Published online Nov 25, 2014. doi: 10.5495/wjcid.v4.i4.16
Revised: August 29, 2014
Accepted: September 23, 2014
Published online: November 25, 2014
Processing time: 160 Days and 23.6 Hours
Negative-sense RNA viruses comprise several zoonotic pathogens that mutate rapidly and frequently emerge in people including Influenza, Ebola, Rabies, Hendra and Nipah viruses. Acute respiratory distress syndrome, encephalitis and vasculitis are common disease outcomes in people as a result of pathogenic viral infection, and are also associated with high case fatality rates. Viral spread from exposure sites to systemic tissues and organs is mediated by virulence factors, including viral attachment glycoproteins and accessory proteins, and their contribution to infection and disease have been delineated by reverse genetics; a molecular approach that enables researchers to experimentally produce recombinant and reassortant viruses from cloned cDNA. Through reverse genetics we have developed a deeper understanding of virulence factors key to disease causation thereby enabling development of targeted antiviral therapies and well-defined live attenuated vaccines. Despite the value of reverse genetics for virulence factor discovery, classical reverse genetic approaches may not provide sufficient resolution for characterization of heterogeneous viral populations, because current techniques recover clonal virus, representing a consensus sequence. In this review the contribution of reverse genetics to virulence factor characterization is outlined, while the limitation of the technique is discussed with reference to new technologies that may be utilized to improve reverse genetic approaches.
Core tip: Several negative sense RNA viruses are capricious, pandemic threats and give no quarter to their human hosts. Reverse genetic approaches have been valuable for discovery of key virulence factors mediating disease with the aim of treatment and vaccine development, and knowledge acquisition to genetically map pathogenic potential. Despite the value of the reverse genetics approach current systems are limited by molecular cloning procedures that do not enable reproduction of genetically heterogeneous virus populations that circulate in nature. Advances in molecular biology may facilitate production of genetically diverse viral populations that better represent natural isolates.
- Citation: Edenborough K, Marsh GA. Reverse genetics: Unlocking the secrets of negative sense RNA viral pathogens. World J Clin Infect Dis 2014; 4(4): 16-26
- URL: https://www.wjgnet.com/2220-3176/full/v4/i4/16.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v4.i4.16
Rational design of vaccines and antiviral therapies is facilitated by discovery of viral pathogenicity factors, the viral genes and proteins producing disease. Negative-sense RNA viruses are comprised of formidable human and zoonotic pathogens consisting of seven viral families; four are characterized by non-segmented genomes (Filoviridae, Rhabdoviridae, Paramyxoviridae and Bornaviridae), while the remaining three are distinguished by segmented genomes (Orthomyxoviridae, Bunyaviridae and Arenaviridae)[1]. Before 1994, when pioneering experiments enabled recovery of the first negative-sense RNA virus from cloned cDNA[2], in vivo serial virus passage, often at suboptimal temperatures, was the main method utilized to generate pathogenic variants[3] and retrospective sequence analysis of viral genes enabled associations between genes and pathogenesis[4]. Today reverse genetics is routinely employed to manipulate viral genomes for the purpose of viral pathogenesis research. Briefly cDNAs, representing the full-length RNA genome/genome segments, are cloned into vectors containing T7 RNA polymerase (T7) or RNA polymerase I (polI) transcriptional units. Transfection of these plasmids, in concert with viral polymerase complex expression, into permissive cells facilitates transcription of viral mRNAs, full length vRNA and recovery of infectious virus[5].
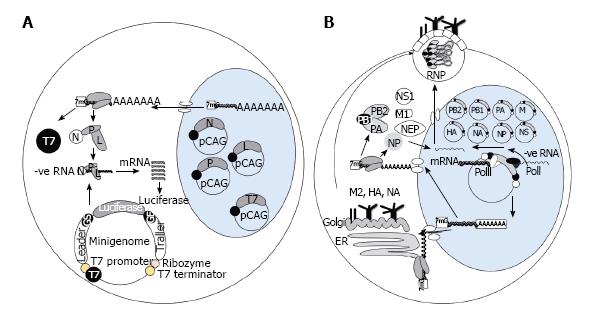
Reverse genetic systems have been optimized for individual viruses by use of minigenomes[6]; open reading frames (ORFs) of reporter constructs encoding bioluminescent enzymes or fluorescent proteins are inserted in between viral noncoding sequences that are sufficient for the transcription and replication activity of the viral polymerase[7] (Figure 1A). Leader and trailer sequences at the respective 3’ and 5’ ends of the vRNA are critical for viral polymerase activity, and hence reporter expression, as demonstrated in the case of Marburg virus[8] and Ebola virus[9,10]. Likewise minigenome constructs for Influenza virus, Orthomyxoviridae, include the 5’ and 3’ noncoding regions of one of the eight vRNAs such as nucleoprotein (NP)[11] or non-structural[12] segments. In the case of Paramyxoviruses, however, the addition of gene start and gene end sequences in combination with leader and trailer sequences have been shown to enhance mRNA production[13], while it may also be important for the number of nucleotides of the minigenome to be perfectly divisible by six as each nucleocapsid (N) protein is thought to interact with six nucleotides[14].
For expression of reporter genes from RNA transcripts, produced from minigenome constructs, viral polymerase complexes are supplied in trans. Eukaryotic expression vectors such as pCAGGS contain strong promoters such as CAG, chicken β actin fused to a cytomegalovirus enhancer, and transient transfection of polymerase constructs promote sufficient viral protein expression to elucidate the minimum number of viral proteins required for reporter expression. For Influenza virus minigenome activity proteins, which form ribonucleoprotein complexes (RNPs), are required including polymerase basic 2 (PB2), PB1, acidic polymerase (PA) and NP proteins[15]. Likewise for Rhabdoviridae[16] and Paramyxoviridae[14] members plasmids encoding N, phosphoprotein (P) and large polymerase (L) are co-transfected with the minigenome to enable reporter expression. The minimal number of proteins required for minigenome activity may vary considerably even within one virus family, e.g., RNA transcription of Respiratory syncytial virus, Paramyxoviridae, was augmented with the inclusion of matrix (M) 2 protein[17], while addition of M protein to Measles virus (MV) minigenome assays reduced reporter expression by reducing vRNA synthesis[18]. Similarly, the expression of accessory proteins may inhibit minigenome activity and repression of some of these proteins may be required for measurement of any polymerase activity[19]. The need for different protein combinations for reporter activity underscores the importance of minigenome assays in determining functional associations between viral proteins for viral mRNA transcription.
Selection of promoters that drive RNA transcription from minigenome constructs is dependent upon whether viral transcription occurs in the nucleus or the cytoplasm during natural replication of the virus. T7-dependent systems may be more suitable for viruses that replicate RNA within the cytoplasm, while polI systems may better mimic viral replication cycles that involve transcription in the nucleus, however recent studies have indicated some exceptions to this view. Infectious Uukuniemi[20], Influenza[21], Thogoto[22], Borna disease virus, MV[23] and Ebola virus[10] have all been successfully recovered from cloned cDNAs by the use of cellular RNA polymerases such as polI (Figure 1B). The conventional role of polI is to transcribe ribosomal RNAs without addition of 5’caps and 3’ poly-A tails[24], therefore it is a suitable host enzyme for the processing of viral RNA molecules generating well defined vRNA 3’ and 5’ termini[25]. To employ RNA polI a cDNA copy corresponding to each viral segment, or a full length cDNA molecule, is placed between a polI truncated promoter and a polI terminator enabling synthesis of vRNA[26]. Interaction between RNA polI and its promoter is species-specific, therefore promoter sequence is carefully selected to suit the cell line destined for virus rescue[27,28].
RNA polII cytomegalovirus promoters have also been utilized to initiate transcription of viral messenger RNA for Influenza virus rescue systems[26] and also has been shown to enhance cRNA expression for MV recovery in relation to other reverse genetic systems, dependent upon T7, however as polII transcripts may be spliced and polyadenylated the utility of polII for virus rescue of other negative-sense RNA viruses is still to be determined[23].
T7 polymerase has been particularly useful for recovery of negative-sense RNA viruses, which mostly undergo transcription in the cytoplasm, including Hendra[29], Nipah[30], MV[31], Sendai[32], Rabies[2], Ebola[33], Marburg[34], Newcastle disease virus[35], RSV[17], Vesicular stomatitis virus[36] and Lymphocytic choriomeningitis virus[37]. Investigators have had more success in virus recovery by insertion of full-length antigenome, rather than genome sense, between T7 promoter and terminator sequences[2]. The T7 promoter can be modified to enhance transcription initiation and reporter expression in minigenome assays by the addition of more than two G nucleotides[38], while if added in combination with a nuclear localization signal the T7 system has enabled recovery of Influenza virus[39]. The disadvantage of T7 systems, in contrast to polI, includes necessary sequences bordering the 5’ and 3’ antigenome ends to form autocatalytic ribozymes that cleave nonviral terminal nucleotides added during transcription. Early rescue systems focussed on correct processing of the RNA 5’ ends, or trailer sequence[38,40], by insertion of an adjacent Hepatitis delta virus (HDV) sequence before the T7 terminator sequence, and the HDV ribozyme sequence has recently been optimized for more efficient vRNA cleavage[41]. Enhanced recovery of infectious virus has also been documented by addition of a hammerhead ribozyme sequence prior to the 3’ leader[41].
An advantage of T7 dependent systems includes transfectable cell lines of several species can be employed for the purpose of virus recovery, providing supplementation with an exogenous source of T7[39]. Choice of cell may be of value when vaccine approved cell lines must be used, or in the case of zoonotic viruses that have limited cell tropism. In early reverse genetics systems cytoplasmic T7 was supplied by addition of recombinant vaccinia virus[42], however its cytopathic effects have been found to impede virus recovery and necessitates plaque purification for removal of vaccinia from the virus culture[43]. These issues have been overcome by use of modified vaccinia Ankara strains[44], however more practical systems are now accessible such as T7 expression plasmids that can be transiently transfected[45] or stably transfected[46,47] into permissive cell lines.
Influenza A virus, a member of Orthomyxoviridae family, contains eight negative-sense RNA segments each corresponding to one of eight viral genes; two of these, the hemagglutinin (HA) and neuraminidase (NA) encode the surface glycoproteins that protrude from the viral envelope. Currently 17 HA and 10 NA types have been identified and all but the most recently described subtype (H17N10 from bats) have been isolated from aquatic birds such as waterfowl and shorebirds, which act as a natural reservoir for the virus[48,49]. The only subtypes circulating in humans, H3N2 and H1N1, cause mild disease associated with viral replication in the upper respiratory tract and large airways[50,51], while replication in the lower respiratory tract[52] or sites outside the respiratory tract results from infection with virulent isolates, such as highly pathogenic avian isolates that infect humans via inter-species transmission events[53].
The virulence factors enabling viral spread beyond the respiratory tract have been characterized in reverse genetic studies. Influenza virus reverse genetic systems have been thoroughly optimized since their initial iterations when purified RNP and RNA were transfected in vitro and recovered with the addition of helper viruses[54]. In early plasmid-based reverse genetic systems influenza viral RNA synthesis was dependent upon supplementation of additional expression plasmids for NP and the polymerase complex, PB1, PB2 and PA, in trans[21,55], however shrewd optimization by inclusion of an RNA polII transcriptional unit on the same plasmid as the RNA polI promoter, in an ambisense direction, generated viral mRNA molecules in cis enabling production of viral protein and vRNA from a single plasmid[56]. More recently gene segments were concatenated onto a single cassette that encoded multiple segments each separated by a transcriptional unit with the aim to hasten the recovery of vaccine seed viruses[57], which is essential for production of pandemic vaccines.
One particular virulence factor that has been well characterized in reverse genetic studies includes the HA glycoprotein, which interacts with terminal sialic acids for host cell attachment[58] and orchestrates fusion of the viral envelope and endosomal membrane of the cell for release of RNPs into the cytoplasm[59]. For efficient fusion the HA precursor must be cleaved at a prominent loop to form two subunits, HA1 and HA2, cleavage of the HA glycoprotein is a process essential for multiple rounds of viral replication and is carried out by enzymes that are produced by the host[60,61]. For most human seasonal influenza and Low Pathogenicity Avian Influenza viruses the cleavage occurs at the site of a single arginine (R) residue[60,62]. For this reason, these viruses are limited to tissues that contain host enzymes with the corresponding recognition preference for single basic amino acids[60,63].
In the HA of highly virulent subtypes insertions of multiple basic amino acid residues have been found and this region has been coined the multi-basic cleavage site (MBCS, Table 1)[64,65]. The role of the MBCS in pathogenesis for chickens was assessed by generating H5N2 mutant viruses containing variable sequence lengths and combinations of basic amino acid residues in the HA via reverse genetics[66]. In this study they found association between presence of a > 4 basic amino acid residues in the cleavage site, efficient HA cleavage in chicken embryonic fibroblasts and lethality to chickens, which was caused by spread of virus to brain via neurons and systemic organs via the blood stream. More recent plasmid based reverse genetic studies have demonstrated a similar role for the MBCS in viral pathogenesis for mammals, such as mice[67] and ferrets[68,69], which corresponds to the pathological and clinical observations of humans infected with highly pathogenic influenza such as detection of virus in systemic organs of fatal H5N1 cases[70] and detection of vRNA in the blood stream of infected patients[71]. Since the importance of the MBCS for influenza pathogenesis has been established recent research has focused on reducing replication of Influenza virus with substrate-analogue peptide mimetic inhibitors that target host cell proteases, specifically those carrying out HA0 cleavage[72]. This is a successful example of the use of reverse genetics for identification of a virulence factor, the MBCS, and production of an inhibitor based on the understanding of virulence mechanisms.
| Virus | Virulence factors | Role in pathogenesis |
| Influenza A | Multibasic cleavage site | Facilitates viral spread to cells outside the respiratory tract |
| Ebola | Glycoprotein Viral protein 24 and nucleoprotein | Transmembrane form mediates host cell attachment and its soluble forms activate mononuclear phagocytes and endothelial cells Antagonists of IFN responses |
| Viral protein 35 | Viral polymerase cofactor that suppresses RIG-I like receptor signalling | |
| Rabies | Glycoprotein | Neurotropic surface glycoprotein that facilitates spread to the brain |
| Phosphoprotein | Viral polymerase cofactor and antagonist of IFN responses | |
| Hendra and Nipah | Phosphoprotein | Viral polymerase cofactor and antagonist of IFN responses |
| V and W proteins | Antagonists of IFN responses | |
| C protein | Regulates viral transcription and affects activation of innate immune cells |
In 1976 Ebola virus, a single-stranded negative-sense RNA virus of 18.9kb within the Filoviridae, first emerged in humans and thereafter several outbreaks in Sudan, western and central Africa have been documented[73]. Index cases are often associated with butchering, handling or consuming bush meat such as fruit bats[74], the potential natural reservoir of the virus[75], and also close contact with non-human primates[76]. Human-to-human transmission occurs via close contact[77] and long incubation periods, prior to symptom development, facilitate viral spread in the community causing stigmatization of health care workers and relatives of the sick[78]. Ebola viruses isolated from different geographical locations in Africa have caused similar disease symptoms and signs such as headache, myalgia, muscle spasms, fever, malaise, abdominal pain, haemorrhage and maculopapular rash[79], although the latter was more commonly noted in infected patients of the Zaire outbreak, which of all the outbreaks, has the highest documented case fatality rate[80,81]. In contrast humans infected with the Reston Ebola virus isolate, which emerged via importation of infected monkeys from the Philippines into Reston, Virginia, United States, developed antibodies to the virus in the absence of clinical disease[82] indicating this virus isolate was not pathogenic, although only a small number of humans were exposed. Reston Ebola virus has since been detected in piggeries in the Philippines while serological studies suggest a small number of pig farm workers have been infected with the virus[83].
Both T7 and polI dependent systems have been utilized for minigenome assays to characterize the viral proteins mediating transcription for Zaire and Reston Ebola viruses, and to recover infectious viruses for several Ebola virus isolates[84]. Reverse engineered viruses have been used to assess the role of the Ebola virus glycoprotein (GP, Table 1), which forms trimeric spikes on the viral envelope and mediates host cell attachment and entry[85]. Produced through a process of transcriptional RNA editing[86], GP protein expression is regulated in infected cells[87]; secretory GP is produced from unedited transcripts, while transmembrane GP is produced from edited transcripts that preside at lower frequency[88]. GP expression is cytotoxic acting to increase the permeability of venous and arterial blood vessels, compromising vascular function[89]. Recently chimeric Ebola viruses, in which the GP of Zaire and Reston virus isolates were exchanged, have been utilized to clarify isolate-specific differences in virulence[90]. Interferon-α/β receptor knock-out (IFNAR-/-) transgenic mice were selected to characterize in vivo pathogenicity of the chimeric viruses, as the need for virus adaptation via serial passage is unnecessary, unlike immune competent mice. Reverse engineered Reston virus was not pathogenic to IFNAR-/- mice and only replicated to low levels in liver and spleen, which mirrors the absence of human disease[90]. In contrast, Zaire virus caused rapid weight loss and replicated to high titres in spleen and liver, which indicated that the IFNAR-/- mouse model recapitulated human disease observations in relation to both virus isolates[90]. Interestingly, introduction of the Reston GP into the Zaire virus did not attenuate virulence in IFNAR-/- mice, suggesting that Zaire GP is not the only determinant of virulence, and instead the robust replicative capacity of Zaire virus in vitro and in vivo was consisted with its virulence for IFNAR-/- mice[90].
Other factors for Zaire Ebola virus virulence have been identified via serial passage of reverse engineered viruses in mice and include viral protein 24 and nucleoprotein (Table 1), factors that are involved in evasion of host type 1 interferon responses[91]. Other components of the innate immune response, particularly transcription factors such as interferon regulatory factor 3, may be suppressed by viral protein 35 (VP35, Table 1), a cofactor of Ebola viral polymerase[92] and production of reverse genetic viruses containing mutations in VP35 have been used in transcriptome studies[93], which may provide key insights into virus host interactions.
Rabies virus, a member of the Rhabdoviridae, was the first single-stranded negative-sense virus recovered by reverse genetics[2]. Wildlife host reservoirs of rabies virus include bats[94], racoons and foxes[95] although cross-species transmission to non-human primates and domestic animals such as dogs and cats perpetuate human disease[96-98]. Risks for human infection include direct exposure to saliva shed from rabid animals occurring via animal bite or contamination of broken skin[99]. Human disease results from fatal encephalitis that progresses as virus spreads to the central nervous system via retrograde transport along axons of peripheral nerves, which is mediated by rabies glycoprotein (G, Table 1)[100]. The 12kb long genome encodes five proteins; nucleoprotein (N), phosphoprotein (P, Table 1), matrix protein (M), G and the RNA-dependent RNA polymerase (L). Viruses produced by reverse genetics have elucidated that G, M and P proteins play important roles in the severity of rabies disease by either facilitating cell to cell spread[101] or antagonizing host innate immune responses[102].
Currently rabies virus vaccines delivered to humans are beta-Propiolactone inactivated and administered intramuscularly in three doses to generate neutralizing G-specific antibody titres, which wane overtime, as such additional booster shots may be needed[103]. Due to the requirement for multiple doses and the high cost of vaccination and post-exposure prophylaxis (PEP) therapies more than 55000[104] rabies-related deaths are still reported annually predominantly in developing countries. Replication deficient rabies viruses (RDRV) produced via reverse genetics may be a low cost alternative to current vaccines and PEP. RDRV have been produced by T7 driven reverse genetic systems such that the viruses contained neither coding capacity for M nor P, instead the latter encoded two copies of G for protein over-expression and induction of greater G specific immune responses[105]. RDRV vectors have also been shown to be immunogenic demonstrated by the induction of neutralizing G-specific antibodies in non-human primates following a prime boost immunization strategy[106]. Safety is a main concern for the use of live attenuated viral vaccines, but because RDRV vectors replicate in cell culture only in the presence of P or M protein supplementation and are innocuous to immune deficient transgenic mice[106] the risk of reversion may not be as great as other live attenuated vaccine viruses that are attenuated through single amino acid changes. Rabies virus has also been shown to be a safe and excellent vaccine vector with the ability to generate antibody responses targeting human immunodeficiency virus envelope proteins, severe acute respiratory syndrome coronavirus and hepatitis C virus proteins[107]. Collectively, these RDRV studies have highlighted the diverse utility of reverse genetics, not only enabling discovery of virulence determinants, but also applying our understanding of virulence to rationally engineer attenuated viruses for the purposes of vaccination.
Hendra virus (HeV) and Nipah virus (NiV), classified within the Henipavirus genus of the Paramyxoviridae, which are harboured asymptomatically by Pteropid fruit bats[108], were identified as aetiological agents of severe human infections that occurred in the 1990’s. Inter-species transmission occurs via intermediate hosts; infected horses were associated with HeV human cases in Australia[109] and infected pigs were associated with NiV cases in Malaysia[110]. Additional risk factors for contraction of Nipah virus in Bangladesh have included consumption of raw date palm sap, a delicacy in Bangali culture, contaminated by bat urine or saliva[111], while limited human-to-human transmission has been documented in cases of very intimate contact such as preparing an infected corpse for burial[112]. In humans HeV and NiV infections are associated with high case fatality rates and severe disease. NiV infected humans develop respiratory and neurological signs such as dyspnoea, disorientation, confusion and muscle spasms that are associated with the expansive tissue tropism of the virus, which includes infection of neurones of the central nervous system, endothelium, lymphoid and respiratory tissues[113]. In 10% of infected people NiV has been shown to reside in a quiescent form for months or years until the virus reactivates causing fatal neurological disease[114]. HeV disease in humans has not been thoroughly characterized as few human infections have occurred, however from limited reports of post-mortems and disease signs it appears the respiratory and neurological disease caused by HeV is akin to that caused by NiV infection[115,116].
NiV and HeV have non-segmented genomes of -18kb in length that encodes for more than six proteins and NiV and HeV are closely related to each other with amino acid sequence similarities of > 80% for many of the viral proteins[117]. Nucleocapsid protein (N), phosphoprotein (P), matrix protein (M), fusion protein (F), glycoprotein (G) and large polymerase (L) are encoded on discrete transcriptional units[118], while three accessory viral proteins are produced from the P gene (Table 1) including the C protein that is transcribed from an alternate open reading frame (ORF), and also V and W proteins that are produced by the addition of G nucleotides into transcribed mRNAs via RNA editing[119]. P, V, W and C proteins play important roles in infection by impeding activation of host antiviral responses. A conserved feature of the paramyxovirus V protein is its ability to bind melanoma differentiation-association gene 5 (MDA5), a pattern recognition receptor, impeding the recognition of dsRNA resulting in inhibition of IFN-β induction[120,121]. Furthermore, V and W proteins can prevent activation of the type I IFN signalling pathway by sequestration of signal transducer and activator of transcription (STAT) in the cytoplasm or nucleus, reducing STAT mediated induction of interferon stimulating genes key to innate antiviral responses[122]. Inhibition of various components of the antiviral response by several NiV and HeV proteins underscores the role of P, V, W and C for viral pathogenesis.
HeV and NiV reverse genetics systems have been successful in virus recovery with use of T7[29,30] dependent systems and co-transfection of the protein expression plasmids N, P and L, which encapsidate the RNA forming the RNP complex, as these proteins are essential for minigenome function[14]. The transfection ratios of N:P:L require optimization for efficient virus recovery as poor reporter expression in minigenome assays have been noted in the context of high concentrations of P protein[45], which likely results from the C protein inhibiting minigenome expression[19]. To further examine importance of C, V and W proteins for NiV pathogenesis several recombinant viruses were produced; Stop codons were introduced downstream of the C ORF site or the RNA editing site to prevent expression of C or V and W respectively. Despite these changes all viruses expressed functional forms of the P protein[123]. Following confirmation of P but not V, W or C expression in infected cells, it was determined that the recombinant viruses replicated efficiently in vitro, which indicated these proteins were not essential for viral replication. Virus pathogenicity was assessed by use of a hamster infection model, wherein it was demonstrated that suppression of C and V, but not W protein, completely attenuated NiV as weight loss, disease signs and high levels of viral replication in systemic organs were not observed[123]. Another study by a different group compared host responses in human endothelial cell lines infected with wildtype NiV verses one of the attenuated viruses, NiV lacking C protein expression (NiVΔC). With microarray analysis they established that compared to wildtype NiV, NiVΔC induced higher levels of cytokines and chemokines such as interleukin 1 beta (IL-1β), IL-8, CXCL2, CXCL3, CXCL6 and CCL20[124]. These findings indicated that NiV C plays a role in inhibiting induction of proinflammatory cytokines and recruitment of leucocytes and lymphocytes into sites of infection such as the lung. This study also examined the pathogenesis of NiVΔC in hamsters and the researchers were only able to partially replicate the attenuated phenotype of NiVΔC, as 30%-90% of animals required euthanasia following infection. The reason for the variation in results between the two studies is yet to be ascertained, however it highlights the limitations associated with insertion of silent restriction sites for the purposes of engineering mutations into plasmids, as laboratories will insert a different variety of silent mutations that may have unknown effects on the virulence of the virus.
RNA viruses, HIV and Influenza virus in particular, spread between and within hosts as genetically heterogeneous virus populations, or quasispecies, clustering around a dominant virus sequence[125]. Quasispecies are perpetuated by spontaneous mutations afforded by low fidelity viral RNA polymerases, and although exact mutation rates may differ for each virus[126], they are in the range of 10-4 mutations per nucleotide copied[127], therefore 1.3 mutations would be expected to occur with every replication of the influenza genome of -13kb. Quasispecies are thought to act cooperatively with the aim of facilitating viral persistence within hosts[125,128]. Genetic heterogeneity has been found important for poliovirus pathogenesis as demonstrated by the ability of a genetically diverse, but not a homogenous, virus population for invasion into the CNS[129]. HIV genetic diversity may also influence viral tropism and larger sequence diversity has been associated with faster disease progression[128]. With this in mind we are faced with a technological drawback; the requirement of producing infectious clones for the purpose of virus rescue also removes population heterogeneity that may play pivotal roles in pathogenesis. Overall, care should be taken in the selection of a consensus sequence to produce infectious clones representative of dominant and also subdominant variants.
Reverse genetic technologies have proven critical to study the contribution of viral genetic factors to disease severity by enabling production of well-defined, recombinant negative-sense RNA viruses, such as a mutant and wildtype viruses, which can be compared for the purpose of identifying chief virulence determinants in the context of host-pathogen systems. For several negative-sense RNA viruses effective rescue methods have been developed, which may be dependent upon either T7 or polI and II transcriptional units. Furthermore, inclusion of polymerase proteins or 5’ and 3’ cleavage sequences for correct vRNA processing may also be necessary for rescue, although these conditions are optimized for each virus and minigenome assays have proved useful for this purpose.
Recombinant viruses, however, are produced by selection of a consensus sequence that forms the basis of the infectious clone and therefore recombinant viruses are likely to constitute only the dominant viral species of a potentially diverse natural virus population. Reduction and alteration in viral heterogeneity, as a consequence of reverse genetics, is a limitation not often taken into account in the context of pathogenesis studies. However, with the advent of next generation sequence technologies for thorough characterization of virus populations we stand in good stead to gather a better grasp of viral heterogeneity in a field isolate and molecular biologists may be capable of recapitulating diverse viral populations via reverse genetics. Recent technologies such as Gibson cloning[130] and barcoding virus populations[131] are likely to enable researchers to produce heterogeneous virus populations that can be studied for characterization of the pathogenic potential of diverse viral populations, with a particular focus on the importance of subdominant viruses for severe disease outcomes. Despite this limitation, reverse genetics enables production of viruses that may be utilized for various future applications such as live-attenuated vaccines, mapping neural pathways in the brain, oncolytic virus production and delivery of microRNAs as a therapy for viral infections.
P- Reviewer: Bener A S- Editor: Ji FF L- Editor: A E- Editor: Lu YJ
| 1. | Neumann G, Whitt MA, Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J Gen Virol. 2002;83:2635-2662. [PubMed] |
| 2. | Schnell MJ, Mebatsion T, Conzelmann KK. Infectious rabies viruses from cloned cDNA. EMBO J. 1994;13:4195-4203. [PubMed] |
| 3. | Pearce JM. Louis Pasteur and rabies: a brief note. J Neurol Neurosurg Psychiatry. 2002;73:82. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 20] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 4. | Hiromoto Y, Saito T, Lindstrom S, Nerome K. Characterization of low virulent strains of highly pathogenic A/Hong Kong/156/97 (H5N1) virus in mice after passage in embryonated hens’ eggs. Virology. 2000;272:429-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 31] [Cited by in RCA: 29] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 5. | Bridgen A, Elliott RM. Rescue of a segmented negative-strand RNA virus entirely from cloned complementary DNAs. Proc Natl Acad Sci USA. 1996;93:15400-15404. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 180] [Cited by in RCA: 181] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 6. | Conzelmann KK. Reverse genetics of mononegavirales. Curr Top Microbiol Immunol. 2004;283:1-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 59] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 7. | Hoenen T, Groseth A, de Kok-Mercado F, Kuhn JH, Wahl-Jensen V. Minigenomes, transcription and replication competent virus-like particles and beyond: reverse genetics systems for filoviruses and other negative stranded hemorrhagic fever viruses. Antiviral Res. 2011;91:195-208. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 96] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 8. | Mühlberger E, Lötfering B, Klenk HD, Becker S. Three of the four nucleocapsid proteins of Marburg virus, NP, VP35, and L, are sufficient to mediate replication and transcription of Marburg virus-specific monocistronic minigenomes. J Virol. 1998;72:8756-8764. [PubMed] |
| 9. | Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. Comparison of the transcription and replication strategies of marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333-2342. [PubMed] |
| 10. | Groseth A, Feldmann H, Theriault S, Mehmetoglu G, Flick R. RNA polymerase I-driven minigenome system for Ebola viruses. J Virol. 2005;79:4425-4433. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 11. | Ozawa M, Shimojima M, Goto H, Watanabe S, Hatta Y, Kiso M, Furuta Y, Horimoto T, Peters NR, Hoffmann FM. A cell-based screening system for influenza A viral RNA transcription/replication inhibitors. Sci Rep. 2013;3:1106. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 19] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 12. | Moncorgé O, Long JS, Cauldwell AV, Zhou H, Lycett SJ, Barclay WS. Investigation of influenza virus polymerase activity in pig cells. J Virol. 2013;87:384-394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 13. | Kuo L, Grosfeld H, Cristina J, Hill MG, Collins PL. Effects of mutations in the gene-start and gene-end sequence motifs on transcription of monocistronic and dicistronic minigenomes of respiratory syncytial virus. J Virol. 1996;70:6892-6901. [PubMed] |
| 14. | Halpin K, Bankamp B, Harcourt BH, Bellini WJ, Rota PA. Nipah virus conforms to the rule of six in a minigenome replication assay. J Gen Virol. 2004;85:701-707. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 97] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 15. | Huang TS, Palese P, Krystal M. Determination of influenza virus proteins required for genome replication. J Virol. 1990;64:5669-5673. [PubMed] |
| 16. | Conzelmann KK, Schnell M. Rescue of synthetic genomic RNA analogs of rabies virus by plasmid-encoded proteins. J Virol. 1994;68:713-719. [PubMed] |
| 17. | Collins PL, Hill MG, Camargo E, Grosfeld H, Chanock RM, Murphy BR. Production of infectious human respiratory syncytial virus from cloned cDNA confirms an essential role for the transcription elongation factor from the 5’ proximal open reading frame of the M2 mRNA in gene expression and provides a capability for vaccine development. Proc Natl Acad Sci USA. 1995;92:11563-11567. [PubMed] |
| 18. | Iwasaki M, Takeda M, Shirogane Y, Nakatsu Y, Nakamura T, Yanagi Y. The matrix protein of measles virus regulates viral RNA synthesis and assembly by interacting with the nucleocapsid protein. J Virol. 2009;83:10374-10383. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 113] [Cited by in RCA: 116] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 19. | Sleeman K, Bankamp B, Hummel KB, Lo MK, Bellini WJ, Rota PA. The C, V and W proteins of Nipah virus inhibit minigenome replication. J Gen Virol. 2008;89:1300-1308. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 52] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Flick R, Pettersson RF. Reverse genetics system for Uukuniemi virus (Bunyaviridae): RNA polymerase I-catalyzed expression of chimeric viral RNAs. J Virol. 2001;75:1643-1655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 100] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 21. | Fodor E, Devenish L, Engelhardt OG, Palese P, Brownlee GG, García-Sastre A. Rescue of influenza A virus from recombinant DNA. J Virol. 1999;73:9679-9682. [PubMed] |
| 22. | Wagner E, Engelhardt OG, Gruber S, Haller O, Kochs G. Rescue of recombinant Thogoto virus from cloned cDNA. J Virol. 2001;75:9282-9286. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 25] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 23. | Martin A, Staeheli P, Schneider U. RNA polymerase II-controlled expression of antigenomic RNA enhances the rescue efficacies of two different members of the Mononegavirales independently of the site of viral genome replication. J Virol. 2006;80:5708-5715. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 104] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 24. | Paule MR, White RJ. Survey and summary: transcription by RNA polymerases I and III. Nucleic Acids Res. 2000;28:1283-1298. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 355] [Cited by in RCA: 361] [Article Influence: 14.4] [Reference Citation Analysis (0)] |
| 25. | Neumann G, Zobel A, Hobom G. RNA polymerase I-mediated expression of influenza viral RNA molecules. Virology. 1994;202:477-479. [PubMed] |
| 26. | Hoffmann E, Neumann G, Kawaoka Y, Hobom G, Webster RG. A DNA transfection system for generation of influenza A virus from eight plasmids. Proc Natl Acad Sci USA. 2000;97:6108-6113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1213] [Cited by in RCA: 1298] [Article Influence: 51.9] [Reference Citation Analysis (0)] |
| 27. | Murakami S, Horimoto T, Yamada S, Kakugawa S, Goto H, Kawaoka Y. Establishment of canine RNA polymerase I-driven reverse genetics for influenza A virus: its application for H5N1 vaccine production. J Virol. 2008;82:1605-1609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 28. | Song MS, Baek YH, Pascua PN, Kwon HI, Park SJ, Kim EH, Lim GJ, Choi YK. Establishment of Vero cell RNA polymerase I-driven reverse genetics for Influenza A virus and its application for pandemic (H1N1) 2009 influenza virus vaccine production. J Gen Virol. 2013;94:1230-1235. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 20] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 29. | Marsh GA, Virtue ER, Smith I, Todd S, Arkinstall R, Frazer L, Monaghan P, Smith GA, Broder CC, Middleton D. Recombinant Hendra viruses expressing a reporter gene retain pathogenicity in ferrets. Virol J. 2013;10:95. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 26] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 30. | Yoneda M, Guillaume V, Ikeda F, Sakuma Y, Sato H, Wild TF, Kai C. Establishment of a Nipah virus rescue system. Proc Natl Acad Sci USA. 2006;103:16508-16513. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 86] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 31. | Takeda M, Takeuchi K, Miyajima N, Kobune F, Ami Y, Nagata N, Suzaki Y, Nagai Y, Tashiro M. Recovery of pathogenic measles virus from cloned cDNA. J Virol. 2000;74:6643-6647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 142] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 32. | Leyrer S, Neubert WJ, Sedlmeier R. Rapid and efficient recovery of Sendai virus from cDNA: factors influencing recombinant virus rescue. J Virol Methods. 1998;75:47-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 29] [Cited by in RCA: 31] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 33. | Neumann G, Feldmann H, Watanabe S, Lukashevich I, Kawaoka Y. Reverse genetics demonstrates that proteolytic processing of the Ebola virus glycoprotein is not essential for replication in cell culture. J Virol. 2002;76:406-410. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 169] [Cited by in RCA: 164] [Article Influence: 7.1] [Reference Citation Analysis (0)] |
| 34. | Albariño CG, Uebelhoer LS, Vincent JP, Khristova ML, Chakrabarti AK, McElroy A, Nichol ST, Towner JS. Development of a reverse genetics system to generate recombinant Marburg virus derived from a bat isolate. Virology. 2013;446:230-237. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 35. | Jiang Y, Liu H, Liu P, Kong X. Plasmids driven minigenome rescue system for Newcastle disease virus V4 strain. Mol Biol Rep. 2009;36:1909-1914. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 12] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 36. | Harty RN, Brown ME, Hayes FP, Wright NT, Schnell MJ. Vaccinia virus-free recovery of vesicular stomatitis virus. J Mol Microbiol Biotechnol. 2001;3:513-517. [PubMed] |
| 37. | Sánchez AB, de la Torre JC. Rescue of the prototypic Arenavirus LCMV entirely from plasmid. Virology. 2006;350:370-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 108] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 38. | Whelan SP, Barr JN, Wertz GW. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:61-119. [PubMed] |
| 39. | de Wit E, Spronken MI, Vervaet G, Rimmelzwaan GF, Osterhaus AD, Fouchier RA. A reverse-genetics system for Influenza A virus using T7 RNA polymerase. J Gen Virol. 2007;88:1281-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 40. | Collins PL, Mink MA, Stec DS. Rescue of synthetic analogs of respiratory syncytial virus genomic RNA and effect of truncations and mutations on the expression of a foreign reporter gene. Proc Natl Acad Sci USA. 1991;88:9663-9667. [PubMed] |
| 41. | Ghanem A, Kern A, Conzelmann KK. Significantly improved rescue of rabies virus from cDNA plasmids. Eur J Cell Biol. 2012;91:10-16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 56] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 42. | Fuerst TR, Niles EG, Studier FW, Moss B. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc Natl Acad Sci USA. 1986;83:8122-8126. [PubMed] |
| 43. | Ito N, Takayama-Ito M, Yamada K, Hosokawa J, Sugiyama M, Minamoto N. Improved recovery of rabies virus from cloned cDNA using a vaccinia virus-free reverse genetics system. Microbiol Immunol. 2003;47:613-617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 169] [Article Influence: 14.1] [Reference Citation Analysis (0)] |
| 44. | Wyatt LS, Moss B, Rozenblatt S. Replication-deficient vaccinia virus encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 233] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 45. | Freiberg A, Dolores LK, Enterlein S, Flick R. Establishment and characterization of plasmid-driven minigenome rescue systems for Nipah virus: RNA polymerase I- and T7-catalyzed generation of functional paramyxoviral RNA. Virology. 2008;370:33-44. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 37] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 46. | Radecke F, Spielhofer P, Schneider H, Kaelin K, Huber M, Dötsch C, Christiansen G, Billeter MA. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773-5784. [PubMed] |
| 47. | Lowen AC, Noonan C, McLees A, Elliott RM. Efficient bunyavirus rescue from cloned cDNA. Virology. 2004;330:493-500. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 81] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 48. | Tong S, Li Y, Rivailler P, Conrardy C, Castillo DA, Chen LM, Recuenco S, Ellison JA, Davis CT, York IA. A distinct lineage of influenza A virus from bats. Proc Natl Acad Sci USA. 2012;109:4269-4274. [PubMed] |
| 49. | Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev. 1992;56:152-179. [PubMed] |
| 50. | Ibricevic A, Pekosz A, Walter MJ, Newby C, Battaile JT, Brown EG, Holtzman MJ, Brody SL. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J Virol. 2006;80:7469-7480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 274] [Cited by in RCA: 319] [Article Influence: 16.8] [Reference Citation Analysis (0)] |
| 51. | Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza a virus. J Virol. 2006;80:8060-8068. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 204] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 52. | Nicholls JM, Chan MC, Chan WY, Wong HK, Cheung CY, Kwong DL, Wong MP, Chui WH, Poon LL, Tsao SW. Tropism of avian influenza A (H5N1) in the upper and lower respiratory tract. Nat Med. 2007;13:147-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 257] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 53. | Kuiken T, Taubenberger JK. Pathology of human influenza revisited. Vaccine. 2008;26 Suppl 4:D59-D66. [PubMed] |
| 54. | Neumann G, Kawaoka Y. Reverse genetics of influenza virus. Virology. 2001;287:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 55. | Pleschka S, Jaskunas R, Engelhardt OG, Zürcher T, Palese P, García-Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188-4192. [PubMed] |
| 56. | Hoffmann E, Zhou N, Webster RG. Eight-plasmid rescue system for Influenza A virus. Int Congr. 2001;1219:1007-1013. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 57. | Neumann G, Fujii K, Kino Y, Kawaoka Y. An improved reverse genetics system for influenza A virus generation and its implications for vaccine production. Proc Natl Acad Sci USA. 2005;102:16825-16829. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 129] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 58. | Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem. 1987;56:365-394. [PubMed] |
| 59. | Han X, Bushweller JH, Cafiso DS, Tamm LK. Membrane structure and fusion-triggering conformational change of the fusion domain from influenza hemagglutinin. Nat Struct Biol. 2001;8:715-720. [PubMed] |
| 60. | Bertram S, Glowacka I, Steffen I, Kühl A, Pöhlmann S. Novel insights into proteolytic cleavage of influenza virus hemagglutinin. Rev Med Virol. 2010;20:298-310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 121] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 61. | Garten W, Bosch FX, Linder D, Rott R, Klenk HD. Proteolytic activation of the influenza virus hemagglutinin: The structure of the cleavage site and the enzymes involved in cleavage. Virology. 1981;115:361-374. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 127] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 62. | Böttcher E, Matrosovich T, Beyerle M, Klenk HD, Garten W, Matrosovich M. Proteolytic activation of influenza viruses by serine proteases TMPRSS2 and HAT from human airway epithelium. J Virol. 2006;80:9896-9898. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 351] [Cited by in RCA: 389] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 63. | Zhirnov OP, Ikizler MR, Wright PF. Cleavage of influenza a virus hemagglutinin in human respiratory epithelium is cell associated and sensitive to exogenous antiproteases. J Virol. 2002;76:8682-8689. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 66] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 64. | Horimoto T, Rivera E, Pearson J, Senne D, Krauss S, Kawaoka Y, Webster RG. Origin and molecular changes associated with emergence of a highly pathogenic H5N2 influenza virus in Mexico. Virology. 1995;213:223-230. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 172] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 65. | Senne DA, Panigrahy B, Kawaoka Y, Pearson JE, Süss J, Lipkind M, Kida H, Webster RG. Survey of the hemagglutinin (HA) cleavage site sequence of H5 and H7 avian influenza viruses: amino acid sequence at the HA cleavage site as a marker of pathogenicity potential. Avian Dis. 1996;40:425-437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 259] [Cited by in RCA: 252] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 66. | Horimoto T, Kawaoka Y. Reverse genetics provides direct evidence for a correlation of hemagglutinin cleavability and virulence of an avian influenza A virus. J Virol. 1994;68:3120-3128. [PubMed] |
| 67. | Hatta M, Gao P, Halfmann P, Kawaoka Y. Molecular basis for high virulence of Hong Kong H5N1 influenza A viruses. Science. 2001;293:1840-1842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1046] [Cited by in RCA: 1075] [Article Influence: 44.8] [Reference Citation Analysis (0)] |
| 68. | Schrauwen EJ, Herfst S, Leijten LM, van Run P, Bestebroer TM, Linster M, Bodewes R, Kreijtz JH, Rimmelzwaan GF, Osterhaus AD. The multibasic cleavage site in H5N1 virus is critical for systemic spread along the olfactory and hematogenous routes in ferrets. J Virol. 2012;86:3975-3984. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 122] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 69. | Suguitan AL, Matsuoka Y, Lau YF, Santos CP, Vogel L, Cheng LI, Orandle M, Subbarao K. The multibasic cleavage site of the hemagglutinin of highly pathogenic A/Vietnam/1203/2004 (H5N1) avian influenza virus acts as a virulence factor in a host-specific manner in mammals. J Virol. 2012;86:2706-2714. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 73] [Cited by in RCA: 77] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 70. | Gao R, Dong L, Dong J, Wen L, Zhang Y, Yu H, Feng Z, Chen M, Tan Y, Mo Z. A systematic molecular pathology study of a laboratory confirmed H5N1 human case. PLoS One. 2010;5:e13315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 71. | de Jong MD, Simmons CP, Thanh TT, Hien VM, Smith GJ, Chau TN, Hoang DM, Chau NV, Khanh TH, Dong VC. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat Med. 2006;12:1203-1207. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1545] [Cited by in RCA: 1492] [Article Influence: 78.5] [Reference Citation Analysis (0)] |
| 72. | Böttcher-Friebertshäuser E, Lu Y, Meyer D, Sielaff F, Steinmetzer T, Klenk HD, Garten W. Hemagglutinin activating host cell proteases provide promising drug targets for the treatment of influenza A and B virus infections. Vaccine. 2012;30:7374-7380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 73. | Feldmann H, Jones S, Klenk HD, Schnittler HJ. Ebola virus: from discovery to vaccine. Nat Rev Immunol. 2003;3:677-685. [PubMed] |
| 74. | Leroy EM, Epelboin A, Mondonge V, Pourrut X, Gonzalez JP, Muyembe-Tamfum JJ, Formenty P. Human Ebola outbreak resulting from direct exposure to fruit bats in Luebo, Democratic Republic of Congo, 2007. Vector Borne Zoonotic Dis. 2009;9:723-728. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 344] [Cited by in RCA: 338] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 75. | Leroy EM, Kumulungui B, Pourrut X, Rouquet P, Hassanin A, Yaba P, Délicat A, Paweska JT, Gonzalez JP, Swanepoel R. Fruit bats as reservoirs of Ebola virus. Nature. 2005;438:575-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1051] [Cited by in RCA: 1020] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 76. | Brown DW. Threat to Humans from Virus Infections of Non-human Primates. Rev Med Virol. 1997;7:239-246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 77. | Baron RC, McCormick JB, Zubeir OA. Ebola virus disease in southern Sudan: hospital dissemination and intrafamilial spread. Bull World Health Organ. 1983;61:997-1003. [PubMed] |
| 78. | Kinsman J. “A time of fear”: local, national, and international responses to a large Ebola outbreak in Uganda. Global Health. 2012;8:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 87] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 79. | MacNeil A, Farnon EC, Wamala J, Okware S, Cannon DL, Reed Z, Towner JS, Tappero JW, Lutwama J, Downing R. Proportion of deaths and clinical features in Bundibugyo Ebola virus infection, Uganda. Emerg Infect Dis. 2010;16:1969-1972. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 96] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 80. | Mahanty S, Bray M. Pathogenesis of filoviral haemorrhagic fevers. Lancet Infect Dis. 2004;4:487-498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 206] [Cited by in RCA: 198] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 81. | Towner JS, Sealy TK, Khristova ML, Albariño CG, Conlan S, Reeder SA, Quan PL, Lipkin WI, Downing R, Tappero JW. Newly discovered ebola virus associated with hemorrhagic fever outbreak in Uganda. PLoS Pathog. 2008;4:e1000212. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 389] [Cited by in RCA: 371] [Article Influence: 21.8] [Reference Citation Analysis (0)] |
| 82. | Filovirus infections among persons with occupational exposure to nonhuman primates. Update. Wkly Epidemiol Rec. 1990;65:185-186. [PubMed] |
| 83. | Miranda ME, Miranda NL. Reston ebolavirus in humans and animals in the Philippines: a review. J Infect Dis. 2011;204 Suppl 3:S757-S760. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 102] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 84. | Theriault S, Groseth A, Artsob H, Feldmann H. The role of reverse genetics systems in determining filovirus pathogenicity. Arch Virol Suppl. 2005;157-177. [PubMed] |
| 85. | Takada A, Robison C, Goto H, Sanchez A, Murti KG, Whitt MA, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764-14769. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 86. | Göringer H. RNA editing. Heidelberg: Springer 2008; . [DOI] [Full Text] |
| 87. | Volchkov VE, Volchkova VA, Muhlberger E, Kolesnikova LV, Weik M, Dolnik O, Klenk HD. Recovery of infectious Ebola virus from complementary DNA: RNA editing of the GP gene and viral cytotoxicity. Science. 2001;291:1965-1969. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 230] [Cited by in RCA: 226] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 88. | Feldmann H, Volchkov VE, Volchkova VA, Ströher U, Klenk HD. Biosynthesis and role of filoviral glycoproteins. J Gen Virol. 2001;82:2839-2848. [PubMed] |
| 89. | Yang ZY, Duckers HJ, Sullivan NJ, Sanchez A, Nabel EG, Nabel GJ. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 2000;6:886-889. [PubMed] |
| 90. | Groseth A, Marzi A, Hoenen T, Herwig A, Gardner D, Becker S, Ebihara H, Feldmann H. The Ebola virus glycoprotein contributes to but is not sufficient for virulence in vivo. PLoS Pathog. 2012;8:e1002847. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 79] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 91. | Ebihara H, Takada A, Kobasa D, Jones S, Neumann G, Theriault S, Bray M, Feldmann H, Kawaoka Y. Molecular determinants of Ebola virus virulence in mice. PLoS Pathog. 2006;2:e73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 184] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 92. | Hartman AL, Dover JE, Towner JS, Nichol ST. Reverse genetic generation of recombinant Zaire Ebola viruses containing disrupted IRF-3 inhibitory domains results in attenuated virus growth in vitro and higher levels of IRF-3 activation without inhibiting viral transcription or replication. J Virol. 2006;80:6430-6440. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 80] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 93. | Hartman AL, Ling L, Nichol ST, Hibberd ML. Whole-genome expression profiling reveals that inhibition of host innate immune response pathways by Ebola virus can be reversed by a single amino acid change in the VP35 protein. J Virol. 2008;82:5348-5358. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 94. | Blackwood JC, Streicker DG, Altizer S, Rohani P. Resolving the roles of immunity, pathogenesis, and immigration for rabies persistence in vampire bats. Proc Natl Acad Sci USA. 2013;110:20837-20842. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 116] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 95. | Real LA, Henderson JC, Biek R, Snaman J, Jack TL, Childs JE, Stahl E, Waller L, Tinline R, Nadin-Davis S. Unifying the spatial population dynamics and molecular evolution of epidemic rabies virus. Proc Natl Acad Sci USA. 2005;102:12107-12111. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 71] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 96. | Knobel DL, Cleaveland S, Coleman PG, Fèvre EM, Meltzer MI, Miranda ME, Shaw A, Zinsstag J, Meslin FX. Re-evaluating the burden of rabies in Africa and Asia. Bull World Health Organ. 2005;83:360-368. [PubMed] |
| 97. | Chomel BB, Belotto A, Meslin FX. Wildlife, exotic pets, and emerging zoonoses. Emerg Infect Dis. 2007;13:6-11. [PubMed] |
| 98. | Gautret P, Blanton J, Dacheux L, Ribadeau-Dumas F, Brouqui P, Parola P, Esposito DH, Bourhy H. Rabies in nonhuman primates and potential for transmission to humans: a literature review and examination of selected French national data. PLoS Negl Trop Dis. 2014;8:e2863. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 24] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 99. | Piyaphanee W, Shantavasinkul P, Phumratanaprapin W, Udomchaisakul P, Wichianprasat P, Benjavongkulchai M, Ponam T, Tantawichian T. Rabies exposure risk among foreign backpackers in Southeast Asia. Am J Trop Med Hyg. 2010;82:1168-1171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 100. | Mazarakis ND, Azzouz M, Rohll JB, Ellard FM, Wilkes FJ, Olsen AL, Carter EE, Barber RD, Baban DF, Kingsman SM. Rabies virus glycoprotein pseudotyping of lentiviral vectors enables retrograde axonal transport and access to the nervous system after peripheral delivery. Hum Mol Genet. 2001;10:2109-2121. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 326] [Cited by in RCA: 313] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 101. | Pulmanausahakul R, Li J, Schnell MJ, Dietzschold B. The glycoprotein and the matrix protein of rabies virus affect pathogenicity by regulating viral replication and facilitating cell-to-cell spread. J Virol. 2008;82:2330-2338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 70] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 102. | Wiltzer L, Okada K, Yamaoka S, Larrous F, Kuusisto HV, Sugiyama M, Blondel D, Bourhy H, Jans DA, Ito N. Interaction of rabies virus P-protein with STAT proteins is critical to lethal rabies disease. J Infect Dis. 2014;209:1744-1753. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 61] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 103. | Manning SE, Rupprecht CE, Fishbein D, Hanlon CA, Lumlertdacha B, Guerra M, Meltzer MI, Dhankhar P, Vaidya SA, Jenkins SR. Human rabies prevention--United States, 2008: recommendations of the Advisory Committee on Immunization Practices. MMWR Recomm Rep. 2008;57:1-28. [PubMed] |
| 104. | WHO. Rabies vaccines: WHO position paper--recommendations. Vaccine. 2010;28:7140-7142. [PubMed] |
| 105. | Faber M, Pulmanausahakul R, Hodawadekar SS, Spitsin S, McGettigan JP, Schnell MJ, Dietzschold B. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J Virol. 2002;76:3374-3381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 158] [Cited by in RCA: 156] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 106. | Cenna J, Hunter M, Tan GS, Papaneri AB, Ribka EP, Schnell MJ, Marx PA, McGettigan JP. Replication-deficient rabies virus-based vaccines are safe and immunogenic in mice and nonhuman primates. J Infect Dis. 2009;200:1251-1260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 44] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 107. | Blaney JE, Wirblich C, Papaneri AB, Johnson RF, Myers CJ, Juelich TL, Holbrook MR, Freiberg AN, Bernbaum JG, Jahrling PB. Inactivated or live-attenuated bivalent vaccines that confer protection against rabies and Ebola viruses. J Virol. 2011;85:10605-10616. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 70] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 108. | Halpin K, Young PL, Field HE, Mackenzie JS. Isolation of Hendra virus from pteropid bats: a natural reservoir of Hendra virus. J Gen Virol. 2000;81:1927-1932. [PubMed] |
| 109. | Dhondt KP, Horvat B. Hendra and Nipah viruses. Mononegaviruses of veterinary importance. Volume 1, Pathobiology and molecular diagnosis. Oxfordshire, UK: CAB International 2013; 38. |
| 110. | Chua KB. Nipah virus outbreak in Malaysia. J Clin Virol. 2003;26:265-275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 277] [Article Influence: 12.6] [Reference Citation Analysis (0)] |
| 111. | Rahman MA, Hossain MJ, Sultana S, Homaira N, Khan SU, Rahman M, Gurley ES, Rollin PE, Lo MK, Comer JA. Date palm sap linked to Nipah virus outbreak in Bangladesh, 2008. Vector Borne Zoonotic Dis. 2012;12:65-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 136] [Cited by in RCA: 152] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 112. | Sazzad HM, Hossain MJ, Gurley ES, Ameen KM, Parveen S, Islam MS, Faruque LI, Podder G, Banu SS, Lo MK. Nipah virus infection outbreak with nosocomial and corpse-to-human transmission, Bangladesh. Emerg Infect Dis. 2013;19:210-217. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 104] [Article Influence: 8.7] [Reference Citation Analysis (1)] |
| 113. | Wong KT, Shieh WJ, Kumar S, Norain K, Abdullah W, Guarner J, Goldsmith CS, Chua KB, Lam SK, Tan CT. Nipah virus infection: pathology and pathogenesis of an emerging paramyxoviral zoonosis. Am J Pathol. 2002;161:2153-2167. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 266] [Cited by in RCA: 287] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 114. | Eaton BT, Broder CC, Middleton D, Wang LF. Hendra and Nipah viruses: different and dangerous. Nat Rev Microbiol. 2006;4:23-35. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 312] [Cited by in RCA: 316] [Article Influence: 16.6] [Reference Citation Analysis (0)] |
| 115. | O’Sullivan JD, Allworth AM, Paterson DL, Snow TM, Boots R, Gleeson LJ, Gould AR, Hyatt AD, Bradfield J. Fatal encephalitis due to novel paramyxovirus transmitted from horses. Lancet. 1997;349:93-95. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 254] [Cited by in RCA: 227] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 116. | Murray K, Selleck P, Hooper P, Hyatt A, Gould A, Gleeson L, Westbury H, Hiley L, Selvey L, Rodwell B. A morbillivirus that caused fatal disease in horses and humans. Science. 1995;268:94-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 551] [Cited by in RCA: 528] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 117. | Rockx B, Winegar R, Freiberg AN. Recent progress in henipavirus research: molecular biology, genetic diversity, animal models. Antiviral Res. 2012;95:135-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 45] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 118. | Rota PA, Lo MK. Molecular virology of the henipaviruses. Curr Top Microbiol Immunol. 2012;359:41-58. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 14] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 119. | Kulkarni S, Volchkova V, Basler CF, Palese P, Volchkov VE, Shaw ML. Nipah virus edits its P gene at high frequency to express the V and W proteins. J Virol. 2009;83:3982-3987. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 73] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 120. | Childs KS, Andrejeva J, Randall RE, Goodbourn S. Mechanism of mda-5 Inhibition by paramyxovirus V proteins. J Virol. 2009;83:1465-1473. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 112] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 121. | Andrejeva J, Childs KS, Young DF, Carlos TS, Stock N, Goodbourn S, Randall RE. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc Natl Acad Sci USA. 2004;101:17264-17269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 788] [Cited by in RCA: 807] [Article Influence: 38.4] [Reference Citation Analysis (0)] |
| 122. | Shaw ML. Henipaviruses employ a multifaceted approach to evade the antiviral interferon response. Viruses. 2009;1:1190-1203. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 23] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 123. | Yoneda M, Guillaume V, Sato H, Fujita K, Georges-Courbot MC, Ikeda F, Omi M, Muto-Terao Y, Wild TF, Kai C. The nonstructural proteins of Nipah virus play a key role in pathogenicity in experimentally infected animals. PLoS One. 2010;5:e12709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 60] [Cited by in RCA: 78] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 124. | Mathieu C, Guillaume V, Volchkova VA, Pohl C, Jacquot F, Looi RY, Wong KT, Legras-Lachuer C, Volchkov VE, Lachuer J. Nonstructural Nipah virus C protein regulates both the early host proinflammatory response and viral virulence. J Virol. 2012;86:10766-10775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 53] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 125. | Lauring AS, Andino R. Quasispecies theory and the behavior of RNA viruses. PLoS Pathog. 2010;6:e1001005. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 698] [Cited by in RCA: 587] [Article Influence: 39.1] [Reference Citation Analysis (0)] |
| 126. | Gutiérrez RA, Viari A, Godelle B, Frutos R, Buchy P. Biased mutational pattern and quasispecies hypothesis in H5N1 virus. Infect Genet Evol. 2013;15:69-76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 10] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 127. | Biebricher CK, Eigen M. The error threshold. Virus Res. 2005;107:117-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 131] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 128. | Tsai L, Tasovski I, Leda AR, Chin MP, Cheng-Mayer C. The number and genetic relatedness of transmitted/founder virus impact clinical outcome in vaginal R5 SHIVSF162P3N infection. Retrovirology. 2014;11:22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 129. | Vignuzzi M, Stone JK, Arnold JJ, Cameron CE, Andino R. Quasispecies diversity determines pathogenesis through cooperative interactions in a viral population. Nature. 2006;439:344-348. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 954] [Cited by in RCA: 831] [Article Influence: 43.7] [Reference Citation Analysis (0)] |
| 130. | Gibson DG, Benders GA, Andrews-Pfannkoch C, Denisova EA, Baden-Tillson H, Zaveri J, Stockwell TB, Brownley A, Thomas DW, Algire MA. Complete chemical synthesis, assembly, and cloning of a Mycoplasma genitalium genome. Science. 2008;319:1215-1220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 809] [Article Influence: 47.6] [Reference Citation Analysis (0)] |
| 131. | Lauring AS, Andino R. Exploring the fitness landscape of an RNA virus by using a universal barcode microarray. J Virol. 2011;85:3780-3791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 34] [Article Influence: 2.4] [Reference Citation Analysis (0)] |