Published online Aug 25, 2012. doi: 10.5495/wjcid.v2.i4.63
Revised: June 8, 2012
Accepted: July 4, 2012
Published online: August 25, 2012
Staphylococcus aureus (S. aureus) is an important human pathogen capable of causing a diverse range of infections. Once regarded as an opportunistic pathogen causing primarily nosocomial infections, recent years have seen the emergence of S. aureus strains capable of causing serious infection even in otherwise healthy human hosts. There has been much debate about whether this transition is a function of unique genotypic characteristics or differences in the expression of conserved virulence factors, but irrespective of this debate it is clear that the ability of S. aureus to cause infection in all of its diverse forms is heavily influenced by its ability to modulate gene expression in response to changing conditions within the human host. Indeed, the S. aureus genome encodes more than 100 transcriptional regulators that modulate the production of virulence factors either directly via interactions with cis elements associated with genes encoding virulence factors or indirectly through their complex interactions with each other. The goal of this review is to summarize recent work describing these regulators and their contribution to defining S. aureus as a human pathogen.
-
Citation: Junecko JM, Zielinska AK, Mrak LN, Ryan DC, Graham JW, Smeltzer MS, Lee CY. Transcribing virulence in
Staphylococcus aureus . World J Clin Infect Dis 2012; 2(4): 63-76 - URL: https://www.wjgnet.com/2220-3176/full/v2/i4/63.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v2.i4.63
Staphylococcus aureus (S. aureus) is a rapidly evolving human pathogen that is a leading cause of both chronic, biofilm-associated infections and acute, life-threatening toxemias. Its ability to cause these infections is dependent on its ability to coordinate the production of a myriad of virulence factors. These include exopolysaccharides, surface-associated protein adhesins, immune modulators, and extracellular proteins including a plethora of toxins. S. aureus employs an equally remarkable array of regulatory elements to coordinate the production of these virulence factors. These elements include (1) small, non-coding RNAs[1]; (2) alternative sigma factors (σB, σH, and σs) responsive to various stress conditions[2]; and (3) trans-acting transcriptional regulators. It is the latter two groups that is the focus of this review. Indeed, one of the first reports of a full S. aureus genome sequence identified 124 open-reading frames likely to encode transcriptional regulators, 89 of which had not been previously identified[3]. Generally speaking, these are DNA-binding proteins, although some have also been shown to modulate virulence phenotypes via direct interactions with mRNA[4]. In general, these factors, some upon activation, are capable of binding a specific sequence associated with their target genes and thereby either enhance or inhibit transcription, although in many cases it has proven difficult to identify a definitive recognition site. Many of these targets are themselves regulatory factors, thus creating a complex network of virulence gene expression.
Because DNA-binding proteins are located in the cytoplasm, it is imperative to have a mechanism of sensing the external environment and translating that information into an intracellular change in gene expression. As with other pathogens, this is often accomplished via two-component systems. In some cases, the activating signals for these systems are known, but in most cases they have not been defined. S. aureus also produces many transcriptional regulators that are not associated with a recognized two-component system, and the activating signals for most of these also remain undefined. Nevertheless, an important role in virulence has been established for many of these regulators, and summarizing these is the focus of this review.
Bacterial two-component systems create a communication bridge to the external environment, allowing the cell to translate an external stimulus into an intracellular change in gene expression. The defining components are a membrane-associated sensor histidine kinase (HK) and a cytoplasmic response regulator (RR). The genes encoding these components are often arranged in an operon, and therefore co-transcribed, with other genes involved in the same signaling pathway. After activation from an external signal, the HK typically dimerizes and trans-autophosphorylates[5]. This leads to phosphorylation of the RR, characteristically at a conserved aspartic acid residue. Phosphorylation of the RR induces a conformational change allowing it to bind DNA at a specific consensus sequence in a manner that alters transcription of the target gene. Based on homology with recognized sensors and response regulators, S. aureus has at least 16 two-component systems[6]. In addition to their role in pathogenesis, at least three of these systems have been shown to modulate resistance to antibacterial agents[7], thus further emphasizing their important role in pathogenesis of S. aureus infection.
The most definitively characterized two-component system in S. aureus is the accessory gene regulator (agr), which was first identified as a transposon-insertion mutant with a reduced capacity to produce multiple exotoxins[8]. AgrC is the sensor kinase, and it is responsive to the accumulation of an extracellular auto-inducing peptide (AIP) that is encoded by agrD and processed for export by AgrB. Induction occurs in vitro as cultures enter the post-exponential growth phase as the AIP accumulates, thus making agr a prototype quorum-sensing regulatory system. This peptide recognition is a unique feature of Gram-positive quorum-sensing systems, as Gram-negative organisms sense small molecules, typically homoserine lactones[9], rather than peptides. While agr itself is highly conserved, variation in the AIP and its AgrC receptor define interference groups, with the AIP of each group inducing the expression upon interaction with its cognate receptor but inhibiting induction upon interaction with the receptor from each of the other groups[10].
After induction by AIP, AgrC autophosphorylates[11] and then phosphorylates AgrA, which is the cytoplasmic response regulator that until recently was thought to bind and activate only the agr-associated P2 and P3 promoters. Induction of the P2 promoter leads to increased transcription of the agr operon (agrABCD), resulting in a positive feedback loop, while induction of the P3 promoter results in increased transcription of RNAIII, with the latter being a primary downstream effector of the agr system[12].
AgrA was also recently shown to bind the promoter region of the gene clusters encoding phenol-soluble modulins (PSMs)[13]. PSMs are small toxins that lyse human neutrophils[14], a key host defense against staphylococcal infection. While PSMs are found in virtually all S. aureus isolates, the levels in which they are produced vary widely among different strains due to differences in the level of agr expression. This has been correlated with increased virulence in several animal models of S. aureus infection, although not necessarily owing to the increased production of PSMs alone[15-17].
RNAIII is the effector molecule of the agr regulatory system. Although RNAIII includes the gene encoding delta-toxin, which is itself a PSM[18], its primary contribution to virulence is regulatory. It is a stable RNA characterized by 14 stem-loop structures and two long helices separating two independent domains[19], and it is these stem-loops that are responsible for its regulatory effects[1]. RNAIII production is induced by the binding of phosphorylated AgrA to the P3 promoter, thus accounting for its increased production in vitro as cultures enter the post-exponential growth phase and AIP accumulates to a critical threshold. In general, induction results in reduced production of surface-associated proteins and enhanced production of exotoxins[20].
The phenotype of an RNAIII mutant is characterized by major changes at the transcriptional level. However, RNAIII itself functions primarily at a post-transcriptional level to affect accessory transcription factors leading to changes in virulence gene expression. For instance, transcription of the gene encoding staphylococcal protein A (spa) is increased in the absence of RNAIII, but this is due to the fact that RNAIII normally represses production of other transcription factors (e.g., SarT, Rot, and ultimately SarS) that would otherwise promote spa transcription[21]. Thus, in the absence of RNAIII, this repression does not occur, which results in the continued high level expression of spa. Additionally, RNAIII binds spa mRNA in a manner that both limits translation and promotes RNase III-mediated degradation[22].
This latter mechanism also plays a primary role in the RNAIII-mediated induction of toxin production[20]. This occurs via both direct and indirect pathways. For instance, in the case of hla, which encodes α toxin, the hla transcript forms a stem-loop structure that sequesters the Shine-Delgarno sequence, thus limiting translation. RNAIII overcomes this limitation by binding the hla transcript and relieving this stem-loop structure[23]. The translation of hla is thus upregulated in the presence of RNAIII via a direct interaction between RNAIII and hla mRNA. In other cases, the regulatory functions are mediated indirectly via the interaction between RNAIII and the rot transcript. Specifically, the regulatory functions of Rot (repressor of toxins) and Agr are antagonistic, with RNAIII limiting the production of Rot by binding to the Shine-Dalgarno sequence of the rot transcript and, as with the spa transcript, both inhibiting translation and targeting the existing transcript for degradation by RNaseIII[1]. In addition to its regulation of the genes encoding individual virulence factors, RNAIII also modulates the expression of other two-component systems including ArlRS, SaeRS and SrrAB, but the mechanism by which this occurs is not known[24-26].
Taken together, these results imply that agr plays a central role in S. aureus regulatory circuits. This is also reflected in the observation that mutation of agr has been consistently associated with a reduced capacity to cause infection[27]. Indeed, a primary determinant of the hypervirulence of isolates of the USA300 clonal lineage is their high level expression of agr and consequent high level production of critical exotoxins including α toxin and PSMs[27]. At the same time, this does not mean that agr expression is critical in all forms of S. aureus infection. One specific phenotype that may be particularly important in this regard is biofilm formation, with the high-level expression of agr generally being associated with a reduced capacity to form a biofilm[28-30]. It has been proposed that induction of agr expression may be important in promoting dispersal of S. aureus cells from an established biofilm, perhaps by inducing the production of extracellular proteases and/or nucleases[31]. This suggests that the expression of agr needs to be carefully controlled in the cells during biofilm development. However, several reports have documented the isolation of agr mutants directly from patients suffering from S. aureus infection[32,33], and it has even been suggested that agr dysfunction may be adaptive for survival within an infected host[34]. In fact, this is one specific context in which agr mutants have been shown to preferentially accumulate[34], perhaps owing to both the negative impact of agr on biofilm formation and the fact that its expression is metabolically expensive[35].
The saeRS two-component system was first identified as a transposon-insertion mutant deficient in exoprotein production[36]. saePQRS is transcribed as a 4-gene operon (saePQRS), with SaeS and SaeR being the sensor and response regulator respectively. A definitive role for SaeP and SaeQ has not yet been determined, although they may be involved in stabilization of SaeS in the membrane and/or modulating its return to the dephosphorylated state[37]. Once phosphorylated, SaeR binds to a specific target sequence (GTTAAN6GTTAA) to activate transcription of saePQRS itself[38]. This is very similar, although not identical, to the AT-rich consensus binding site identified by Nygaard et al[39] based on alignments with additional SaeR-regulated target genes[39].
Several studies have demonstrated that sae also modulates the production of virulence factors other than toxins including surface proteins and capsule biosynthesis components[38,40-43]. Several experimental observations suggest that saePQRS is downstream of agr, as well as other regulatory loci. Transcription of saePQRS is activated by agr but is repressed by SigB, while SaeRS does not seem to affect transcription of agr, sigB or sarA, suggesting that SaeRS acts as an important downstream regulator within the S. aureus global regulatory network[36,42,44,45]. Genetic experiments on exoprotein production also suggest that saePQRS is downstream of and epistatic to agr[42]. Furthermore, inactivation of either agr or sae had a comparable impact on the virulence of a USA300 isolate in a murine pneumonia model[46]. However, while inactivation of agr or sae results in reduced production of extracellular proteins, the exoprotein profiles of the two mutants are not identical[36,47]. It is also clear from several studies that the two regulons are not equivalent[39,41,43]. For instance, inactivation of sae results in decreased transcription of the fnbA and fnbB genes[39,41], both of which encode fibronectin-binding proteins, while inactivation of agr has the opposite effect[48]. Thus, while sae seems to function downstream of agr, it is also capable of regulating its target genes independent of agr.
One of the most commonly studied strains of S. aureus is Newman, which has a naturally occurring point mutation in saeS resulting in substitution of a leucine with a proline (L18P). This results in increased kinase activity leading to constitutive activation of SaeR and increased transcription of the saePQRS genes[49]. However, only certain target genes within the SaeRS regulon are differentially regulated in Newman due to the polymorphism of SaeS. Class I target genes are sensitive to the SaeSP allele and Class II genes are not. Although the mechanistic basis for this difference is not clear, it does not appear to be due to a gene dosage effect[41]. When SaeSL is cloned into wild-type Newman, it is dominant over SaeSP, suggesting instability of the system upon over-production, perhaps due to SaeS phosphatase rather than kinase activity[41].
Inactivation of sae is associated with increased transcription of several genes encoding extracellular proteases and increased accumulation of the corresponding proteases themselves[50], and this may well have an indirect effect on other virulence phenotypes of S. aureus. For instance, Newman is one of the few strains in which inactivation of the staphylococcal accessory regulator (sarA) does not result in an α toxin-deficient phenotype, and it was recently demonstrated that this is due to the hyperactivity of SaeSP leading to the reduced production of extracellular proteases, and consequent reduced degradation of the toxin, rather than transcriptional changes associated with hla[51].
The environmental cues modulating SaeRS activity have not been clearly defined but are associated with stress conditions including high salt, low pH, and subinhibitory concentrations of antibiotic[44]. Because SaeRS is induced by hydrogen peroxide and α-defensins, and because many toxins are SaeRS-regulated, it has been hypothesized that this system could promote escape from polymorphonuclear leukocytes after phagocytosis[44]. Indeed, it has been demonstrated that an saeRS mutant strain has an impaired ability to survive in human neutrophils after phagocytosis[43].
Fournier et al[52] used transposon mutagenesis to identify genes involved in the regulation of the multidrug efflux pump NorA and identified arlS, inactivation of which resulted in increased resistance to quinolones. ArlS is the sensor and ArlR is the response regulator of this two-component system. A subsequent study confirmed that ArlRS also modulates the production of exoproteins, but in this case the phenotype was opposite to that of an agr mutant, with an arlRS mutant exhibiting increased production of multiple exoproteins[52]. Additionally, an agr/arl double mutant exhibited an exoprotein phenotype comparable to the isogenic arl mutant, suggesting that arlRS is upstream rather than downstream of agr. In contrast, arlRS induces expression of sarA. To the extent that sarA is a major repressor of protease production, this is consistent with the observation that protease activity is increased in an arlRS mutant[53]. Whether these effects are direct or indirect remains unclear.
Together, these results suggest that arlRS may be a key regulatory element that defines the “balance” between agr and sarA. Both of these regulatory elements have been implicated in biofilm formation, and arlRS has also been shown to have an impact in this regard. Specifically, inactivation of arlRS results in increased autolysis and an enhanced capacity to form a biofilm[52]. The fact that the biofilm phenotype appears to be independent of any effect on production of the ica-encoded poly-N-acetylglucosamine (PNAG)[54], together with the demonstration that extracellular DNA released from lysed S. aureus cells contributes to biofilm formation[55], suggest that increased autolysis may be responsible for the biofilm phenotype. However, the biofilm formed by an arlRS mutant is sensitive to exogenous proteinases, suggesting that this biofilm is also at least partially dependent on protein-protein interactions[54]. This is consistent with the observation that inactivation of arlRS results in dramatically increased amounts of extracellular and surface-associated protein A[53], both of which have been shown to contribute to S. aureus biofilm formation[56]. Finally, arlRS has been shown to promote the production of additional virulence factors including the exfoliative toxin and capsular polysaccharides, the latter being an indirect effect mediated through its positive regulation of MgrA production[57-59].
Like arlRS, the lytSR two-component system is a negative regulator of S. aureus autolysis[60] and biofilm formation[61], and in fact arlRS is an activator of lytSR transcription[62]. These phenotypes are likely to be connected in that current models suggest that lytSR, together with CidR, collectively control the release of extracellular DNA (eDNA) by influencing expression of the lrgAB and cidABC operons, respectively, in modulating the production of murein hydrolases and consequently cell lysis[63]. Specifically, CidR activation of cid operon results in increased production of murein hydrolases, increased release of eDNA, and an increased capacity to form a biofilm, while activation of the lrgAB operon by LytSR has the opposite effects[55,64]. Although a cause-and-effect relationship between these phenotypes has not been proven, extracellular nuclease, whether applied exogenously or produced by S. aureus, has been shown to limit biofilm formation at least under certain in vitro conditions[29,64,65].
The srrAB two-component system was first identified based on homology with the ResDE two-component system in B. subtilis[66]. In response to oxygen stress, SrrAB represses expression of agr and the genes encoding certain exotoxins, including TSST-1[66]. However, it also represses transcription of spa and the production of protein A, which suggests that the impact of srrAB is not mediated directly through its regulation of agr but rather by direct interactions between the SrrA response regulator and the target genes themselves[67,68]. SrrAB also positively regulates expression of the icaADBC operon and production of PNAG, apparently by repressing transcription of the icaR-encoded repressor[67]. Whether the effect of SrrAB on the production of protein A or PNAG affects biofilm formation remains unknown, but the latter has been correlated with increased resistance to phagocytosis[47,69]. The link between oxygen availability, the activity of SrrAB, and the production of multiple types of virulence factors provides an important example of the link between central metabolic processes and virulence in S. aureus, a link that is also increasingly being made in the context of other S. aureus regulatory elements[70].
The hssRS two-component system is an iron-responsive system that is highly conserved among Gram-positive pathogens including B. anthracis, L. monocytogenes, S. epidermidis and E. faecalis, suggesting a conserved mechanism of iron acquisition among these organisms[71]. Iron is an essential nutrient for many bacterial species during infection[71,72]. However, free iron is severely limited in the human body but rather is complexed with a variety of iron-binding proteins. Therefore, in order for bacterial organisms to acquire iron they must have a mechanism for freeing complexed iron. In vivo, S. aureus can acquire iron in the form of heme, likely accessed via lysis of erythrocytes, using highly efficient transport systems that can move heme into the bacterial cytoplasm[73-75]. However, a high level of heme is toxic to the bacterial cell. To avoid toxicity, S. aureus senses heme by HssS resulting in HssR phosphorylation and binding to the promoter of hrtAB, which encodes an iron efflux pump that maintains intracellular heme homeostasis[76]. Whether there is cross-talk between this system and heme uptake systems, however, has not been demonstrated. In the absence of HrtAB, intracellular iron builds up causing a stress response characterized by the increased production of multiple virulence factors. Indeed, an hrtAB mutant is more virulent than the wild-type[71], likely due to the stress response induced by increasing intracellular heme.
The preceding discussion of two-component systems in S. aureus is by no means comprehensive, but it does summarize the impact of some of the best characterized systems. However, in the interest of inclusivity, we would note the existence of other, less well-characterized systems including KdpDE, which has been shown to link the AI-2/LuxS quorum-sensing system with capsule production[77]; VraSR, which induces a stress response to cell-wall inhibitors such as β-lactams and vancomycin[78]; GraSR, which aids in resistance to oxidative stress, heat stress, and vancomycin resistance[79]; BceAB, which is associated with altered susceptibility to bacitracin[7], and NsaRS, which plays a role in biofilm formation as well as cell envelope stability in response to cell wall and membrane disruption[80]. An additional two-component system that stands out from the others because it is the only one that is essential in S. aureus is WalKR (YycGF), which has been shown to be involved in peptidoglycan crosslinking and biofilm formation[81-83].
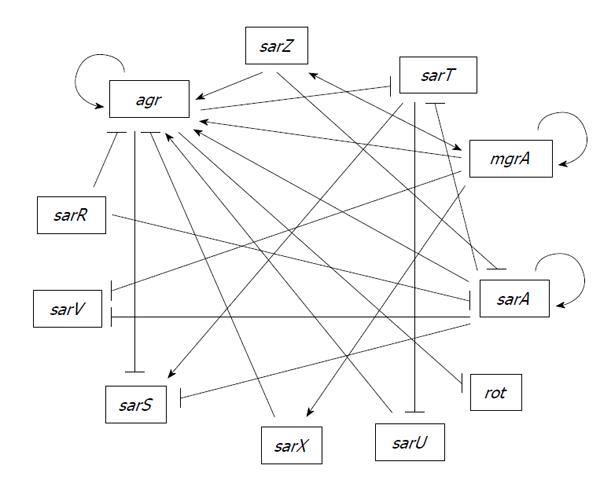
A primary class of transcriptional regulators that are not part of a two-component regulatory system, but do interact in multiple pathways with such systems, is the SarA family. The first gene encoding a member of this family, also identified in a screen of a transposon mutant library based on altered production of exotoxins, was designated the sar[84], which was subsequently changed to sarA based on identification of additional homologs now totaling 11[85]. Members of the SarA family have been shown to interact with each other forming part of a complex regulatory network controlling virulence factors (Figure 1). While all are winged-helix DNA-binding proteins, they can be divided into three structural families consisting of (1) relatively small, single-domain proteins (SarA, SarR, SarT, SarV, SarX and Rot); (2) larger, two-domain proteins (SarS, SarU, and SarY) in which each domain shares homology with the smaller homologs; and (3) small homologs with similarity to the MarR protein of Gram-negative bacteria (SarZ and MgrA)[85].
SarA, the prototype member of the SarA family, was identified in a screen of transposon-insertion mutants in the S. aureus strain DB based in part on its increased production of multiple exoproteins, a phenotype which clearly distinguished sarA from agr[84]. Based on this, it was proposed that sarA may function as a “counter-regulatory system to that of agr”. This is consistent with the hypothesis that many regulatory functions in S. aureus are defined by the “balance” between agr and sarA, an issue that is discussed in more detail below in the specific context of biofilm formation. However, subsequent studies also confirmed that SarA binds to intergenic region between the agr P2 and P3 promoters[86,87] and is required for maximal transcription of agr[88]. Thus, it has become clear that SarA modulates the production of S. aureus virulence factors via both agr-dependent and agr-independent pathways.
The sarA locus is complex and includes three promoters (P1, P2, and P3) that drive the production of three transcripts (sarB, sarC and sarA respectively), with all three sharing the same termination site just downstream of the gene encoding SarA. The upstream sarA P2 and P3 promoters modulate the production of SarA by an unknown mechanism[89]. Specifically, inactivation of P2 and P3 results in reduced production of SarA from the P1 promoter, and while the effect is relatively modest (approximately 2-fold), it appears to be functionally relevant with respect to both the agr-dependent and agr-independent pathways of SarA-mediated regulation[89]. This may account for the inability to demonstrate a difference between the three transcripts in complementation studies using a multi-copy vector targeting the cna promoter[90].
Both the signals that modulate SarA production and/or activity and the binding site for SarA are poorly defined. The DNA-binding activity of SarA is altered by redox state in vitro[91]. A recent report also demonstrated that SarA is phosphorylated by at least two serine/threonine kinases (Stk1/PknB and a poorly defined kinase encoded by SA0077) and that this also alters its DNA-binding capacity[92]. Two approaches have been taken to identifying the SarA binding site, with the first being alignment of the promoter regions of genes whose RNA products are altered in sarA mutants[93] and the second being the relatively unbiased approach of selective enhancement of systematic evolution of ligands by exponential enrichment (SELEX)[94], but generally speaking both approaches failed to define the characteristics of a definitive binding site beyond it being AT rich. It has been suggested on this basis that SarA may act as an architectural accessory protein rather than a classic transcription factor, a suggestion that is supported by the observation that SarA supports E. coli lambda phage integrase mediated site-specific recombination[91]. A recent report describing the interaction between SarA, SarR (see below) and cis elements within the agr promoter region also suggested that SarA binding may locally bend DNA in a fashion that brings AgrA into a favorable conformation to initiate transcription, particularly at the agr P2 promoter[87].
Irrespective of the mechanism involved, it is clear that inactivation of sarA results in major changes in the production and/or persistence of multiple RNA transcripts and that this has a global impact on the S. aureus virulon. Transcriptional profiling comparisons between the 8325-4 strain RN6390 and the clinical osteomyelitis isolate UAMS-1 demonstrate that this is somewhat strain-dependent, with the impact of mutating sarA in the latter being comparatively greater than the impact in the former[24]. RN6390 has defects in at least two genes that also have a major impact on global regulatory circuits. One of these is in rsbU, which results in reduced activity of the sigB regulon[47]. This is consistent with the observation that sigB has been shown to increase expression of sarA and reduce the level of RNAIII[95]. In this respect it is important to note that 8325-4 strains also have a mutation in tcaR, which results in reduced production of SarS[96], a SarA homolog also shown to influence the regulatory functions of SarA (see below).
As with agr, inactivation of sarA has been shown to attenuate virulence in multiple animal models of S. aureus infection including endopthalmitis, septic arthritis, osteomyelitis, and endocarditis[97-99]. Interestingly, all of these infections can arguably be said to be biofilm associated, an important observation given that inactivation of sarA has been consistently shown to result in a reduced capacity to form a biofilm[28]. In this respect it is also important to note that the impact of sarA on biofilm formation is opposite to that of agr[29], thus suggesting that the role of sarA in biofilm formation is independent of its regulation of agr. Indeed, in RN6390, a strain which expresses agr at high levels, inactivation of agr enhances biofilm formation in a manner that is reversed by concomitant inactivation of sarA, thus demonstrating that the impact of sarA is epistatic to agr in this context. Taken together, such results are consistent with the hypothesis that a primary determinant of the overall patterns of S. aureus virulence factor production is the “balance” between expression of agr and sarA[24], perhaps to the point of determining the relative capacity of different S. aureus strains to cause chronic, biofilm-associated infections vs acute, toxin-mediated disease.
There are several possible explanations for the biofilm-deficient phenotype of sarA mutants including the reduced expression of the icaADBC operon resulting in decreased production of PNAG, and the increased production of extracellular nucleases and proteases[29,31,64,65,100,101]. It seems unlikely that the decreased production of PNAG plays a predominant role given that inactivation of sarA has a greater impact on biofilm formation than inactivation of icaADBC[28]. Similarly, extracellular DNA has been shown to contribute to biofilm formation, but inactivation of the genes encoding S. aureus exonucleases has relatively little impact on the biofilm-deficient phenotype of sarA mutants[64,65]. In contrast, inactivation of the genes encoding extracellular proteases has a significant impact on the ability of sarA mutants to form a biofilm[24,29,102]. The effect of these proteases is presumably mediated via degradation of surface-associated proteins including FnbA, FnbB and protein A (Spa), all of which contribute to biofilm formation in S. aureus and have been shown to be produced in reduced amounts in sarA mutants owing to protease-mediated degradation[51,103,104].
Interestingly, the increased production of extracellular proteases has also been shown to result in the reduced accumulation of critical extracellular toxins in sarA mutants, at least under in vitro conditions[51]. These include α toxin and PSMs, both of which have been implicated as primary determinants of the hypervirulence of USA300 isolates[105]. Thus, while agr and sarA have opposite effects on the production of extracellular proteases, inactivation of either generally results in a toxin-deficient phenotype. This also suggests an alternative explanation for the reduced virulence of sarA mutants, although the relative contribution of these two sarA-dependent phenotypes in this respect remains to be determined.
Finally, the impact of SarA on exotoxin production has been shown to be heavily influenced by SaeRS, with the hyperactivity of SaeRS in Newman attenuating the increased production of extracellular proteases to a degree that impacts the α toxin and PSM phenotypes of a Newman sarA mutant[51]. This, together with the impact of SarA on expression of agr, provides direct indications of the interactive role of SarA in S. aureus regulatory circuits. Additional interactions involving other SarA homologs are described below.
SarR was discovered based on its affinity for the sarAP2 promoter, with the binding of SarR repressing sarA[106]. In contrast, SarA binds its own promoters to enhance transcription, thus providing an example in which SarA and SarR serve competitive roles in modulating gene transcription. These two proteins also serve the opposing roles with respect to Agr since binding of SarA, together with AgrA, promote transcription from the agr P2 promoter whereas binding of SarR has the opposite effect[87].It should be noted that SarR was originally shown to activate agr P2 promoter[107,108]. This discrepancy was attributed to the difference in sigB[87] but it is unclear how sigB reverses the effect. SarR promotes transcription of the proteases Aur and SspA, and may be involved in the SarA-dependent repression of these proteases, presumably also via competition for binding sites in the promoter regions[109]. In addition, SarR also binds to the rot promoter[107,108] but it is not known how SarR affects Rot. Thus, SarR plays an important but opposing role to SarA in both the agr-dependent and agr-independent pathways of SarA-mediated regulation. Based on these findings, SarR likely plays an important role in virulence due to its regulation of the well-characterized regulators SarA, Rot and Agr.
SarS (previously designated SarH1) was discovered using a search for proteins with affinity for the promoter region of agr-P3, spa, hla and ssp. However, SarS only affects expression of spa and hla but not RNAIII or ssp[110]. SarS is an activator of spa and a repressor of hla but it is repressed by Agr and SarA and activated by SigB and TcaR[96,111]. The fact that SarS is regulated by SigB and TcaR may explain why SarA affects hla transcription differently in RN6390, in which both SigB and TcaR are defective, than in other clinical isolates[111,112].
SarT was originally described by Schmidt et al[113] following a search for SarA homologs[113]. The sarT gene encodes a 118-residue protein and is present in certain strains of S. aureus including members of Clonal Complex 8 (CC8), to which CA-MRSA strains of the USA300 lineage and RN6390 belong, but absent in other clinically-relevant strains such as UAMS-1 (CC30 lineage)[113,114]. In the RN6390 background, sarT and agr are mutually repressive thus forming a negative feedback loop. In addition, sarT is also repressed by SarA[115]. Repression of agr by SarT was thought to explain the repression of hla by SarT in RN6390[115,116]. However, a later study shows that SarT represses hla via sae independently of agr and rot in strain COL[117]. A high level of agr in RN6390[118] may account for this difference. In RN6390, SarT also induces expression of protein A but indirectly through activation of sarS.
Adjacent to but divergently transcribed from sarT is sarU, whose expression is repressed by SarT[116]. Additionally, inactivation of sarU results in a reduction of both RNAII and RNAIII expression, suggesting a positive effect of SarU on agr. Because sarT has been shown to be repressed by agr, these relationships implicate a feedback loop involving SarT, SarU and RNAIII[116].
Recent studies have concluded that sarT and sarU are expressed at undetectable levels by northern blot[119]. However, deletion studies have revealed downstream effects of these genes, suggesting that they are expressed at very low but relevant levels[113,115,116]. For instance, it has been shown that a significant number of spontaneous non-hemolytic variants arise in biofilms that are phenotypically but not genotypically agr deficient. Transcriptional profiling of these variants found a 6-fold reduction in sarU suggesting SarU may be responsible for the agr deficiency[120]. These results imply that SarU may play a key role during biofilm-associated infections by modulating agr.
SarV was identified based on homology to SarA family. Both SarA and MgrA repress sarV gene expression. SarV is involved in regulation of autolysis, which may be part of the common pathway through which SarA and MgrA control autolysis[121]. Under laboratory conditions, sarV is poorly transcribed and the protein is not detectable in various strains in all phases of growth, likely due to repression by SarA and MgrA[119].
SarX was also identified based on sequence homology with the SarA family of transcription regulators. SarX has been shown to have maximal expression during the stationary phase of growth[122]. MgrA positively regulates sarX gene expression. SarX also acts as a repressor of the agr locus and can therefore regulate other genes via Agr[122]. SarX is highly expressed in RN6390 but is only expressed at very low levels in several tested strains[122] possibly due to difference in SigB in these strains. SarX has been shown to activate biofilm formation in S. epidermidis[123] raising the possibility that it may also have an effect on biofilm in S. aureus.
SarZ was originally identified as a regulator of hemolysins and promotes virulence in both silkworm and mouse infection models[124]. SarZ positively regulates agr and mgrA expression but negatively regulates expression of sarA. SarZ affects surface proteins, toxins and biofilm through modulating the aforementioned global regulators as well as direct activation on SspA protease[125]. However, SarZ binding to the promoters of various target genes is nonspecific[124,126]. In addition, SarZ activates SarS and is activated by MgrA[125]. SarZ and MgrA therefore interact in a positive feedback loop. SarZ expression is growth phase dependent, with maximum expression during early exponential phase[126]. Like MgrA, SarZ senses oxidative stress via a conserved cysteine residue providing another connection between metabolism and virulence[127,128].
MgrA, also referred to as Rat and NorR, was identified in three independent laboratories[129-131]. MgrA regulates a multitude of virulence factors as well as antibiotic resistance[130,131]. Truong-Bolduc et al[130,132] initially described MgrA (NorR) as a regulator of NorA, which is a multi-drug efflux pump providing quinolone resistance by direct DNA binding to the NorA promoter[130,132]. Binding of MgrA to the NorA promoter is dependent on phosphorylation by the kinase Stk1 (PknB), and RsbU is involved in dephosphorylation of MgrA leading to strain-dependent differences in MgrA function[133,134].
MgrA has been shown to up-regulate expression of 175 genes and down-regulate expression of 180 genes[59]. It was later shown that MgrA regulates hla and spa expression through agr-dependent and independent pathways[135]. In addition, MgrA has been found to repress biofilm formation, which is dependent on surface proteins, in part, through agr-dependent pathway and DNA release, probably by affecting LytSR and LrgAB[129,135,136].
MgrA has been shown to affect virulence in animal models of infection[59,137]. MgrA acts as a redox-switch as oxidation of the unique cysteine residue leads to its dissociation of MgrA from DNA[137]. Small molecules that disrupt the DNA-binding of MgrA had been shown to attenuate S. aureus virulence in mice[138] suggesting that MgrA could be a potential drug target.
Rot, repressor of toxins, is yet another SarA homolog[139]. It shares a high degree of sequence similarity to other members of the SarA family, but differs by its pI value (pI 5.0), which is more acidic than other SarA- homologs (pI values ranging from 8.5 to 10.7). Rot was first identified using transposon mutagenesis and screening for mutants capable of restoring the expression of α toxin and proteases in an agr-negative background[139]. Transcription of rot originates from at least three promoters and is growth-phase dependent[140,141]. Rot has an opposing effect on virulence gene expression by comparison to agr[139,142]. RNAIII blocks the translation of rot mRNA via an antisense mechanism, which explains why the regulatory function of Rot is only detected in agr deficient strains[20,143]. ClpX, a molecular chaperone, has also been shown to modulate Rot expression, likely by stimulating translation of the rot mRNA via a mechanism independent of RNAIII[144]. Rot has also been shown to repress α-toxin production by repressing the SaeRS two-component system[117]. In addition to toxins, Rot also positively regulates protein A[21,112,144,145].
The S. aureus genome contains 6 ORFs with homology to the AraC/XylS family of transcriptional regulators. Of these, two have been characterized and demonstrated to play a role in biofilm formation[146,147]. Rbf was first identified using transposon mutagenesis in a screening for biofilm-deficient mutants and demonstrated to control biofilm in response to NaCl and glucose[147]. It was later determined that Rbf promotes biofilm formation via repression of icaR, a repressor of the icaADBC operon whose gene products synthesize PNAG. However, Rbf is unable to bind specifically at the icaR promoter[148,149] suggesting other regulators or post-translational modification of Rbf may be involved. Rbf has been shown to promote virulence in a murine foreign-body infection model[150].
Rsp, another AraC family regulator, has recently been characterized and shown to regulate biofilm formation[146]. However, Rsp differs from Rbf by repressing biofilm by negatively regulating surface proteins; in particular, FnbA, which has been shown to promote biofilm formation in the cell accumulation phase[151]. Interestingly, Rsp inhibits biofilm through repression of FnbA at the stage of primary attachment by direct binding to the promoter of fnbA[146].
CodY functions as a highly conserved regulatory transcription factor in low-GC Gram-positive bacteria and has recently been identified as a regulator of several virulence factors in S. aureus[152-154]. CodY acts in response to metabolite effectors such as GTP and the branched-chain amino acids isoleucine, leucine, and valine[153], all of which interact with a branched-chain amino acid domain on CodY, facilitating the direct binding of CodY to several promoters associated with virulence genes[155]. It is thought that CodY primarily acts as a negative regulator of virulence genes in S. aureus[156]. Microarray analysis and DNase footprinting of codY mutant clinical isolates have recently identified several negatively regulated targets of CodY including agr, ica, and hla[153,155]. The agr locus is responsible for regulation of many virulence factors and thus, the repression of this locus by CodY has profound phenotypic effects on the expression of virulence genes in S. aureus. For example, capsular polysaccharide production is repressed by CodY through agr as well as by direct promoter binding[155].
Apart from direct regulation of virulence genes, CodY also affects metabolic regulation in S. aureus via amino acid synthesis, carbon flow, nitrogen assimilation, and transport systems[153]. CodY is activated in nutrient-replete environments, repressing virulence factors and metabolic synthesis genes. For example, CodY is associated with repression of a lactate dehydrogenase (ldh1), which interconverts pyruvate and lactate[157]. S. aureus strains lacking ldh1 show significant attenuation of virulence[157]. Thus, CodY is able to regulate virulence via direct binding of virulence gene promoters and via inhibition of metabolic regulatory pathways providing another regulatory link between metabolism and virulence[70,156].
CodY is repressed by the intracellular chaperone ClpC[40], possibly via ClpC-induced proteolytic degradation in association with ClpP. Although CodY acts as a repressor of virulence genes, it can also be negatively regulated under various environmental conditions, eliminating the repressive effect of CodY on virulence genes.
Sigma factors are highly conserved among bacterial species. They provide promoter specificity to the RNA polymerase, and are highly regulated by anti-sigma factors via direct binding of the protein[158]. There are currently four identified Sigma factors in S. aureus: SigA, which is responsible for transcription of housekeeping genes; SigB, which is responsible for the transcription of stress-response genes; SigS, which controls expression of genes required for overall fitness and survival[2]; and SigH, which has a demonstrated involvement in competence and more recently, prophage integration and excision[159,160].
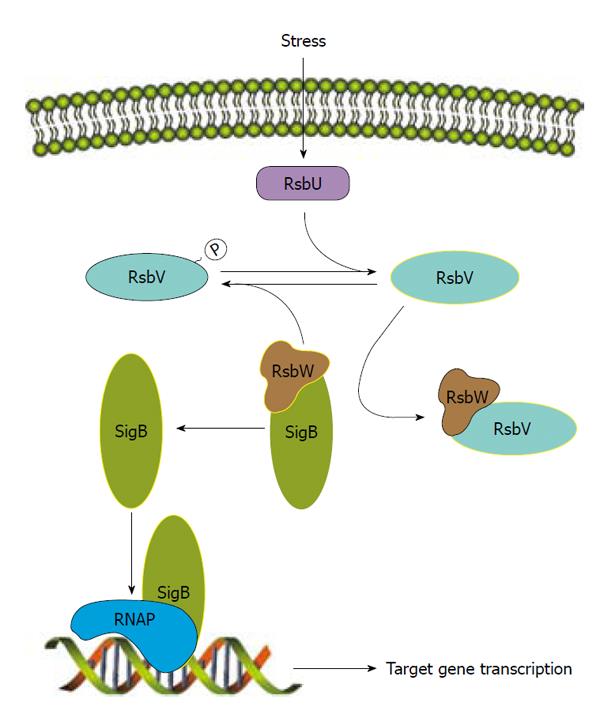
The most thoroughly studied of these is SigB, which is transcribed from the four-gene operon rsbUVWsigB that encodes an anti-sigma factor (RsbW), anti-anti-sigma factor (RsbV) and RsbU, an anti-RsbV phosphatase[95,161].The regulation of SigB is very tightly controlled by RsbW, RsbV and RsbU (Figure 2). SigB controls expression of an array of genes responsible for the survival of hydrogen peroxide-induced stress and desiccation as well as production of the carotenoid staphyloxanthin and extracellular proteases[161-164]. SigB has also been demonstrated to aid in heat tolerance and resistance to cell-wall active antibiotics[165,166]. The repressive effect of SigB on V8 proteases positively regulates biofilm formation[162] because the presence of extracellular proteases has been correlated with the inability to form a biofilm[164]. SigB regulates its target genes either by recognizing a conserved sequence or by downstream regulators. For example, SigB effect on sarA or agr expression has been reported[95,167]. More recently, SigB has been shown to regulate several extracellular virulence factors and capsule through SpoVG[168,169], demonstrating a role for SigB in virulence as a response to stress.
In this review, we describe several regulators involved in virulence regulation. These represent only a fraction of all regulators encoded in the S. aureus genome. S. aureus is a pathogen that can cause a wide range of diseases and can infect almost every tissue. It is thus not surprising that a large number of regulators are needed to modulate the production of various virulence factors in different environmental conditions in the host. What is surprising is the high degree of complexity of the interactions among the regulators. Compounding the complexity is the finding that virulence genes in different strains often are regulated differently. The molecular mechanisms underlying some of the strain differences have been illustrated but most have not. Nonetheless, significant progress has been made toward understanding virulence gene regulation. However, most of the results have been obtained by in vitro studies. The big challenge that lies ahead would be to test the in vitro results in suitable animal models to better understand virulence gene regulation in pathogenesis. With the rise of antibiotic resistance and the prevalence of multi-drug resistant isolates, fully understanding the virulence regulation in pathogenesis may provide sound rationale for identifying regulators as potential targets for anti-staphylococcal drug therapies. Targeting a cellular factor not absolutely required for survival, such as a virulence regulator, may lessen selective pressures, and therefore resistance, while still attenuating virulence of the organism[170,171].
Peer reviewer: Dr. Janne Kudsk Klitgaard, Institute of Clinical Microbiology, University of Southern Denmark, Campusvej 55, Odense M 5230, Denmark
S- Editor Wu X L- Editor A E- Editor Zheng XM
| 1. | Felden B, Vandenesch F, Bouloc P, Romby P. The Staphylococcus aureus RNome and its commitment to virulence. PLoS Pathog. 2011;7:e1002006. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 105] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 2. | Shaw LN, Lindholm C, Prajsnar TK, Miller HK, Brown MC, Golonka E, Stewart GC, Tarkowski A, Potempa J. Identification and characterization of sigma, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS One. 2008;3:e3844. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 50] [Cited by in RCA: 58] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 3. | Kuroda M, Ohta T, Uchiyama I, Baba T, Yuzawa H, Kobayashi I, Cui L, Oguchi A, Aoki K, Nagai Y. Whole genome sequencing of meticillin-resistant Staphylococcus aureus. Lancet. 2001;357:1225-1240. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1492] [Cited by in RCA: 1444] [Article Influence: 60.2] [Reference Citation Analysis (0)] |
| 4. | Morrison JM, Dunman PM. The modulation of Staphylococcus aureus mRNA turnover. Future Microbiol. 2011;6:1141-1150. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 5. | Dutta R, Qin L, Inouye M. Histidine kinases: diversity of domain organization. Mol Microbiol. 1999;34:633-640. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 190] [Cited by in RCA: 188] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 6. | Cheung AL, Bayer AS, Zhang G, Gresham H, Xiong YQ. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol Med Microbiol. 2004;40:1-9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 345] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 7. | Kawada-Matsuo M, Yoshida Y, Nakamura N, Komatsuzawa H. Role of two-component systems in the resistance of Staphylococcus aureus to antibacterial agents. Virulence. 2011;2:427-430. [PubMed] |
| 8. | Peng HL, Novick RP, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365-4372. [PubMed] |
| 9. | Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197-222. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1301] [Cited by in RCA: 1108] [Article Influence: 69.3] [Reference Citation Analysis (0)] |
| 10. | Geisinger E, George EA, Chen J, Muir TW, Novick RP. Identification of ligand specificity determinants in AgrC, the Staphylococcus aureus quorum-sensing receptor. J Biol Chem. 2008;283:8930-8938. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 74] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 11. | George Cisar EA, Geisinger E, Muir TW, Novick RP. Symmetric signalling within asymmetric dimers of the Staphylococcus aureus receptor histidine kinase AgrC. Mol Microbiol. 2009;74:44-57. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 12. | Novick RP. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol Microbiol. 2003;48:1429-1449. [PubMed] |
| 13. | Queck SY, Jameson-Lee M, Villaruz AE, Bach TH, Khan BA, Sturdevant DE, Ricklefs SM, Li M, Otto M. RNAIII-independent target gene control by the agr quorum-sensing system: insight into the evolution of virulence regulation in Staphylococcus aureus. Mol Cell. 2008;32:150-158. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 474] [Cited by in RCA: 444] [Article Influence: 26.1] [Reference Citation Analysis (0)] |
| 14. | Wang R, Braughton KR, Kretschmer D, Bach TH, Queck SY, Li M, Kennedy AD, Dorward DW, Klebanoff SJ, Peschel A. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat Med. 2007;13:1510-1514. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 763] [Cited by in RCA: 824] [Article Influence: 45.8] [Reference Citation Analysis (0)] |
| 15. | Li M, Diep BA, Villaruz AE, Braughton KR, Jiang X, DeLeo FR, Chambers HF, Lu Y, Otto M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc Natl Acad Sci U S A. 2009;106:5883-5888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 302] [Cited by in RCA: 319] [Article Influence: 19.9] [Reference Citation Analysis (0)] |
| 16. | Otto M. Staphylococcus aureus toxin gene hitchhikes on a transferable antibiotic resistance element. Virulence. 2010;1:49-51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 17. | Queck SY, Khan BA, Wang R, Bach TH, Kretschmer D, Chen L, Kreiswirth BN, Peschel A, Deleo FR, Otto M. Mobile genetic element-encoded cytolysin connects virulence to methicillin resistance in MRSA. PLoS Pathog. 2009;5:e1000533. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 152] [Cited by in RCA: 161] [Article Influence: 10.1] [Reference Citation Analysis (0)] |
| 18. | Novick RP, Geisinger E. Quorum sensing in staphylococci. Annu Rev Genet. 2008;42:541-564. [PubMed] |
| 19. | Benito Y, Kolb FA, Romby P, Lina G, Etienne J, Vandenesch F. Probing the structure of RNAIII, the Staphylococcus aureus agr regulatory RNA, and identification of the RNA domain involved in repression of protein A expression. RNA. 2000;6:668-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 134] [Cited by in RCA: 124] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 20. | Boisset S, Geissmann T, Huntzinger E, Fechter P, Bendridi N, Possedko M, Chevalier C, Helfer AC, Benito Y, Jacquier A. Staphylococcus aureus RNAIII coordinately represses the synthesis of virulence factors and the transcription regulator Rot by an antisense mechanism. Genes Dev. 2007;21:1353-1366. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 356] [Cited by in RCA: 362] [Article Influence: 20.1] [Reference Citation Analysis (0)] |
| 21. | Gao J, Stewart GC. Regulatory elements of the Staphylococcus aureus protein A (Spa) promoter. J Bacteriol. 2004;186:3738-3748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 65] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 22. | Huntzinger E, Boisset S, Saveanu C, Benito Y, Geissmann T, Namane A, Lina G, Etienne J, Ehresmann B, Ehresmann C. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824-835. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 267] [Cited by in RCA: 267] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 23. | Morfeldt E, Taylor D, von Gabain A, Arvidson S. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 1995;14:4569-4577. [PubMed] |
| 24. | Cassat J, Dunman PM, Murphy E, Projan SJ, Beenken KE, Palm KJ, Yang SJ, Rice KC, Bayles KW, Smeltzer MS. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology. 2006;152:3075-3090. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 121] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 25. | Dunman PM, Murphy E, Haney S, Palacios D, Tucker-Kellogg G, Wu S, Brown EL, Zagursky RJ, Shlaes D, Projan SJ. Transcription profiling-based identification of Staphylococcus aureus genes regulated by the agr and/or sarA loci. J Bacteriol. 2001;183:7341-7353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 26. | Jones RC, Deck J, Edmondson RD, Hart ME. Relative quantitative comparisons of the extracellular protein profiles of Staphylococcus aureus UAMS-1 and its sarA, agr, and sarA agr regulatory mutants using one-dimensional polyacrylamide gel electrophoresis and nanocapillary liquid chromatography coupled with tandem mass spectrometry. J Bacteriol. 2008;190:5265-5278. [PubMed] |
| 27. | Cheung GY, Wang R, Khan BA, Sturdevant DE, Otto M. Role of the accessory gene regulator agr in community-associated methicillin-resistant Staphylococcus aureus pathogenesis. Infect Immun. 2011;79:1927-1935. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 215] [Cited by in RCA: 252] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 28. | Beenken KE, Blevins JS, Smeltzer MS. Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun. 2003;71:4206-4211. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 312] [Cited by in RCA: 340] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 29. | Beenken KE, Mrak LN, Griffin LM, Zielinska AK, Shaw LN, Rice KC, Horswill AR, Bayles KW, Smeltzer MS. Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10790. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 129] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 30. | Vuong C, Saenz HL, Götz F, Otto M. Impact of the agr quorum-sensing system on adherence to polystyrene in Staphylococcus aureus. J Infect Dis. 2000;182:1688-1693. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 348] [Cited by in RCA: 355] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 31. | Boles BR, Thoendel M, Roth AJ, Horswill AR. Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS One. 2010;5:e10146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 260] [Cited by in RCA: 321] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 32. | Shopsin B, Drlica-Wagner A, Mathema B, Adhikari RP, Kreiswirth BN, Novick RP. Prevalence of agr dysfunction among colonizing Staphylococcus aureus strains. J Infect Dis. 2008;198:1171-1174. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 33. | Traber KE, Lee E, Benson S, Corrigan R, Cantera M, Shopsin B, Novick RP. agr function in clinical Staphylococcus aureus isolates. Microbiology. 2008;154:2265-2274. [PubMed] |
| 34. | Shopsin B, Eaton C, Wasserman GA, Mathema B, Adhikari RP, Agolory S, Altman DR, Holzman RS, Kreiswirth BN, Novick RP. Mutations in agr do not persist in natural populations of methicillin-resistant Staphylococcus aureus. J Infect Dis. 2010;202:1593-1599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 82] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 35. | Sakoulas G, Moise PA, Rybak MJ. Accessory gene regulator dysfunction: an advantage for Staphylococcus aureus in health-care settings? J Infect Dis. 2009;199:1558-1559. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 20] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 36. | Giraudo AT, Raspanti CG, Calzolari A, Nagel R. Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol. 1994;40:677-681. [PubMed] |
| 37. | Jeong DW, Cho H, Lee H, Li C, Garza J, Fried M, Bae T. Identification of the P3 promoter and distinct roles of the two promoters of the SaeRS two-component system in Staphylococcus aureus. J Bacteriol. 2011;193:4672-4684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 55] [Cited by in RCA: 67] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 38. | Sun F, Li C, Jeong D, Sohn C, He C, Bae T. In the Staphylococcus aureus two-component system sae, the response regulator SaeR binds to a direct repeat sequence and DNA binding requires phosphorylation by the sensor kinase SaeS. J Bacteriol. 2010;192:2111-2127. [PubMed] |
| 39. | Nygaard TK, Pallister KB, Ruzevich P, Griffith S, Vuong C, Voyich JM. SaeR binds a consensus sequence within virulence gene promoters to advance USA300 pathogenesis. J Infect Dis. 2010;201:241-254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 134] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 40. | Luong TT, Sau K, Roux C, Sau S, Dunman PM, Lee CY. Staphylococcus aureus ClpC divergently regulates capsule via sae and codY in strain newman but activates capsule via codY in strain UAMS-1 and in strain Newman with repaired saeS. J Bacteriol. 2011;193:686-694. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 50] [Cited by in RCA: 51] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 41. | Mainiero M, Goerke C, Geiger T, Gonser C, Herbert S, Wolz C. Differential target gene activation by the Staphylococcus aureus two-component system saeRS. J Bacteriol. 2010;192:613-623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 104] [Cited by in RCA: 123] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 42. | Novick RP, Jiang D. The staphylococcal saeRS system coordinates environmental signals with agr quorum sensing. Microbiology. 2003;149:2709-2717. [PubMed] |
| 43. | Voyich JM, Vuong C, DeWald M, Nygaard TK, Kocianova S, Griffith S, Jones J, Iverson C, Sturdevant DE, Braughton KR. The SaeR/S gene regulatory system is essential for innate immune evasion by Staphylococcus aureus. J Infect Dis. 2009;199:1698-1706. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 170] [Cited by in RCA: 160] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 44. | Geiger T, Goerke C, Mainiero M, Kraus D, Wolz C. The virulence regulator Sae of Staphylococcus aureus: promoter activities and response to phagocytosis-related signals. J Bacteriol. 2008;190:3419-3428. [PubMed] |
| 45. | Goerke C, Fluckiger U, Steinhuber A, Zimmerli W, Wolz C. Impact of the regulatory loci agr, sarA and sae of Staphylococcus aureus on the induction of alpha-toxin during device-related infection resolved by direct quantitative transcript analysis. Mol Microbiol. 2001;40:1439-1447. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 124] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 46. | Montgomery CP, Boyle-Vavra S, Daum RS. Importance of the global regulators Agr and SaeRS in the pathogenesis of CA-MRSA USA300 infection. PLoS One. 2010;5:e15177. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 154] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 47. | Herbert S, Ziebandt AK, Ohlsen K, Schäfer T, Hecker M, Albrecht D, Novick R, Götz F. Repair of global regulators in Staphylococcus aureus 8325 and comparative analysis with other clinical isolates. Infect Immun. 2010;78:2877-2889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 272] [Cited by in RCA: 308] [Article Influence: 20.5] [Reference Citation Analysis (0)] |
| 48. | Saravia-Otten P, Müller HP, Arvidson S. Transcription of Staphylococcus aureus fibronectin binding protein genes is negatively regulated by agr and an agr-independent mechanism. J Bacteriol. 1997;179:5259-5263. [PubMed] |
| 49. | Adhikari RP, Novick RP. Regulatory organization of the staphylococcal sae locus. Microbiology. 2008;154:949-959. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 79] [Cited by in RCA: 85] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 50. | Rogasch K, Rühmling V, Pané-Farré J, Höper D, Weinberg C, Fuchs S, Schmudde M, Bröker BM, Wolz C, Hecker M. Influence of the two-component system SaeRS on global gene expression in two different Staphylococcus aureus strains. J Bacteriol. 2006;188:7742-7758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 151] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 51. | Zielinska AK, Beenken KE, Joo HS, Mrak LN, Griffin LM, Luong TT, Lee CY, Otto M, Shaw LN, Smeltzer MS. Defining the strain-dependent impact of the Staphylococcal accessory regulator (sarA) on the alpha-toxin phenotype of Staphylococcus aureus. J Bacteriol. 2011;193:2948-2958. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 52. | Fournier B, Hooper DC. A new two-component regulatory system involved in adhesion, autolysis, and extracellular proteolytic activity of Staphylococcus aureus. J Bacteriol. 2000;182:3955-3964. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 193] [Cited by in RCA: 196] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 53. | Fournier B, Klier A, Rapoport G. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2001;41:247-261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 185] [Cited by in RCA: 186] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 54. | Toledo-Arana A, Merino N, Vergara-Irigaray M, Débarbouillé M, Penadés JR, Lasa I. Staphylococcus aureus develops an alternative, ica-independent biofilm in the absence of the arlRS two-component system. J Bacteriol. 2005;187:5318-5329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 146] [Cited by in RCA: 162] [Article Influence: 8.1] [Reference Citation Analysis (0)] |
| 55. | Rice KC, Mann EE, Endres JL, Weiss EC, Cassat JE, Smeltzer MS, Bayles KW. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113-8118. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 488] [Cited by in RCA: 534] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 56. | Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penadés JR, Lasa I. Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol. 2009;191:832-843. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 205] [Cited by in RCA: 248] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 57. | Fournier B, Klier A. Protein A gene expression is regulated by DNA supercoiling which is modified by the ArlS-ArlR two-component system of Staphylococcus aureus. Microbiology. 2004;150:3807-3819. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 58. | Kato F, Kadomoto N, Iwamoto Y, Bunai K, Komatsuzawa H, Sugai M. Regulatory mechanism for exfoliative toxin production in Staphylococcus aureus. Infect Immun. 2011;79:1660-1670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 32] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 59. | Luong TT, Lee CY. The arl locus positively regulates Staphylococcus aureus type 5 capsule via an mgrA-dependent pathway. Microbiology. 2006;152:3123-3131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 77] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 60. | Brunskill EW, Bayles KW. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol. 1996;178:611-618. [PubMed] |
| 61. | Sharma-Kuinkel BK, Mann EE, Ahn JS, Kuechenmeister LJ, Dunman PM, Bayles KW. The Staphylococcus aureus LytSR two-component regulatory system affects biofilm formation. J Bacteriol. 2009;191:4767-4775. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 110] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 62. | Liang X, Zheng L, Landwehr C, Lunsford D, Holmes D, Ji Y. Global regulation of gene expression by ArlRS, a two-component signal transduction regulatory system of Staphylococcus aureus. J Bacteriol. 2005;187:5486-5492. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 116] [Cited by in RCA: 118] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 63. | Rice KC, Bayles KW. Molecular control of bacterial death and lysis. Microbiol Mol Biol Rev. 2008;72:85-109, table of contents. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 291] [Cited by in RCA: 275] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 64. | Mann EE, Rice KC, Boles BR, Endres JL, Ranjit D, Chandramohan L, Tsang LH, Smeltzer MS, Horswill AR, Bayles KW. Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS One. 2009;4:e5822. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 335] [Cited by in RCA: 374] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 65. | Tsang LH, Cassat JE, Shaw LN, Beenken KE, Smeltzer MS. Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS One. 2008;3:e3361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 85] [Cited by in RCA: 103] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 66. | Yarwood JM, McCormick JK, Schlievert PM. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol. 2001;183:1113-1123. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 227] [Cited by in RCA: 229] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 67. | Pragman AA, Ji Y, Schlievert PM. Repression of Staphylococcus aureus SrrAB using inducible antisense srrA alters growth and virulence factor transcript levels. Biochemistry. 2007;46:314-321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 32] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 68. | Pragman AA, Yarwood JM, Tripp TJ, Schlievert PM. Characterization of virulence factor regulation by SrrAB, a two-component system in Staphylococcus aureus. J Bacteriol. 2004;186:2430-2438. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 133] [Cited by in RCA: 152] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 69. | Ulrich M, Bastian M, Cramton SE, Ziegler K, Pragman AA, Bragonzi A, Memmi G, Wolz C, Schlievert PM, Cheung A. The staphylococcal respiratory response regulator SrrAB induces ica gene transcription and polysaccharide intercellular adhesin expression, protecting Staphylococcus aureus from neutrophil killing under anaerobic growth conditions. Mol Microbiol. 2007;65:1276-1287. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 70. | Somerville GA, Proctor RA. At the crossroads of bacterial metabolism and virulence factor synthesis in Staphylococci. Microbiol Mol Biol Rev. 2009;73:233-248. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 297] [Article Influence: 18.6] [Reference Citation Analysis (0)] |
| 71. | Torres VJ, Stauff DL, Pishchany G, Bezbradica JS, Gordy LE, Iturregui J, Anderson KL, Dunman PM, Joyce S, Skaar EP. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe. 2007;1:109-119. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 193] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 72. | Bullen JJ, Griffiths E. Iron and infection: molecular, physiological and clinical aspects. New York: John Wiley and Sons 1999; . |
| 73. | Skaar EP, Humayun M, Bae T, DeBord KL, Schneewind O. Iron-source preference of Staphylococcus aureus infections. Science. 2004;305:1626-1628. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 315] [Cited by in RCA: 331] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 74. | Skaar EP, Schneewind O. Iron-regulated surface determinants (Isd) of Staphylococcus aureus: stealing iron from heme. Microbes Infect. 2004;6:390-397. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 167] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 75. | Torres VJ, Pishchany G, Humayun M, Schneewind O, Skaar EP. Staphylococcus aureus IsdB is a hemoglobin receptor required for heme iron utilization. J Bacteriol. 2006;188:8421-8429. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 227] [Article Influence: 11.9] [Reference Citation Analysis (0)] |
| 76. | Hammer ND, Skaar EP. Molecular mechanisms of Staphylococcus aureus iron acquisition. Annu Rev Microbiol. 2011;65:129-147. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 231] [Cited by in RCA: 268] [Article Influence: 20.6] [Reference Citation Analysis (0)] |
| 77. | Zhao L, Xue T, Shang F, Sun H, Sun B. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect Immun. 2010;78:3506-3515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 105] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 78. | Gardete S, Wu SW, Gill S, Tomasz A. Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother. 2006;50:3424-3434. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 147] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 79. | Falord M, Mäder U, Hiron A, Débarbouillé M, Msadek T. Investigation of the Staphylococcus aureus GraSR regulon reveals novel links to virulence, stress response and cell wall signal transduction pathways. PLoS One. 2011;6:e21323. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 122] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 80. | Kolar SL, Nagarajan V, Oszmiana A, Rivera FE, Miller HK, Davenport JE, Riordan JT, Potempa J, Barber DS, Koziel J. NsaRS is a cell-envelope-stress-sensing two-component system of Staphylococcus aureus. Microbiology. 2011;157:2206-2219. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 81] [Cited by in RCA: 78] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 81. | Delaune A, Poupel O, Mallet A, Coic YM, Msadek T, Dubrac S. Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS One. 2011;6:e17054. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 71] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 82. | Dubrac S, Boneca IG, Poupel O, Msadek T. New insights into the WalK/WalR (YycG/YycF) essential signal transduction pathway reveal a major role in controlling cell wall metabolism and biofilm formation in Staphylococcus aureus. J Bacteriol. 2007;189:8257-8269. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 237] [Cited by in RCA: 286] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 83. | Dubrac S, Msadek T. Identification of genes controlled by the essential YycG/YycF two-component system of Staphylococcus aureus. J Bacteriol. 2004;186:1175-1181. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 197] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 84. | Cheung AL, Koomey JM, Butler CA, Projan SJ, Fischetti VA. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci U S A. 1992;89:6462-6466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 295] [Cited by in RCA: 277] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 85. | Cheung AL, Nishina KA, Trotonda MP, Tamber S. The SarA protein family of Staphylococcus aureus. Int J Biochem Cell Biol. 2008;40:355-361. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 160] [Cited by in RCA: 161] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 86. | Rechtin TM, Gillaspy AF, Schumacher MA, Brennan RG, Smeltzer MS, Hurlburt BK. Characterization of the SarA virulence gene regulator of Staphylococcus aureus. Mol Microbiol. 1999;33:307-316. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 79] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 87. | Reyes D, Andrey DO, Monod A, Kelley WL, Zhang G, Cheung AL. Coordinated regulation by AgrA, SarA, and SarR to control agr expression in Staphylococcus aureus. J Bacteriol. 2011;193:6020-6031. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 98] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 88. | Cheung AL, Projan SJ. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168-4172. [PubMed] |
| 89. | Cheung AL, Manna AC. Role of the distal sarA promoters in SarA expression in Staphylococcus aureus. Infect Immun. 2005;73:4391-4394. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 90. | Blevins JS, Gillaspy AF, Rechtin TM, Hurlburt BK, Smeltzer MS. The Staphylococcal accessory regulator (sar) represses transcription of the Staphylococcus aureus collagen adhesin gene (cna) in an agr-independent manner. Mol Microbiol. 1999;33:317-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 77] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 91. | Fujimoto DF, Higginbotham RH, Sterba KM, Maleki SJ, Segall AM, Smeltzer MS, Hurlburt BK. Staphylococcus aureus SarA is a regulatory protein responsive to redox and pH that can support bacteriophage lambda integrase-mediated excision/recombination. Mol Microbiol. 2009;74:1445-1458. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 48] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 92. | Didier JP, Cozzone AJ, Duclos B. Phosphorylation of the virulence regulator SarA modulates its ability to bind DNA in Staphylococcus aureus. FEMS Microbiol Lett. 2010;306:30-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 42] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 93. | Chien Y, Manna AC, Projan SJ, Cheung AL. SarA, a global regulator of virulence determinants in Staphylococcus aureus, binds to a conserved motif essential for sar-dependent gene regulation. J Biol Chem. 1999;274:37169-37176. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 186] [Cited by in RCA: 188] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 94. | Sterba KM, Mackintosh SG, Blevins JS, Hurlburt BK, Smeltzer MS. Characterization of Staphylococcus aureus SarA binding sites. J Bacteriol. 2003;185:4410-4417. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 51] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 95. | Bischoff M, Entenza JM, Giachino P. Influence of a functional sigB operon on the global regulators sar and agr in Staphylococcus aureus. J Bacteriol. 2001;183:5171-5179. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 171] [Cited by in RCA: 177] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 96. | McCallum N, Bischoff M, Maki H, Wada A, Berger-Bächi B. TcaR, a putative MarR-like regulator of sarS expression. J Bacteriol. 2004;186:2966-2972. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 44] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 97. | Blevins JS, Elasri MO, Allmendinger SD, Beenken KE, Skinner RA, Thomas JR, Smeltzer MS. Role of sarA in the pathogenesis of Staphylococcus aureus musculoskeletal infection. Infect Immun. 2003;71:516-523. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 98. | Booth MC, Cheung AL, Hatter KL, Jett BD, Callegan MC, Gilmore MS. Staphylococcal accessory regulator (sar) in conjunction with agr contributes to Staphylococcus aureus virulence in endophthalmitis. Infect Immun. 1997;65:1550-1556. [PubMed] |
| 99. | Xiong YQ, Willard J, Yeaman MR, Cheung AL, Bayer AS. Regulation of Staphylococcus aureus alpha-toxin gene (hla) expression by agr, sarA, and sae in vitro and in experimental infective endocarditis. J Infect Dis. 2006;194:1267-1275. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 131] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 100. | Boles BR, Horswill AR. Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog. 2008;4:e1000052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 602] [Cited by in RCA: 667] [Article Influence: 39.2] [Reference Citation Analysis (0)] |
| 101. | O'Neill E, Pozzi C, Houston P, Humphreys H, Robinson DA, Loughman A, Foster TJ, O'Gara JP. A novel Staphylococcus aureus biofilm phenotype mediated by the fibronectin-binding proteins, FnBPA and FnBPB. J Bacteriol. 2008;190:3835-3850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 341] [Cited by in RCA: 387] [Article Influence: 22.8] [Reference Citation Analysis (0)] |
| 102. | Beenken KE, Dunman PM, McAleese F, Macapagal D, Murphy E, Projan SJ, Blevins JS, Smeltzer MS. Global gene expression in Staphylococcus aureus biofilms. J Bacteriol. 2004;186:4665-4684. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 435] [Cited by in RCA: 450] [Article Influence: 21.4] [Reference Citation Analysis (0)] |
| 103. | Karlsson A, Saravia-Otten P, Tegmark K, Morfeldt E, Arvidson S. Decreased amounts of cell wall-associated protein A and fibronectin-binding proteins in Staphylococcus aureus sarA mutants due to up-regulation of extracellular proteases. Infect Immun. 2001;69:4742-4748. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 115] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 104. | McGavin MJ, Zahradka C, Rice K, Scott JE. Modification of the Staphylococcus aureus fibronectin binding phenotype by V8 protease. Infect Immun. 1997;65:2621-2628. [PubMed] |
| 105. | Joo HS, Cheung GY, Otto M. Antimicrobial activity of community-associated methicillin-resistant Staphylococcus aureus is caused by phenol-soluble modulin derivatives. J Biol Chem. 2011;286:8933-8940. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 106. | Manna A, Cheung AL. Characterization of sarR, a modulator of sar expression in Staphylococcus aureus. Infect Immun. 2001;69:885-896. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 100] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 107. | Hsieh HY, Tseng CW, Stewart GC. Regulation of Rot expression in Staphylococcus aureus. J Bacteriol. 2008;190:546-554. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 35] [Cited by in RCA: 39] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 108. | Manna AC, Cheung AL. Transcriptional regulation of the agr locus and the identification of DNA binding residues of the global regulatory protein SarR in Staphylococcus aureus. Mol Microbiol. 2006;60:1289-1301. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 36] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 109. | Gustafsson E, Oscarsson J. Maximal transcription of aur (aureolysin) and sspA (serine protease) in Staphylococcus aureus requires staphylococcal accessory regulator R (sarR) activity. FEMS Microbiol Lett. 2008;284:158-164. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 110. | Tegmark K, Karlsson A, Arvidson S. Identification and characterization of SarH1, a new global regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol. 2000;37:398-409. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 128] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 111. | Cheung AL, Schmidt K, Bateman B, Manna AC. SarS, a SarA homolog repressible by agr, is an activator of protein A synthesis in Staphylococcus aureus. Infect Immun. 2001;69:2448-2455. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 112. | Oscarsson J, Kanth A, Tegmark-Wisell K, Arvidson S. SarA is a repressor of hla (alpha-hemolysin) transcription in Staphylococcus aureus: its apparent role as an activator of hla in the prototype strain NCTC 8325 depends on reduced expression of sarS. J Bacteriol. 2006;188:8526-8533. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 46] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 113. | Schmidt KA, Manna AC, Gill S, Cheung AL. SarT, a repressor of alpha-hemolysin in Staphylococcus aureus. Infect Immun. 2001;69:4749-4758. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 114. | Cassat JE, Dunman PM, McAleese F, Murphy E, Projan SJ, Smeltzer MS. Comparative genomics of Staphylococcus aureus musculoskeletal isolates. J Bacteriol. 2005;187:576-592. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 95] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 115. | Schmidt KA, Manna AC, Cheung AL. SarT influences sarS expression in Staphylococcus aureus. Infect Immun. 2003;71:5139-5148. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 116. | Manna AC, Cheung AL. sarU, a sarA homolog, is repressed by SarT and regulates virulence genes in Staphylococcus aureus. Infect Immun. 2003;71:343-353. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 82] [Cited by in RCA: 75] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 117. | Li D, Cheung A. Repression of hla by rot is dependent on sae in Staphylococcus aureus. Infect Immun. 2008;76:1068-1075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 57] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 118. | Blevins JS, Beenken KE, Elasri MO, Hurlburt BK, Smeltzer MS. Strain-dependent differences in the regulatory roles of sarA and agr in Staphylococcus aureus. Infect Immun. 2002;70:470-480. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 149] [Cited by in RCA: 161] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 119. | Ballal A, Manna AC. Expression of the sarA family of genes in different strains of Staphylococcus aureus. Microbiology. 2009;155:2342-2352. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 40] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 120. | Yarwood JM, Paquette KM, Tikh IB, Volper EM, Greenberg EP. Generation of virulence factor variants in Staphylococcus aureus biofilms. J Bacteriol. 2007;189:7961-7967. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 121. | Manna AC, Ingavale SS, Maloney M, van Wamel W, Cheung AL. Identification of sarV (SA2062), a new transcriptional regulator, is repressed by SarA and MgrA (SA0641) and involved in the regulation of autolysis in Staphylococcus aureus. J Bacteriol. 2004;186:5267-5280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 91] [Cited by in RCA: 102] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 122. | Manna AC, Cheung AL. Expression of SarX, a negative regulator of agr and exoprotein synthesis, is activated by MgrA in Staphylococcus aureus. J Bacteriol. 2006;188:4288-4299. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 54] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 123. | Rowe SE, Mahon V, Smith SG, O'Gara JP. A novel role for SarX in Staphylococcus epidermidis biofilm regulation. Microbiology. 2011;157:1042-1049. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 22] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 124. | Kaito C, Morishita D, Matsumoto Y, Kurokawa K, Sekimizu K. Novel DNA binding protein SarZ contributes to virulence in Staphylococcus aureus. Mol Microbiol. 2006;62:1601-1617. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 75] [Cited by in RCA: 82] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 125. | Tamber S, Cheung AL. SarZ promotes the expression of virulence factors and represses biofilm formation by modulating SarA and agr in Staphylococcus aureus. Infect Immun. 2009;77:419-428. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 126. | Ballal A, Ray B, Manna AC. sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus. J Bacteriol. 2009;191:1656-1665. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 127. | Poor CB, Chen PR, Duguid E, Rice PA, He C. Crystal structures of the reduced, sulfenic acid, and mixed disulfide forms of SarZ, a redox active global regulator in Staphylococcus aureus. J Biol Chem. 2009;284:23517-23524. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 78] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 128. | Chen PR, Nishida S, Poor CB, Cheng A, Bae T, Kuechenmeister L, Dunman PM, Missiakas D, He C. A new oxidative sensing and regulation pathway mediated by the MgrA homologue SarZ in Staphylococcus aureus. Mol Microbiol. 2009;71:198-211. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 114] [Cited by in RCA: 101] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 129. | Ingavale SS, Van Wamel W, Cheung AL. Characterization of RAT, an autolysis regulator in Staphylococcus aureus. Mol Microbiol. 2003;48:1451-1466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 109] [Cited by in RCA: 109] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 130. | Truong-Bolduc QC, Zhang X, Hooper DC. Characterization of NorR protein, a multifunctional regulator of norA expression in Staphylococcus aureus. J Bacteriol. 2003;185:3127-3138. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 99] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 131. | Luong TT, Newell SW, Lee CY. Mgr, a novel global regulator in Staphylococcus aureus. J Bacteriol. 2003;185:3703-3710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 130] [Cited by in RCA: 127] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 132. | Ng EY, Trucksis M, Hooper DC. Quinolone resistance mediated by norA: physiologic characterization and relationship to flqB, a quinolone resistance locus on the Staphylococcus aureus chromosome. Antimicrob Agents Chemother. 1994;38:1345-1355. [PubMed] |
| 133. | Truong-Bolduc QC, Ding Y, Hooper DC. Posttranslational modification influences the effects of MgrA on norA expression in Staphylococcus aureus. J Bacteriol. 2008;190:7375-7381. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 62] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 134. | Truong-Bolduc QC, Hooper DC. Phosphorylation of MgrA and its effect on expression of the NorA and NorB efflux pumps of Staphylococcus aureus. J Bacteriol. 2010;192:2525-2534. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 69] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 135. | Ingavale S, van Wamel W, Luong TT, Lee CY, Cheung AL. Rat/MgrA, a regulator of autolysis, is a regulator of virulence genes in Staphylococcus aureus. Infect Immun. 2005;73:1423-1431. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 125] [Cited by in RCA: 130] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 136. | Trotonda MP, Tamber S, Memmi G, Cheung AL. MgrA represses biofilm formation in Staphylococcus aureus. Infect Immun. 2008;76:5645-5654. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 78] [Article Influence: 4.6] [Reference Citation Analysis (0)] |
| 137. | Chen PR, Bae T, Williams WA, Duguid EM, Rice PA, Schneewind O, He C. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591-595. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 164] [Cited by in RCA: 169] [Article Influence: 8.9] [Reference Citation Analysis (0)] |
| 138. | Sun F, Zhou L, Zhao BC, Deng X, Cho H, Yi C, Jian X, Song CX, Luan CH, Bae T. Targeting MgrA-mediated virulence regulation in Staphylococcus aureus. Chem Biol. 2011;18:1032-1041. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 139. | McNamara PJ, Milligan-Monroe KC, Khalili S, Proctor RA. Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol. 2000;182:3197-3203. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 151] [Cited by in RCA: 146] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 140. | Jelsbak L, Hemmingsen L, Donat S, Ohlsen K, Boye K, Westh H, Ingmer H, Frees D. Growth phase-dependent regulation of the global virulence regulator Rot in clinical isolates of Staphylococcus aureus. Int J Med Microbiol. 2010;300:229-236. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 141. | Manna AC, Ray B. Regulation and characterization of rot transcription in Staphylococcus aureus. Microbiology. 2007;153:1538-1545. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 142. | Saïd-Salim B, Dunman PM, McAleese FM, Macapagal D, Murphy E, McNamara PJ, Arvidson S, Foster TJ, Projan SJ, Kreiswirth BN. Global regulation of Staphylococcus aureus genes by Rot. J Bacteriol. 2003;185:610-619. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 213] [Cited by in RCA: 215] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 143. | Geisinger E, Adhikari RP, Jin R, Ross HF, Novick RP. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol Microbiol. 2006;61:1038-1048. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 187] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 144. | Jelsbak L, Ingmer H, Valihrach L, Cohn MT, Christiansen MH, Kallipolitis BH, Frees D. The chaperone ClpX stimulates expression of Staphylococcus aureus protein A by Rot dependent and independent pathways. PLoS One. 2010;5:e12752. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 145. | Oscarsson J, Harlos C, Arvidson S. Regulatory role of proteins binding to the spa (protein A) and sarS (staphylococcal accessory regulator) promoter regions in Staphylococcus aureus NTCC 8325-4. Int J Med Microbiol. 2005;295:253-266. [PubMed] |
| 146. | Lei MG, Cue D, Roux CM, Dunman PM, Lee CY. Rsp inhibits attachment and biofilm formation by repressing fnbA in Staphylococcus aureus MW2. J Bacteriol. 2011;193:5231-5241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 147. | Lim Y, Jana M, Luong TT, Lee CY. Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol. 2004;186:722-729. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 145] [Cited by in RCA: 147] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 148. | Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O'Donnell S, Rowe S, O'Gara JP, Lee CY. Rbf promotes biofilm formation by Staphylococcus aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol. 2009;191:6363-6373. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 75] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 149. | Cramton SE, Gerke C, Schnell NF, Nichols WW, Götz F. The intercellular adhesion (ica) locus is present in Staphylococcus aureus and is required for biofilm formation. Infect Immun. 1999;67:5427-5433. [PubMed] |
| 150. | Luong TT, Lei MG, Lee CY. Staphylococcus aureus Rbf activates biofilm formation in vitro and promotes virulence in a murine foreign body infection model. Infect Immun. 2009;77:335-340. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 26] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 151. | Vergara-Irigaray M, Valle J, Merino N, Latasa C, García B, Ruiz de Los Mozos I, Solano C, Toledo-Arana A, Penadés JR, Lasa I. Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun. 2009;77:3978-3991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 127] [Cited by in RCA: 147] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 152. | Majerczyk CD, Sadykov MR, Luong TT, Lee C, Somerville GA, Sonenshein AL. Staphylococcus aureus CodY negatively regulates virulence gene expression. J Bacteriol. 2008;190:2257-2265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 162] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 153. | Pohl K, Francois P, Stenz L, Schlink F, Geiger T, Herbert S, Goerke C, Schrenzel J, Wolz C. CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J Bacteriol. 2009;191:2953-2963. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 182] [Article Influence: 11.4] [Reference Citation Analysis (0)] |
| 154. | Tu Quoc PH, Genevaux P, Pajunen M, Savilahti H, Georgopoulos C, Schrenzel J, Kelley WL. Isolation and characterization of biofilm formation-defective mutants of Staphylococcus aureus. Infect Immun. 2007;75:1079-1088. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 108] [Cited by in RCA: 122] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 155. | Majerczyk CD, Dunman PM, Luong TT, Lee CY, Sadykov MR, Somerville GA, Bodi K, Sonenshein AL. Direct targets of CodY in Staphylococcus aureus. J Bacteriol. 2010;192:2861-2877. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 155] [Cited by in RCA: 176] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 156. | Stenz L, Francois P, Whiteson K, Wolz C, Linder P, Schrenzel J. The CodY pleiotropic repressor controls virulence in gram-positive pathogens. FEMS Immunol Med Microbiol. 2011;62:123-139. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 81] [Cited by in RCA: 91] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 157. | Richardson AR, Libby SJ, Fang FC. A nitric oxide-inducible lactate dehydrogenase enables Staphylococcus aureus to resist innate immunity. Science. 2008;319:1672-1676. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 208] [Cited by in RCA: 225] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 158. | Campbell EA, Westblade LF, Darst SA. Regulation of bacterial RNA polymerase sigma factor activity: a structural perspective. Curr Opin Microbiol. 2008;11:121-127. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 159. | Morikawa K, Inose Y, Okamura H, Maruyama A, Hayashi H, Takeyasu K, Ohta T. A new staphylococcal sigma factor in the conserved gene cassette: functional significance and implication for the evolutionary processes. Genes Cells. 2003;8:699-712. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 64] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 160. | Tao L, Wu X, Sun B. Alternative sigma factor sigmaH modulates prophage integration and excision in Staphylococcus aureus. PLoS Pathog. 2010;6:e1000888. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 32] [Cited by in RCA: 32] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 161. | Senn MM, Giachino P, Homerova D, Steinhuber A, Strassner J, Kormanec J, Flückiger U, Berger-Bächi B, Bischoff M. Molecular analysis and organization of the sigmaB operon in Staphylococcus aureus. J Bacteriol. 2005;187:8006-8019. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 87] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 162. | Martí M, Trotonda MP, Tormo-Más MA, Vergara-Irigaray M, Cheung AL, Lasa I, Penadés JR. Extracellular proteases inhibit protein-dependent biofilm formation in Staphylococcus aureus. Microbes Infect. 2010;12:55-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 86] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 163. | Chaibenjawong P, Foster SJ. Desiccation tolerance in Staphylococcus aureus. Arch Microbiol. 2011;193:125-135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 84] [Cited by in RCA: 89] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 164. | Lauderdale KJ, Boles BR, Cheung AL, Horswill AR. Interconnections between Sigma B, agr, and proteolytic activity in Staphylococcus aureus biofilm maturation. Infect Immun. 2009;77:1623-1635. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 215] [Article Influence: 13.4] [Reference Citation Analysis (0)] |
| 165. | Cebrián G, Condón S, Mañas P. Heat-adaptation induced thermotolerance in Staphylococcus aureus: Influence of the alternative factor sigmaB. Int J Food Microbiol. 2009;135:274-280. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 166. | Chen HY, Chen CC, Fang CS, Hsieh YT, Lin MH, Shu JC. Vancomycin activates σ(B) in vancomycin-resistant Staphylococcus aureus resulting in the enhancement of cytotoxicity. PLoS One. 2011;6:e24472. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 35] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 167. | Cheung AL, Chien YT, Bayer AS. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect Immun. 1999;67:1331-1337. [PubMed] |
| 168. | Meier S, Goerke C, Wolz C, Seidl K, Homerova D, Schulthess B, Kormanec J, Berger-Bächi B, Bischoff M. sigmaB and the sigmaB-dependent arlRS and yabJ-spoVG loci affect capsule formation in Staphylococcus aureus. Infect Immun. 2007;75:4562-4571. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 65] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 169. | Schulthess B, Bloes DA, François P, Girard M, Schrenzel J, Bischoff M, Berger-Bächi B. The σB-dependent yabJ-spoVG operon is involved in the regulation of extracellular nuclease, lipase, and protease expression in Staphylococcus aureus. J Bacteriol. 2011;193:4954-4962. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 170. | Bowser TE, Bartlett VJ, Grier MC, Verma AK, Warchol T, Levy SB, Alekshun MN. Novel anti-infection agents: small-molecule inhibitors of bacterial transcription factors. Bioorg Med Chem Lett. 2007;17:5652-5655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 171. | Clatworthy AE, Pierson E, Hung DT. Targeting virulence: a new paradigm for antimicrobial therapy. Nat Chem Biol. 2007;3:541-548. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 908] [Cited by in RCA: 983] [Article Influence: 54.6] [Reference Citation Analysis (0)] |