urine)[12,15,16]. If RNA markers can be used to assess pathogen viability for BSI, the application of PCR based methods on RNA (cDNA) would be of great significance.
BSI can be caused by numerous pathogens[20]. In this study, the most commonly detected Gram-positive bacterium; Staphylococcus aureus (S. aureus) was chosen for reconstruction experiments. To investigate which nucleic acid molecule most favourably correlates to the viability status of S. aureus, DNA and rRNA of 16S rRNA gene, and DNA and mRNA levels of the tuf gene (elongation factor Tu) were measured in response to antibiotic therapy. Both the 16S rRNA and tuf gene are household genes with relatively high expression levels and therefore indicative of protein expression and thus most likely bacterial viability[21]. The aim of this work was to find a suitable marker for S. aureus viability to be able to improve BSI diagnostics.
S. aureus (ATCC 25923) was used for reconstruction (spiking) experiments. Todd Hewitt (TH) broth was inoculated with S. aureus and cultured overnight at 35 °C. Subsequently, a 1:100 dilution was made in fresh TH broth (5 mL) for additional culturing to exponential phase (optical density 0.2 at 600 nm; approximately 1 × 107 cells/mL).
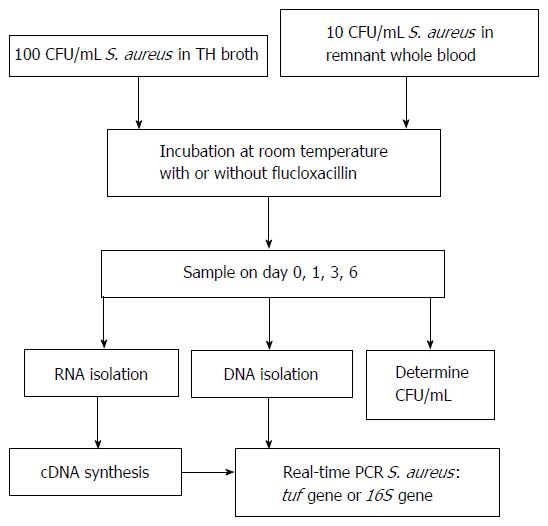
See Figure 1 for an overview of the experimental setup. S. aureus bacteria grown in exponential phase were diluted in either TH broth or pooled (of similar blood type, i.e., O+), 1 d old, residual whole blood from healthy volunteers. The 100 and 10 colony forming units (CFU)/mL dilutions were made in 2 tubes with an end volume of 10 mL TH broth or whole blood, respectively. To one tube an overdose of flucloxacillin (floxapen 5 μg/mL, 1 mL, Actavis, Baarn, the Netherlands) was added (to kill the bacteria) and to the other 1 mL physiological salt solution, this was used as a control. Both tubes were placed on a shaker at room temperature. On days 0, 1, 3, and 6, 200 μL samples were taken from both tubes (flucloxacillin treated and untreated) for DNA and RNA isolation. Additionally, 100 μL was taken to determine CFU/mL on blood agar plates (Tryptone Soya Agar with sheep blood, Oxoid Deutschland GmbH, Wesel, Germany). Bacterial death was defined as the inability of producing colonies on bloodagar.
The obtained 200 μL samples (TH broth and whole blood) were centrifuged at 14000 rpm for 2 min. The supernatant was removed and the pellet was washed once with 200 μL ultra-pure water and centrifuged for 2 min at 14000 rpm. The obtained pellet was resuspended in 20 μL lysozym (12.5%) and 75 μL lysostaphin (100 μg/mL) and incubated for 30 min at 37 °C while shaking (1000 rpm). RLT buffer with β-mercaptoethanol (1:100) (Qiagen RNA blood mini kit) was added and the samples were stored at -80 °C until all time points were collected. The EasyMAG (BioMérieux, Marcy L’Etoile, France) was used for DNA isolation by using the specific B protocol. RNA was isolated by using the RNA blood mini kit (Qiagen), according to manufacturer’s instructions. DNAse treatment was performed as described in the manual provided (Qiagen RNA blood mini kit) using columns to degrade the DNA in the samples.
Reverse transcription was performed on RNA samples using the SuperScript™ II First-Strand Synthesis System for real-time-PCR (Invitrogen, Carlsbad, CA, United States, according to manufacturer’s protocol). Each sample was split in two for the plus and minus reverse transcriptase reaction to check DNA degradation (DNAse treatment on column).
Table 1 for an overview of the used primers and probes (tuf and 16S rDNA). The 16S rDNA primers, specific for most clinically relevant staphylococci, were described by Matsuda et al[22]. However, the 16S rDNA forward primer was slightly modified to adapt to proper annealing temperature. An XS- probe (Biolegio, Nijmegen, The Netherlands) for Staphylococcus spp. detection based on 16S was specifically designed. The PCR mix used has been described previously[23]. Additionally, tuf or 16S primers (900 nmol/L), tuf or 16S probe (200 nmol/L), and 5 μL sample were added to obtain an final volume of 20 μL. A positive and negative control (nuclease free water) were added in each PCR run. The tuf PCR and program used were described previously[24]. Both PCRs were run in white plates on the LightCycler 480 II (Roche Diagnostics).
| Gene | Forward primer 5'-3' | Reverse primer 5'-3' | Probe 5'-3' | Ref. |
| tuf | tcctggttcaattacaccacatactg | ggaaatagaattgtggacgatagtttga | FAM-tgataatacrtawacttctgc-BHQ1 | [24] |
| 16S | acggtcttgctgtcactta | tacacatatgttcttccctaataa | VIC-gtaacggcttaccaaggc-BHQ1 | [22] |
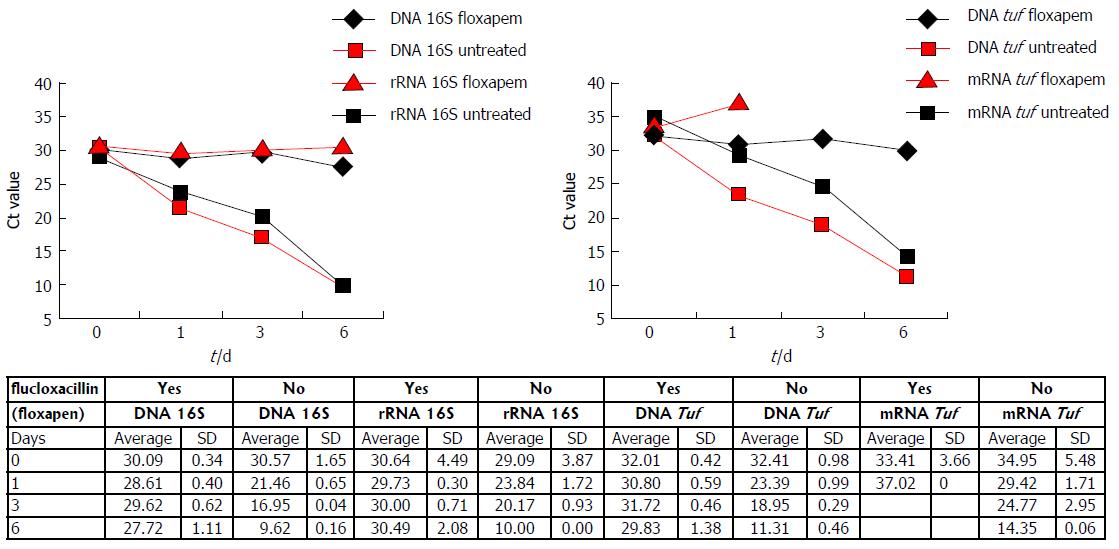
In absence of flucloxacillin the S. aureus bacteria continued to grow. At day 0, on average 330 ± 28 (average ± SD) CFU/mL were detected on bloodagar plates (Table 2). At days 1, 3, and 6 the CFU/mL increased to > 1000 CFU/mL. In contrast, bacterial growth was arrested in flucloxacillin (antibiotic) treated samples and no colonies were detected on bloodagar at days 3 and 6.
| Flucloxacillin | Yes | No | |||
| Average | SD | Average | SD | ||
| Days | 0 | 290 | 14 | 330 | 28 |
| 1 | 255 | 7 | Infinity | ND | |
| 3 | 0 | 0 | Infinity | ND | |
| 6 | 0 | 0 | Infinity | ND | |
Simultaneously, samples were taken for DNA and RNA isolation. In the absence of flucloxacillin, Ct values of both 16S (DNA and rRNA) and tuf (DNA and mRNA) decreased in time (Figure 2). In the presence of flucloxacillin, DNA of the 16S rDNA gene and the tuf gene were detected until day 6, while bloodagar plates indicated absence of culturable S. aureus on day 3. 16S rRNA also remained detectable up to 6 d of treatment. However, tuf mRNA could not be detected on days 3 and 6. The data indicate that S. aureus DNA and rRNA can still persist in the absence of viable bacteria as demonstrated using culture.
The Ct values obtained on day zero are similar for both 16S DNA and rRNA with(out) flucloxacillin. This is not true for tuf DNA and mRNA. Ct values obtained for tuf mRNA are on average 2 Ct higher as compared to tuf DNA (day 0).
These results demonstrate that tuf mRNA is the nucleic acid target that could only be detected from samples which contain culturable bacteria. DNA and rRNA targets could be detected in flucloxacillin treated S. aureus cultures that do not further contain culturable bacteria. This experiment was performed twice on independent days, and showed similar results.
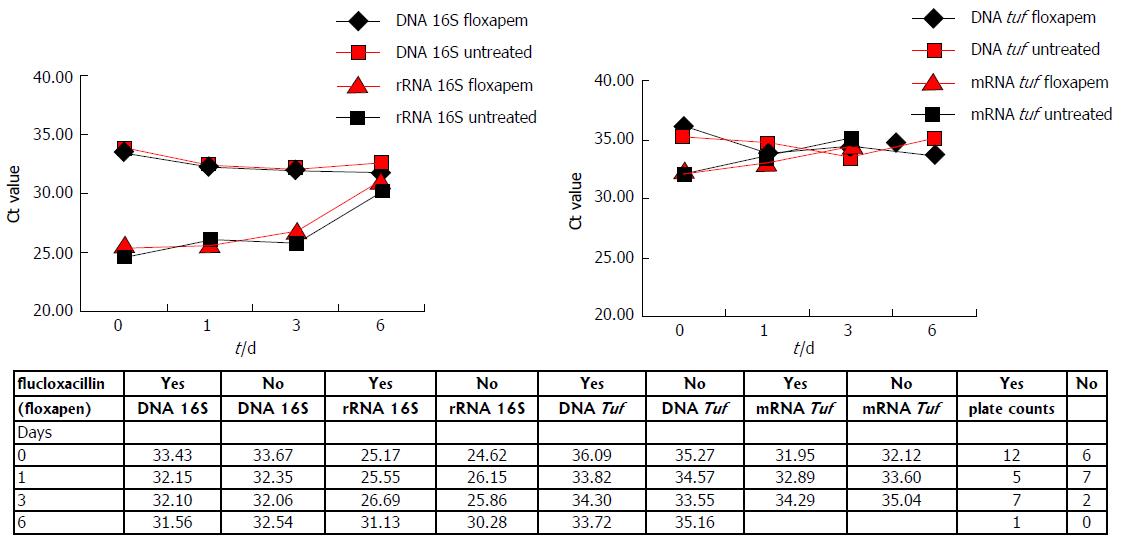
In order to mimic a bloodstream infection, whole blood samples instead of TH broth were spiked with log phase S. aureus and growth was measured both in presence and absence of flucloxacillin. Furthermore, a more clinical significant initial bacterial load was used of 10 CFU/mL (instead of 100 CFU/mL). The plate counts (Figure 3) show that in both presence and absence of flucloxacillin bacterial numbers decrease. DNA and RNA measurements of 16S and tuf also showed growth arrest, both in presence and absence of flucloxacillin, as no decrease in Ct values was observed in time (0-6 d) (Figure 3), this in contrast to the results obtained by using spiked TH broth in absence of flucloxacillin (Figure 2).
The Ct values obtained by detecting 16S DNA and rRNA are lower as compared to the Ct values for tuf DNA and mRNA. A difference of at least 6 Ct was observed when comparing 16S rRNA levels to tuf mRNA levels. When comparing Ct values for DNA detection of both genes the differences were less pronounced, but still significant (approximately 3 Ct). A clear difference was observed between the spiked TH broth samples (Figure 2) and the spiked whole blood samples at day zero (Figure 3). Ct values for 16S DNA and rRNA were comparable in TH broth (day 0), but not in whole blood. For tuf DNA and mRNA this phenomenon was also observed, in whole blood the Ct values for tuf mRNA were lower than for tuf DNA (day 0). Furthermore, there seemed to be a trend towards higher Ct values from day 0-6, independent of flucloxacillin, for both 16S rRNA and tuf mRNA in whole blood. This confirms the culture results obtained from whole blood.
In addition, it was investigated whether fresh (max 1 h) and residual (1 d old) whole blood differed in their performances as medium for bacterial culture. TH broth was used as a control: In the absence of flucloxacillin large amounts of colonies were detected up to 6 d of culture, while in the presence of flucloxacillin colonies were detected on days 0 and 1, and no colonies were detected on bloodagar plates on days 3 and 6 (Table 3). Plate counts from spiked blood samples did not resemble those obtained from spiked TH broth samples. The initial plate count on day 0, while initiated from the same log phase culture, was half or less when compared to the count on day 0 for TH broth samples. However, no significant differences were observed between fresh and residual blood samples both in presence or absence of flucloxacillin.
| TH broth | Fresh whole blood | Remnant whole blood | |||||
| Flucloxacillin | Yes | No | Yes | No | Yes | No | |
| Days | 0 | 20 | 22 | 10 | 4 | 3 | 6 |
| 1 | 14 | Infinity | 2 | 3 | 1 | 1 | |
| 3 | 0 | Infinity | 2 | 1 | 0 | 3 | |
| 6 | 0 | Infinity | 1 | 0 | 0 | 0 | |
For patient survival it is important to provide fast and accurate identification of BSI causing pathogens. Only viable microorganisms can be cultured, and fastidious or damaged organisms can be present in whole blood but often remain culture negative. Molecular diagnostics might provide solutions for these problems, as pathogens in antibiotic treated patients who remain culture negative (due to presence of antibiotics in the bloodstream) can be identified by PCR[25,26]. As BSI is defined as the presence of viable pathogens in the bloodstream, it might be important for a molecular assay to allow pathogen viability measurements from whole blood.
The results that were obtained after spiking S. aureus in TH broth, with and without flucloxacillin, indicated that tuf mRNA might be a more promising marker to measure viability than DNA and rRNA. Tuf mRNA levels correlated with the culture results from both TH broth and whole blood, whereas 16S DNA, 16S rRNA and tuf DNA levels did not. These data confirm results from previous studies[9,10,15,18].
The S. aureus bacteria used in this study seemed to die in whole blood (growth reduction on agar plates in absence of flucloxacillin) or enter a state in which they are viable but non-culturable (VBNC)[27-29]. Bacteria enter the VBNC state in response to stress, such as starvation, incubation outside the growth temperature range, or oxygen concentration[29]. In this study, several stressful conditions might have been present. S. aureus was cultured in whole blood in which white cells might inhibit bacterial growth[30]. Additionally, incubation took place at room temperature (RT) for 6 d, and waste products were not removed from the culture tube. In future studies, it might be useful to remove the white blood cells from whole blood before spiking (buffy coat), and incubate the samples at 35 °C instead of RT to create better growth conditions. Furthermore, different S. aureus strains need to be tested to confirm our results.
In this study, bacteria were considered dead when they were unable to produce colonies on bloodagar. However, as mentioned before, bacteria can enter a VBNC under stressful conditions[27,31]. Bacteria that are not culturable can potentially still be viable and infective. A limitation of this study is that bacterial viability was only measured by colony formation on bloodagar plates. In future studies additional methods to assess bacterial viability might be included, for instance the Live/Dead BacLight Bacterial Viability Kit (Invitrogen). This kit provides two nucleic acid stains [green-fluorescent SYTO 9 dye and red-fluorescent propidium iodide (PI)] to be able distinguish live bacteria (intact membranes) from dead bacteria (compromised membranes). PI is a cell membrane impermeable dye and can only enter compromised pathogens[32]. Another option to differentiate live from dead pathogens is exposure to the dye propidium monoazide (PMA) followed by real-time PCR. PMA cannot penetrate viable cells with intact cytoplasmic membranes[33]. The PMA dye can enter dead pathogens and bind DNA, thereby inhibiting PCR amplification.
The Ct values obtained for S. aureus spiked in TH broth are different from those in the whole blood. Because a lower amount of S. aureus bacteria (10 CFU/mL) was spiked in whole blood, as compared to TH broth (100 CFU/mL), one would expect the Ct value to be 3,3 (1 log) higher in whole blood samples. This difference of approximately 3 Ct was seen in whole blood as compared to TH broth for DNA (both 16S and tuf). However, the Ct values obtained for tuf mRNA and 16S rRNA (t = 0) were higher in TH broth as compared to whole blood (approximately 2 and 5 Ct, respectively). Both RNA targets (tuf and 16S) seem to be expressed at a higher level in whole blood. This unexpected phenomenon might be a result of the difference in environment (blood vs broth). This confirms findings reported by Cenciarini et al[14] who showed that it is difficult to compare RNA viability markers for one pathogen kept in different conditions.
In this study, detection of mRNA and rRNA was performed by using reverse-transcription real-time PCR. Birch et al[34] investigated the use of PCR, real-time-PCR and nucleic acid sequence based amplification (NASBA) for assessment of bacterial viability. They found that NASBA offered the highest sensitivity of the three methods tested. However, presence of residual fliC DNA and mRNA could be detected by NASBA 30 h post-death (culture negative). Other studies have shown that RNA detection by NASBA could be used to monitor infections after antibiotic treatment[12,19]. These contradictory findings again demonstrate that it is important to thoroughly investigate which RNA target is suitable for viability measurement of a certain pathogen.
In this study, results were obtained from as little as 200 μL whole blood. Larger volumes of blood are needed to be able to detect clinical relevant bacterial loads[35]. As bacterial enrichment is a prerequisite to be able to detect bacteria from whole blood, RNA isolation methods should include such an approach. Both Polaris (Biocartis, Mechelen, Belgium) and MolYsis (Molzym GmbH, Bremen, Germany) have developed suitable techniques for pathogen DNA enrichment from large volumes of blood[35]. However, these enrichment strategies are not suitable for RNA isolation. A small pilot study indicated that the first steps, of both the MolYsis and the Polaris pathogen enrichment methods, in which human cells and DNA were removed, did not kill the pathogens present in the whole blood samples as shown by positive cultures (data not shown).
In conclusion, this study clearly demonstrated that detection of S. aureus tuf mRNA, in contrast to DNA and rRNA, correlates to bacterial viability status as determined by culture. Therefore, tuf mRNA might be a promising marker to measure active S. aureus bloodstream infection. After development of RNA isolation procedures from large volumes of whole blood, future clinical studies are needed to validate the preliminary findings obtained in this study.
Bloodstream infections (BSIs) are characterized by high morbidity and mortality and can be caused by a broad variety of microorganisms. Bacteremia is defined as the presence of viable bacteria in the bloodstream. Currently, blood cultures are still the gold standard to detect pathogens from the bloodstream. However, cultures are very time-consuming (24-72 h) and patients need to be treated immediately. Molecular assays (detection of pathogen DNA) can provide results within hours, but the clinical value of DNA detection is still unclear. It might be useful to be able to assess viability of the bacteria in blood samples of patients. Molecular assays that detect the presence of DNA of a specific pathogen can be positive even after viable organisms have been eradicated. The clinical value of pathogen DNA, rRNA and mRNA detection from whole blood needs further investigation.
Molecular diagnostics can provide improved detection and identification of pathogens causing BSI. Implementation of these methods reduces time-to results, offers high sensitivity and specificity, and overall improve the laboratory process for BSI. Detection of DNA by polymerase chain reaction does not provide information about the viability status of a pathogen as the DNA can originate from either living or dead pathogens. The authors attempted to find a marker that enabled their to measure Staphylococcus aureus (S. aureus) viability to further improve BSI diagnostics.
The authors describe a novel approach in molecular diagnostics based on the need to assess bacterial viability and not only detect the presence of bacterial DNA. Clinical relevance of bacterial DNA detection can be limited due to the longer half life of DNA. Antibiotic treatment does not result in the break-down of pathogen DNA. It is important to note that the frequently used 16S rDNA/rRNA target cannot be used to monitor viability of S. aureus bacteria. More research is needed to confirm them data and to find suitable mRNA targets to be able to detect the broad variety of bacteria that are commonly detected in patients.
The study results suggest that Tuf mRNA may represent a suitable marker for the detection of viable S. aureus.
This manuscript describes a novel approach in molecular diagnostics based on the need to monitor viability and not only DNA presence of a microorganism.
P- Reviewer: Fukuda S, Giamarellos-Bourboulis EJ, Schwan WR
S- Editor: Song XX L- Editor: A E- Editor: Liu SQ
| 1. | Spraycar M. Stedman’s Medical Dictionary. Williams and Wilkins. 1995;. |
| 2. | Loonen AJ, Jansz AR, Stalpers J, Wolffs PF, van den Brule AJ. An evaluation of three processing methods and the effect of reduced culture times for faster direct identification of pathogens from BacT/ALERT blood cultures by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis. 2012;31:1575-1583. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 88] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 3. | Riedel S, Carroll KC. Blood cultures: key elements for best practices and future directions. J Infect Chemother. 2010;16:301-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 59] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 4. | van Veen SQ, Claas EC, Kuijper EJ. High-throughput identification of bacteria and yeast by matrix-assisted laser desorption ionization-time of flight mass spectrometry in conventional medical microbiology laboratories. J Clin Microbiol. 2010;48:900-907. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 486] [Cited by in RCA: 459] [Article Influence: 30.6] [Reference Citation Analysis (0)] |
| 5. | Fraser A, Paul M, Almanasreh N, Tacconelli E, Frank U, Cauda R, Borok S, Cohen M, Andreassen S, Nielsen AD. Benefit of appropriate empirical antibiotic treatment: thirty-day mortality and duration of hospital stay. Am J Med. 2006;119:970-976. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 150] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 6. | Gaibani P, Rossini G, Ambretti S, Gelsomino F, Pierro AM, Varani S, Paolucci M, Landini MP, Sambri V. Blood culture systems: rapid detection--how and why. Int J Antimicrob Agents. 2009;34 Suppl 4:S13-S15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 7. | Wallet F, Nseir S, Baumann L, Herwegh S, Sendid B, Boulo M, Roussel-Delvallez M, Durocher AV, Courcol RJ. Preliminary clinical study using a multiplex real-time PCR test for the detection of bacterial and fungal DNA directly in blood. Clin Microbiol Infect. 2010;16:774-779. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 90] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 8. | Yanagihara K, Kitagawa Y, Tomonaga M, Tsukasaki K, Kohno S, Seki M, Sugimoto H, Shimazu T, Tasaki O, Matsushima A. Evaluation of pathogen detection from clinical samples by real-time polymerase chain reaction using a sepsis pathogen DNA detection kit. Crit Care. 2010;14:R159. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 9. | Hellyer TJ, DesJardin LE, Teixeira L, Perkins MD, Cave MD, Eisenach KD. Detection of viable Mycobacterium tuberculosis by reverse transcriptase-strand displacement amplification of mRNA. J Clin Microbiol. 1999;37:518-523. [PubMed] |
| 10. | Iyer R, Mukherjee P, Wang K, Simons J, Wormser GP, Schwartz I. Detection of Borrelia burgdorferi nucleic acids after antibiotic treatment does not confirm viability. J Clin Microbiol. 2013;51:857-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 11. | Josephson KL, Gerba CP, Pepper IL. Polymerase chain reaction detection of nonviable bacterial pathogens. Appl Environ Microbiol. 1993;59:3513-3515. [PubMed] |
| 12. | Morré SA, Sillekens PT, Jacobs MV, de Blok S, Ossewaarde JM, van Aarle P, van Gemen B, Walboomers JM, Meijer CJ, van den Brule AJ. Monitoring of Chlamydia trachomatis infections after antibiotic treatment using RNA detection by nucleic acid sequence based amplification. Mol Pathol. 1998;51:149-154. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 13. | Arraiano CM, Yancey SD, Kushner SR. Stabilization of discrete mRNA breakdown products in ams pnp rnb multiple mutants of Escherichia coli K-12. J Bacteriol. 1988;170:4625-4633. [PubMed] |
| 14. | Cenciarini C, Courtois S, Raoult D, La Scola B. Influence of long time storage in mineral water on RNA stability of Pseudomonas aeruginosa and Escherichia coli after heat inactivation. PLoS One. 2008;3:e3443. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 15. | Jou NT, Yoshimori RB, Mason GR, Louie JS, Liebling MR. Single-tube, nested, reverse transcriptase PCR for detection of viable Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:1161-1165. [PubMed] |
| 16. | Loeffler J, Hebart H, Cox P, Flues N, Schumacher U, Einsele H. Nucleic acid sequence-based amplification of Aspergillus RNA in blood samples. J Clin Microbiol. 2001;39:1626-1629. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 71] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Sheridan GE, Masters CI, Shallcross JA, MacKey BM. Detection of mRNA by reverse transcription-PCR as an indicator of viability in Escherichia coli cells. Appl Environ Microbiol. 1998;64:1313-1318. [PubMed] |
| 18. | Simpkins SA, Chan AB, Hays J, Pöpping B, Cook N. An RNA transcription-based amplification technique (NASBA) for the detection of viable Salmonella enterica. Lett Appl Microbiol. 2000;30:75-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 86] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 19. | van der Vliet GM, Schepers P, Schukkink RA, van Gemen B, Klatser PR. Assessment of mycobacterial viability by RNA amplification. Antimicrob Agents Chemother. 1994;38:1959-1965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 75] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 20. | Wisplinghoff H, Bischoff T, Tallent SM, Seifert H, Wenzel RP, Edmond MB. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin Infect Dis. 2004;39:309-317. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3023] [Cited by in RCA: 3121] [Article Influence: 148.6] [Reference Citation Analysis (0)] |
| 21. | Chaffin DO, Taylor D, Skerrett SJ, Rubens CE. Changes in the Staphylococcus aureus transcriptome during early adaptation to the lung. PLoS One. 2012;7:e41329. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 64] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 22. | Matsuda K, Tsuji H, Asahara T, Kado Y, Nomoto K. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol. 2007;73:32-39. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 220] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 23. | Huijsmans CJ, Damen J, van der Linden JC, Savelkoul PH, Hermans MH. Comparative analysis of four methods to extract DNA from paraffin-embedded tissues: effect on downstream molecular applications. BMC Res Notes. 2010;3:239. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 68] [Cited by in RCA: 76] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 24. | Loonen AJ, Jansz AR, Kreeftenberg H, Bruggeman CA, Wolffs PF, van den Brule AJ. Acceleration of the direct identification of Staphylococcus aureus versus coagulase-negative staphylococci from blood culture material: a comparison of six bacterial DNA extraction methods. Eur J Clin Microbiol Infect Dis. 2011;30:337-342. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 11] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 25. | Mauro MV, Cavalcanti P, Perugini D, Noto A, Sperlì D, Giraldi C. Diagnostic utility of LightCycler SeptiFast and procalcitonin assays in the diagnosis of bloodstream infection in immunocompromised patients. Diagn Microbiol Infect Dis. 2012;73:308-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 26. | Wellinghausen N, Kochem AJ, Disqué C, Mühl H, Gebert S, Winter J, Matten J, Sakka SG. Diagnosis of bacteremia in whole-blood samples by use of a commercial universal 16S rRNA gene-based PCR and sequence analysis. J Clin Microbiol. 2009;47:2759-2765. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 172] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 27. | Davey HM. Life, death, and in-between: meanings and methods in microbiology. Appl Environ Microbiol. 2011;77:5571-5576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 199] [Cited by in RCA: 169] [Article Influence: 12.1] [Reference Citation Analysis (0)] |
| 28. | Keer JT, Birch L. Molecular methods for the assessment of bacterial viability. J Microbiol Methods. 2003;53:175-183. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 278] [Cited by in RCA: 247] [Article Influence: 11.2] [Reference Citation Analysis (0)] |
| 29. | Oliver JD. The viable but nonculturable state in bacteria. J Microbiol. 2005;43 Spec No:93-100. [PubMed] |
| 30. | Högman CF, Gong J, Eriksson L, Hambraeus A, Johansson CS. White cells protect donor blood against bacterial contamination. Transfusion. 1991;31:620-626. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 88] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 31. | Trevors JT. Can dead bacterial cells be defined and are genes expressed after cell death. J Microbiol Methods. 2012;90:25-28. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 25] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 32. | Deligeorgiev TG, Kaloyanova S, Vaquero JJ. Intercalating Cyanine Dyes for Nucleic Acid Detection. Recent Patents on Materials Science. 2009;1-26. [RCA] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 28] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 33. | Nocker A, Cheung CY, Camper AK. Comparison of propidium monoazide with ethidium monoazide for differentiation of live vs. dead bacteria by selective removal of DNA from dead cells. J Microbiol Methods. 2006;67:310-320. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 692] [Cited by in RCA: 713] [Article Influence: 37.5] [Reference Citation Analysis (0)] |
| 34. | Birch L, Dawson CE, Cornett JH, Keer JT. A comparison of nucleic acid amplification techniques for the assessment of bacterial viability. Lett Appl Microbiol. 2001;33:296-301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 73] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 35. | Loonen AJ, Bos MP, van Meerbergen B, Neerken S, Catsburg A, Dobbelaer I, Penterman R, Maertens G, van de Wiel P, Savelkoul P. Comparison of pathogen DNA isolation methods from large volumes of whole blood to improve molecular diagnosis of bloodstream infections. PLoS One. 2013;8:e72349. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 66] [Article Influence: 5.5] [Reference Citation Analysis (0)] |