INTRODUCTION
Antimicrobial selection, dosage and duration of treatment have long-term effects on resistance development, and for this reason, the potential effects should be considered in every antimicrobial treatment decision[1]. In this sense, antimicrobial stewardship has been defined as the optimal selection, dose and duration of an antimicrobial that results in the best clinical outcome for the treatment of infection with minimal toxicity to the patient and minimal impact on subsequent resistance development[1-3]. Because gross tonnage of antimicrobial use in the outpatient population far outweighs that used in hospital patients, the community is a target-rich environment for proactive interventions to improve antimicrobial stewardship[3]. To this end the objective of the development of a new antibiotic for the treatment of community-acquired infections is to define the optimal dose regimen resulting in the best clinical outcome with minimal patient toxicity, minimal impact on human microbiota and minimal resistance development[4].
The analysis of the relationship between pharmacokinetics and pharmacodynamics (PK/PD) is a key instrument to improve antimicrobial stewardship since it supports optimal selection, dosing and duration of therapy[3]. PK/PD allows identification of the drug exposure measure that is closely associated not only with the ability to kill organisms but also to suppress the emergence of resistant subpopulations[5].
PK/PD FOR MICROBIOLOGICAL AND THERAPEUTIC EFFICACY PREDICTION
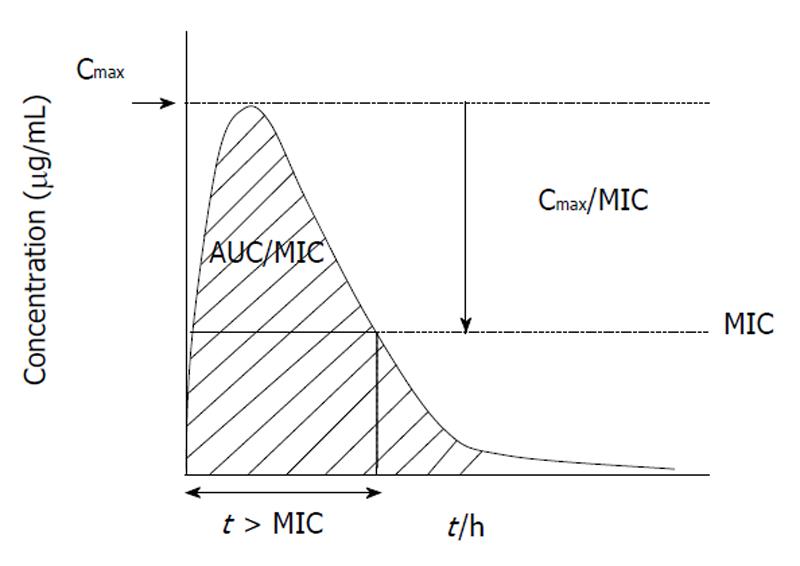
Understanding the relationship of PK/PD parameters and clinical outcome has expanded in the last 15 years allowing more closely correlation of in vitro potency with in vivo efficacy[6]. Nowadays, PK/PD studies are critical during drug development for proof-of-concept, dose and interval selection in clinical trials, and thus for determining susceptibility breakpoints[3]. Once the PK/PD parameter is known, then the PK/PD susceptibility breakpoint up to which in vivo activity is predicted can be determined for a given dose regimen[5]. Figure 1 shows the three PK/PD parameters predictor of antimicrobial efficacy. It is important to recognize that different amounts of drug exposure will translate into different degrees of antimicrobial effect. This means that for β-lactams (time-dependent antibiotics showing the minimal post-antibiotic effect) maximizing the duration of time that bacteria are exposed to effective concentrations increases the probability of clinical and bacteriological response to therapy. However, the whole dosing interval needs not to be covered by drug concentrations over the minimal inhibitory concentration (MIC) to obtain bacteriostasis or bactericidal activity; the classically considered percentages of the dosing interval to be covered for respiratory pathogens being 20% and 40% for carbapenems, 30% and 50% for penicillins and 40% and 65% for cephalosporins, respectively[5]. Values of 40-50 for the percentage of the dosing interval that serum concentrations exceed the MIC (t > MIC) have also been associated with bacteriologic efficacy for macrolides against respiratory pathogens[7]. In concentration-dependent agents showing prolonged post-antibiotic effect, as fluoroquinolones and azalides, maximizing drug concentrations increases the probability of positive clinical and bacteriological responses to therapy, with the area under the concentration-time curve (AUC)/MIC ratio (AUC/MIC) as the adequate PK/PD parameter with values of 20-25 and approx. about 250-300 predicting bacteriostasis and bactericidal activity, respectively, for respiratory pathogens[5]. However for clinical outcome values slightly higher (AUC/MIC approximately 30) than those for stasis are required, as has been shown for fluoroquinolones in community-acquired pneumococcal pneumonia[5].
Figure 1 Three pharmacokinetics and pharmacodynamics parameters predictor of antimicrobial efficacy.
AUC: Diagonal striped area.
Although it is important to address the association between drug exposure and response pre-clinically, it is also of vital importance to define this association in real patients. In recent years the introduction of mathematical models such as Monte Carlo simulations allows optimal combinations of PK/PD data to predict therapeutic results of particular dosing schedules in large populations[8]. To describe the likely range of exposures that will occur clinically, previous information regarding the central tendency and measures of dispersion of the pharmacokinetic parameter value is required. Monte Carlo simulations are adequate for factoring in inter-subject variations in pharmacokinetics or in variations subsequent to illness. Through the generation of a population values of interest using data from small PK studies, and then randomly generating a large number of patient values according to an underlying statistical distribution, Monte Carlo simulations determine the probability of achieving a specific value of a PK/PD parameter for a large population. The fraction of simulated subjects who attain the exposure target (AUC/MIC ratio, t > MIC) intended is calculated for each MIC in the distribution. The target attainment rate should exceed 90%, since when the rate falls below this value, the probability of effectiveness of the dosing regimen is significantly decreased[9].
One of the main problems for predicting in vivo efficacy is that in vivo antibiotics interact with bacteria in a more complex way than in vitro due to the presence of serum proteins acting as an interface. Serum proteins (i.e, protein binding) may affect values of PK/PD indices predicting therapeutic outcome. In the absence of standardised methods for testing the impact of protein binding on the activity of antibiotics[10], classically it is accepted that only the unbound fraction of the compound is active in vitro and presumably in vivo, and the mathematical extrapolation of active drug from total drug by using the reported protein binding rate is the method used for estimating free concentrations to be used in pharmacodynamic calculations. However the reversibility of protein binding implies that limitation of activity may be far from absolute, even in highly protein bound agents[11]. For this reason, studies investigating the effect of protein binding on in vitro and in vivo antibacterial activity are essential for highly bound agents as cefditoren.
PK/PD FOR COUNTERING RESISTANCE SPREAD
Although important advances in the knowledge of PK/PD principles have been gained in the last decade, the accumulated information is far from complete, especially in relation to prevention of resistance emergence and spread. Studies of antibiotic dosing regimens considering the development of resistance have not been of high priority, with most PK/PD indices focused on microbiological efficacy and subsequent clinical outcome[12]. Therefore PK/PD models for the search of PK/PD principles and breakpoints predicting diffusion of resistance are needed[12]. In addition, most PK/PD studies include susceptible wild-type bacteria as target organisms, thus more knowledge of drug PK/PD versus bacteria with different resistance mutations and susceptibility levels would be desirable.
Resistance is a process with two successive steps. The first (emergence) occurs by mutation of the genome (chromosome, plasmids, integrons or transposons) or acquisition of exogenous DNA by transformation or horizontal gene transfer, that is though to occur by chance not relying on the presence of antibiotics in the environment[13]. However it appears that antibiotic-induced stress increases genetic exchange including genes mediating resistance[13] since it has been reported that fluoroquinolones induce competence in Streptococcus pneumoniae (S. pneumoniae) for DNA transformation in the absence of bacterial killing[14]. The second step (dissemination) is the spread of resistant bacteria (clonal spread), plasmid (plasmid spread) or resistance determinants (gene spread), and it is clearly associated with the selective pressure exerted by antibiotic prescription[13]. Resistance resulting from mutations or DNA transformation has fitness associated costs; consequently, resistant bacteria are usually inferior to wild-type bacteria and can easily be uncompeted when antibiotic pressure is not present[12]. However antibiotic pressure may unmask these minor resistant subpopulations. For this reason, PK/PD studies using mixed cultures of susceptible strains with strains harbouring different resistance traits are highly valuable to assess the ability of different dosing regimens to counter resistance clonal spread or intra-strain diffusion of resistant subpopulations (gene spread). Of special interest are studies exploring the effects of antibiotic regimens on human microbiota, as the flora present in the nasopharynx. The nasopharynx is the niche from where resistant bacterial populations of Streptococcus pyogenes (S. pyogenes), Streptococcus pneumoniae and Haemophilus influenzae (H. influenzae) (exclusive human-adapted commensals and the most prevalent bacterial isolates from community-acquired respiratory tract infections when occasionally acting as pathogens) can be selected and further spread.
PK/PD, not only as a weapon for efficacy prediction but also for countering resistance diffusion and reducing collateral effects on commensal flora, should be considered as part of every antimicrobial treatment decision together with classical criteria as safety, route of administration, derived costs and treatment compliance.
CEFDITOREN-PIVOXIL AND CEFDITOREN PHARMACOKINETICS
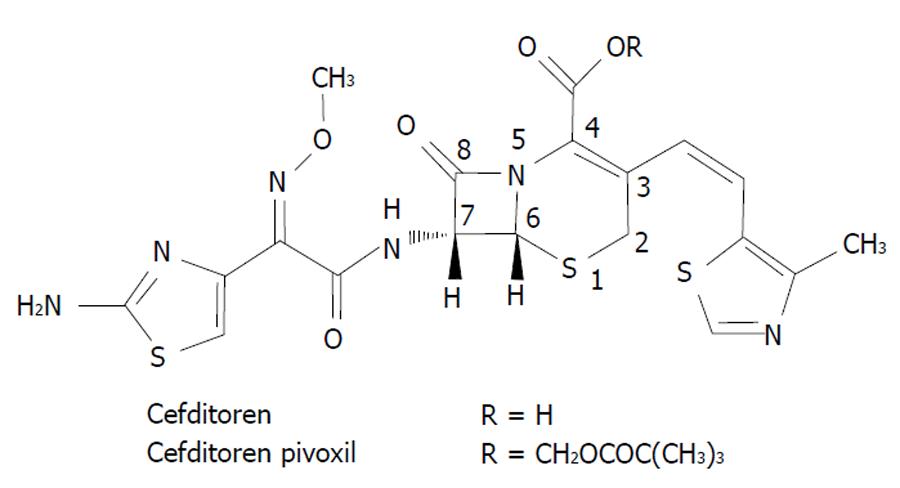
Cefditoren is the active form of cefditoren-pivoxil, an oral third-generation aminothiazolyl cephalosporin with structural components similar to those of first and third generation cephalosporins[15]. Figure 2 shows the chemical structure of cefditoren. The group attached at the C-7 position of the cephem skeleton affords activity against Gram negative microorganisms, whereas the one attached at the C-3 position (not seen in other non-first-generation cephalosporins) affords activity against Gram positive bacteria[15]. According to the study by Yamada et al[16] the structure of the PBP2X of S. pneumoniae complexed with cefditoren revealed that the unique methylthiazole group of the C-3 side chain of the cephem skeleton fits well into the PBP binding pocket, and this feature is likely to play a role in the high activity of cefditoren against S. pneumoniae. Cefditoren is stable to hydrolysis by common β-lactamases as TEM-1, TEM-2 and ROB-1 produced by H. influenzae.
Figure 2 Chemical structure of cefditoren.
When orally administered to humans the prodrug cefditoren-pivoxil is completely and rapidly hydrolyzed to cefditoren by esterases during absorption, and the drug is distributed in the circulation blood as active cefditoren. The oral bioavailability of cefditoren-pivoxil is low in the fasted state (15%-20%), but greatly increases when the drug is administered with high fat meals, with 50% and 70% increase in mean values of Cmax and AUC, respectively, compared to values in the fasted state[15]. In a phase I study where 400 mg cefditoren-pivoxil was administered as single dose in the fed state, values of the pharmacokinetic parameters determined were: Cmax = 3.7 ± 0.7 μg/mL, Tmax = 2 h, AUC∞ = 12.5 ± 1.6 μg × h/mL and t1/2 = 1.54 ± 0.20 h[17]. The mean volume of distribution at steady state of cefditoren is 9.3 L and the binding to plasma proteins (primarily human serum albumin) averages 88%[15]. Concentrations of cefditoren in bronchial mucosa have been determined in patients undergoing fibrobronchoscopy that had received 400 mg cefditoren-pivoxil as single dose 1-4 h prior to sample collection, with values ranging from 0.56 to 1.04 mg/kg[15]. Cefditoren, not appreciably metabolized, is mainly eliminated by excretion into the urine, with a renal clearance of approximately 4.1-5.6 L/h[15].
IN VITRO ACTIVITY OF CEFDITOREN AGAINST HUMAN-ADAPTED PATHOGENS
S. pyogenes: Are there susceptibility problems?
It is well known that S. pyogenes is uniformly susceptible to all β-lactams in contrast to macrolides (with resistance rates varying from 6.9% in USA[18] to 25.6% in Hong Kong[19]) and the emerging resistance to fluoroquinolones in America, Europe and Japan[20-23]. In recent surveillance studies carried out in Europe, the intrinsic activity of cefditoren against S. pyogenes was high, with MIC90 values of ≤ 0.03 - ≤ 0.06 μg/mL[24-26]. These values were lower than those of other oral β-lactams used as comparators that ranged from 0.06-0.06 μg/mL (penicillin), 0.06-0.12 μg/mL (amoxicillin), 0.12-0.12 μg/mL (cefpodoxime and cefuroxime), 0.12-0.25 μg/mL (cefixime) and 0.5-1 μg/mL (ceftibuten) to 1-2 μg/mL for cefaclor[25,26].
H. influenzae: Enzymatic and non-enzymatic resistance traits
While resistance to quinolones among strains of H. influenzae continues to be exceptionally rare, H. influenzae is intrinsically resistant to macrolides, associated with the presence of efflux pumps in virtually all strains[27]. With respect to β-lactams, the classical ampicillin resistance is mediated by β-lactamases, with a prevalence of β-lactamase producing strains of 14.3% in Europe, 19.2% in the Middle East, 21.0% in Latin America, 25.8% in North America and 26.8% in Asia-Pacific regions in a recent multinational study[28]. In addition to this enzymatic-mediated resistance, reports of strains showing mutations in the ftsI gene encoding PBP3 are increasing [β-lactamase negative ampicillin-resistant (BLNAR) strains and β-lactamase positive amoxicillin/clavulanate-resistant (BLPACR) strains]. These mutations confer resistance to amoxicillin/clavulanate, ampicillin/sulbactam, piperacillin/tazobactam, cefaclor, cefonicid, cefprozil, and cefuroxime[29]. In Japan, the reported prevalence of BLNAR in 2005-2008 was up to 42.9%[30], with a polyclonal dissemination[27], and with an increase in the prevalence of BLPACR (with clonal dissemination) from 1999 to 2008[31].
Several reviews and surveillances have worldwide shown the high in vitro activity of cefditoren against H. influenzae, with MIC90 values generally ≤ 0.03 μg/mL[25,26,32-37]. In these studies, among β-lactamase negative or positive strains without mutations in the ftsI gene, high susceptibility rates were found for amoxicillin/clavulanate and cefuroxime. However differences arose when considering strains with mutations in the ftsI gene for which MIC90 values were 4-8 μg/mL for amoxicillin/clavulanate and 2-16 μg/mL for cefuroxime[25,26,32,34,37] in contrast to MIC90 values of cefditoren of ≤ 0.06 μg/mL[25,26,34,37].
S. pneumoniae: The king of evolving resistance traits
Introduction of the 7-valent pneumococcal conjugate vaccine (PCV7) for children immunization has markedly influenced antibiotic susceptibility, with decreases in penicillin nonsusceptibility and erythromycin resistance in countries as Spain when comparing the pre- and post- vaccination periods (1998-1999 vs 2006-2007), from 50.2% to 22.9% and from 34.9% to 21.1%, respectively[38]. This decline in the prevalence of resistance to penicillin, and to a lesser degree to erythromycin is in accordance with the situation described in a pan-European report[39]. Nonsusceptibility to levofloxacin is approx. 2.2% worldwide[40].
In the current decade several studies have been carried out in different countries, regardless the introduction or not of PCV7, to determine the intrinsic activity of cefditoren versus comparators[25,26,35,41-49]. Most of these studies explored the intrinsic activity of cefditoren by distributing strains by penicillin susceptibility category, and showed the high intrinsic activity of cefditoren against penicillin-susceptible strains (MIC90 from ≤ 0.03 to 0.06 μg/mL), penicillin-intermediate strains (MIC90 from 0.25 to 0.5 μg/mL) and penicillin-resistant strains (MIC90 from 0.5 to 1 μg/mL), with values against penicillin-intermediate and resistant strains lower (higher intrinsic activity) than those required for the comparators used in the different studies (amoxicillin, cefdinir, cefprozil, cefuroxime, cefixime, ceftibuten, cefpodoxime, erythromycin, clarithromycin and azithromycin)[25,26,35,43-49]. In most of these studies MIC90 of cefditoren against penicillin-intermediate and resistant strains was one-dilution lower than that of cefotaxime.
It should be noted that since resistance is linked to serotypes, in countries where PCV7 has been introduced penicillin nonsusceptibility is decreasing. The multicentre, multinational Antibiotic Resistance Isolates in South Europe (ARISE) study carried out in Europe in the pre-PCV7 era (including large number of isolates from serotypes 6, 9, 14 and 23, included in PCV7 and traditionally linked to resistance) showed MIC50/MIC90 values of cefditoren of ≤ 0.03/0.5 μg/mL[50]. In the last SAUCE study (2006-2007) carried out in Spain after PCV7 introduction, MIC50/MIC90 values of cefditoren were ≤ 0.015/0.125 μg/mL[38]. However in parallel to the decrease in the prevalence of PCV7 serotypes there has been a high increase in the prevalence of non-PCV7 serotypes. According to a recent (2009) study in Spain, 85.3% of isolates received in the Reference Laboratory for Pneumococci were non-PCV7 serotypes, 79.8% of them susceptible to penicillin[51]. The intrinsic activity of cefditoren against the remaining 20.2% penicillin nonsusceptible isolates belonging to non-PCV7 serotypes varied on serotype basis: MIC90 of 1 μg/mL for penicillin-resistant and 0.5 μg/mL for penicillin-intermediate serotype 19A isolates, 0.5 μg/mL for serotype 11A, 0.25 μg/mL for serotypes 35B, 6A and 15A, 0.12 μg/mL for serotype 24F and 0.06 μg/mL for serotype 23B isolates[51].
CEFDITOREN PK/PD FOR PREDICTION OF THERAPEUTIC EFFICACY
By mathematically relating the mean cefditoren exposure achieved in the healthy volunteers included in the Phase I study with the range of potential MICs for target pathogens, the values of t > MIC determined were: approximately 55% for MIC of 0.5 μg/mL, approximately 68% for MIC of 0.25 μg/mL, approximately 81% for MIC of 0.12 μg/mL and approximately 94% for MIC of 0.06 μg/mL[17]. However, as previously commented, in vivo antibiotics interact with bacteria in a more complex way than in vitro due to the presence of serum proteins acting as an interface. This is especially important for cefditoren that exhibits a high protein binding rate to human albumin (88%)[15]. For this reason to determine if protein binding pharmacodynamically impacts cefditoren activity, experimental studies were required. As above described, H. influenzae and S. pyogenes are uniformly and highly susceptible to cefditoren, with MIC90 of approximately ≤ 0.06 μg/mL. Thus, investigations on the effect of protein binding on cefditoren antibacterial activity have been performed using different S. pneumoniae strains as target bacteria since against this species cefditoren exhibits a wider range of MIC values.
One particular characteristic of cefditoren is that its rate of binding to albumin is similar in mice and humans. This gives the opportunity to extrapolate PK/PD predicting therapeutic efficacy from in vivo mice models to humans. Based on this characteristic, the pharmacodynamic effect of the high protein binding of cefditoren has been studied both in vitro and in vivo.
To explore in vitro the effect of the presence of human albumin on the activity of cefditoren, a one-compartmental computerized pharmacodynamic dynamic model was used. The serum profile of cefditoren (total concentrations) obtained over 24 h with a 400 mg bid regimen was simulated using media containing human albumin physiological concentrations (4.9 g/dL) (75% human serum and 25% broth)[52]. When the protein binding rate of cefditoren in the device was determined, the value was similar (87.1%) to the one described in humans in the literature[15]. Antibacterial activity was determined over time using as target bacteria six penicillin nonsusceptible S. pneumoniae strains exhibiting cefditoren MICs of 0.25 and 0.5 μg/mL and initial inocula ranging from 2 × 107 to 6 × 107 cfu/mL[52]. At 24 h, reductions > 99.9% in the initial inocula (i.e, bactericidal activity) were obtained for strains with MIC of 0.25 μg/mL, with reductions ranging from 52.9% to 96.8% for strains with MIC of 0.5 μg/mL[52].
To explore in vivo the effects of protein binding on cefditoren PK/PD and therapeutic efficacy, a mouse sepsis model was performed[53]. As target infecting strains three S. pneumoniae isolates of index serotypes (6B, 19F and 23F) were chosen, as these serotypes have been identified as independent factors for 30 d mortality in bacteremic disease in humans[54]. In addition, these three strains exhibited exceptional high cefditoren MICs: 1 μg/mL (serotype 6B), 2 μg/mL (serotype 19F) and 4 μg/mL (serotype 23F)[53], favoring measurement of concentrations below MIC values for the pharmacodynamic analysis. Mice were intraperitoneally infected with lethal doses (producing 100% mortality in untreated controls) of approx. 107 cfu/mL of the infecting strain 1 h prior to treatment initiation[53]. Cefditoren was administered every 8h up to a total of six doses, using dose-ranging regimens with doses ranging from 6.25 mg/kg to 100 mg/kg[53]. Animals were followed for 7 d and deaths were recorded. The protein binding rate in mice serum was experimentally measured, showing 86.9% binding rate. The total t > MIC (t > MIC) and free t >MIC (ft > MIC) (considering the 86.9% binding rate) associated with survival were determined. Values of t > MIC of approximately 35% (ft > MIC of approximately 19%) were associated with 100% survival of animals infected by strains with MICs of 1 and 2 μg/mL[53]. In those infected with the strain with MIC of 4 μg/mL and treated with the maximum tested dose (100 mg/kg), 60% survival was obtained with t > MIC of approximately 27% (ft > MIC of approximately 16%)[53].
According to the results obtained in these studies assessing the effect of protein binding on cefditoren activity, limitation of protein binding is far from absolute since the antibacterial activity was higher than the one predicted by ft > MIC, both in vitro and in vivo. In this sense in the animal model, cefditoren t > MIC of approximately 35% (free t > MIC of approximately 19%) produced 100% survival in animals infected by strains with cefditoren MIC of 1-2 μg/mL[53]. A free t > MIC value of approximately 20% was also related to >99.9% reduction in bacterial load of isolates with cefditoren MIC of 0.25 μg/mL in the in vitro simulation[52].
For β-lactams, values of t > MIC of 40% have been related with clinical cure in humans[55,56], and values of 33% with bacteriostasis[57], a term linked to the definition of the “susceptible” category by the Food and Drug Administration (FDA) and Clinical and Laboratory Standards Institute[29,58] as likely inhibition of the pathogen if the antimicrobial compound reaches the concentration usually achievable after administration of the recommended dose. These two cut-offs were applied in a Monte Carlo simulation with cefditoren total (and extrapolated free) concentrations determined in the phase I study[17] for a range of MIC values of 0.015 to 1 μg/mL[59]. Considering total t > MIC of 33% (bacteriostatic end-point) as end-point (approximately the value for total drug linked to 100% survival in the mice model), MICs up to 0.5 μg/mL were covered by cefditoren[59]. When considering free t > MIC of 33% (a value higher than the one with free drug required in the mice model for 100% survival and in the pharmacodynamic simulation for > 99.9% reduction in bacterial load), MICs up to 0.25 μg/mL were covered[59]. When the classical theoretical value of free t > MIC of 40% was considered, strains up to 0.12 μg/mL were covered by cefditoren in the Monte Carlo simulation[59].
According to these data, the cefditoren breakpoint for susceptibility should be within the range between ≤ 0.12 μg/mL and ≤ 0.5 μg/mL. This last value has been proposed by several authors considering cefditoren MIC90 values lower than the breakpoints values for parenteral 3rd generation cephalosporins and the pharmacokinetics of cefditoren[60-62], and was the breakpoint approved by the Reference Member State (Spain) during the Mutual Recognition Procedure (MRP) in Europe (susceptibility ≤ 0.5 μg/mL)[63]. The most conservative breakpoint (≤ 0.12 μg/mL) was proposed by the FDA in the prescribing information[58]. With this strictest breakpoint value, 100% S. pyogenes, nearly 100% H. influenzae, and in Spain in 2006-2007 approximately 95% pneumococci were susceptible to cefditoren[38].
CEFDITOREN PK/PD FOR COUNTERING RESISTANCE SPREAD
As has been previously commented, studies of antibiotic dosing regimens considering the development of resistance have not been of high priority, with most PK/PD indices focused on microbiological efficacy and subsequent clinical outcome[12]. The importance of studies exploring the ability of antimicrobials for countering selection of resistance (resistant subpopulations within a single strain or nonsusceptible strains within polymicrobial niches) lies in the selection of resistance in polymicrobial niches involved in certain pathologies as otitis, acute exacerbations of chronic bronchitis or in pathologies where indirect pathogenicity has been described as streptococcal pharyngitis. Maybe more important are studies exploring selection of resistance within the bacterial microbiota present in the nasopharynx where resistance in human-adapted commensals emerges and is further selected and diffused.
Mucosal surfaces may be simultaneously colonized by multiple species. The success of an organism in colonizing, and maybe in establishing a subsequent infection, might be determined by its ability to compete with cohabitants of its niche. In the oropharynx there is an intricate balance between human-adapted pathogens (S. pyogenes, H. influenzae, S. pneumoniae) and other nasopharyngeal flora[64]. According to different studies, throat carrier rates of S. pyogenes is 15%-20% among school-aged children during seasonal peaks[65,66], carriage of S. pneumoniae ranges from 10% to 40% in an age-dependent manner[67], and up to 80% of healthy persons carry H. influenzae[68], with multiple strains in 50% of positive samples[69] and a high turnover of strains[70]. This is the baseline that antibiotics can alter by selecting specific resistant populations that existed in low numbers in the absence of antibiotic pressure due to the high fitness cost of resistance.
Different studies exploring the ability of cefditoren for countering selection of resistance have been performed, this making a difference between the data available for this third generation cephalosporin and those for other marketed compounds.
Countering intra-strain diffusion of resistance in H. influenzae
H. influenzae is naturally able to take up DNA from the environment. A recent in vitro study found that the mechanism of the in vitro transfer of the ftsI gene involves classical transformation[71]. To explore the ability of cefditoren (compared with amoxicillin/clavulanic acid) to counter intra-strain diffusion of resistance linked to ftsI gene mutations, a dynamic study (using a two-compartment computerized device) was performed. Free concentrations of 400 mg bid cefditoren versus 875/125 mg tid amoxicillin/clavulanic acid were simulated over 24 h, and total and recombined populations derived from intra-strain ftsI diffusion both in β-lactamase positive and negative H. influenzae strains were measured[72]. The mixture of the wild-type and the recombined subpopulation was obtained by allowing transformation through 3h-incubation of extracted DNA from a BLNAR (or BLPACR) strain with the wild-type isolate (recipient strain). This mixture was used as initial inocula (107 cfu/mL) and was introduced into the device[72]. In antibiotic-free simulations the recombined subpopulations of the β-lactamase negative strain increased over time, showing good fitness[72]. These recombined subpopulations neither increased over time in the presence of physiological free concentrations of amoxicillin/clavulanic acid, and were very small or undetectable at 24 h (ft > MIC of 30.7% for the recombined population), suggesting that mutations in the ftsI gene did not offer competitive advantage[72]. In the case of the β-lactamase positive strain, in antibiotic-free simulations, the recombined subpopulations did not increase or even decreased, but in simulations with amoxicillin/clavulanic acid they increased from 0.00095% at 0 h to 6% or 32% at 24 h in simulations with DNA from BLNAR and from BLPACR, respectively (ft > MIC of 0% for the recombined population)[72]. Thus, the accumulation of the two resistance traits (mutations in the ftsI gene + production of β-lactamase) in the recombined population offered competitive advantage in the presence of amoxicillin/clavulanic acid. In contrast, in cefditoren simulations (free concentrations) no recombined populations could be detected at 24 h, with high ft > MIC values (> 83%) against recipient or donor strains[72]. From the pharmacodynamic perspective, this in vitro study showed that ft > MIC values higher than those classically considered for prediction of bacteriological response are needed to counter intra-strain ftsI gene diffusion since they should cover any possible recombined subpopulation[72]. Antibiotics such as cefditoren, with high intrinsic activity against H. influenzae regardless of the resistance genotype, exhibit potential for countering recombinant subpopulations. This suggests epidemiological advantages derived from limitation of intra-strain diffusion and spread of nonenzymatic resistance mechanisms in H. influenzae.
Countering selection of resistance in multibacterial niches of the same species
Once strains with different resistance traits have emerged, bacterial dynamics in polymicrobial niches represent the baseline that antibiotics can alter by selecting specific strains. The influence of different resistance traits on the competitive growth of H. influenzae[73] or of S. pneumoniae[74] has been studied in the absence and in the presence of antibiotics using the previously mentioned in vitro two-compartment computerized device.
The ability of cefditoren (vs amoxicillin/clavulanic acid and cefuroxime) to counter selection of resistant H. influenzae strains (exhibiting mutations in the ftsI gene) within a multi-strain niche was explored using an initial inoculum (4 × 106 cfu/mL) composed of four strains (one β-lactamase negative, one β-lactamase positive, one BLNAR and one BLPACR) in a 1:1:1:1 proportion[73]. Free concentrations obtained after 400 mg bid cefditoren, 875/125 mg tid amoxicillin/clavulanic acid and 500 mg bid cefuroxime were simulated[73]. In antibiotic-free simulations the frequency (among the whole mixed inocula) of the BLPACR strain carrying both resistance traits (mutations in the ftsI gene + β-lactamase production) at 24 h was as low as 1%, contrasting with the frequency (90%) of the strain without resistance traits (the β-lactamase negative strain)[73]. As occurred in the study on subpopulations within a single strain[72], these results evidenced that the accumulation of enzymatic and non-enzymatic resistance traits has negative influence on bacterial fitness in the absence of antibiotics. In the presence of antibiotics the BLPACR strain was the predominant at 24 h, with significantly higher activity of cefditoren versus comparators mainly due to differences in the reduction obtained in the BLNAR and BLPACR populations (highly reduced with cefditoren but not with the other two β-lactams)[73]. These differences were related with ft > MIC values < 20% for amoxicillin/clavulanic acid and 0% for cefuroxime against BLNAR and BLPACR strains, while values were > 67% for cefditoren against all strains[73]. These results suggest that antibiotic treatments unmask populations with specific resistance traits in a magnitude that depended on (1) their capability to reduce the mixed inocula over time; and (2) values of pharmacodynamic parameters obtained for the different strains in mixed inocula simulations, that favored cefditoren which concentrations are not affected by the β-lactamase produced by the β-lactamase producing strains and its activity is not influenced by the presence of ftsI gene mutations[73].
In the case of S. pneumoniae one study was carried out simulating a multi-strain niche of four pneumococcal strains with different resistance genotype determinants: serotype 19A (tetM), serotype 4 (pbp1a, gyrA and parC mutations), a serotype 19F (pbp1a, pbp2x and ermB mutations) and a serotype 23F (pbp1a, pbp2x, pbp2b-10 changes- mutations and CAT)[74]. The initial inoculum was 4-8 × 106 cfu/mL with 1:1:1:1 proportion of strains. Free serum concentrations obtained after 400 mg bid cefditoren, 500 mg bid cefuroxime, 400 mg od cefixime, 500 mg bid cefaclor or 875 mg bid amoxicillin were simulated over 24 h in the two-compartmental computerized device[74]. Population analysis profiles using plates with different amoxicillin concentrations were performed prior and at the end of the simulations[74]. In antibiotic-free simulations the worst fitness corresponded to the strain carrying multiple resistance traits (serotype 23F) that disappeared at 10h, while the majority of the final population corresponded to serotype 4, with marginal populations of serotype 19A and 19F[74]. As with H. influenzae, in the absence of antibiotic pressure, the accumulation of genetic determinants of resistance had fitness cost, influencing the competitive growth of strains in the simulated multibacterial niche of the same species. In antibiotic simulations, all penicillin/amoxillin-susceptible strains were eradicated (serotypes 19A and 4; t > MIC ≥ 43%), with the remaining two penicillin-resistant strains in the mixed inocula showing different evolutions depending on the antibiotic used: a) cefditoren produced > 2 log10 reduction in the total initial inocula (t > MIC ≥ 45%) at 12-24 h, with the remaining population growing in plates with ≥ 4 μg/mL of amoxicillin; b) the other oral cephalosporins did not produce any reduction in the total initial inocula at 12-24 h (t > MIC = 0%) with minor subpopulations growing in plates with ≥ 4 μg/mL of amoxicillin, and c) amoxicillin produced 2.6 log10 decrease at 12 h (t > MIC = 47.5%) but 1.1 log10 increase in initial inocula at 24 h with the majority of the population growing in plates with ≥ 4 μg/mL of amoxicillin[74]. This study showed that antibiotic activity against S. pneumoniae mixed inocula depended, as in studies with H. influenzae, on the pharmacodynamic activity against each of the strains, together with the possible selection of intra-strain resistant subpopulations[74].
Countering selection of resistance in multibacterial niches of different species
An experimental study was carried out using the previously mentioned two-compartment computerised device and 1:1:1:1 mixed inocula (4 × 106 cfu/mL) composed by one S. pyogenes, one penicillin-resistant S. pneumoniae, one β-lactamase positive H. influenzae and one BLPACR H. influenzae[75]. In antibiotic-free simulations up to 96.3% of the population at 24 h corresponded to the S. pyogenes strain, 3.2% to the BLPACR strain and 0.5% to the S. pneumoniae strain, with the β-lactamase positive H. influenzae being undetectable[75]. This mixed inoculum was exposed to free concentrations obtained after 400 mg bid cefditoren, 875 mg tid amoxicillin or 875/125 mg tid amoxicillin/clavulanic acid regimens over 24 h[75]. In simulations with amoxicillin, the β-lactamase production by H. influenzae strains precluded the eradication of streptococcal strains (S. pyogenes and S. pneumoniae) by degradating amoxicillin and decreasing ft > MIC from 100% in bacterial-free simulations to approximately 25% in simulations with the mixed inocula for S. pyogenes and from approximately 43% to approximately 18% for S. pneumoniae[75]. The presence of clavulanic acid in simulations with amoxicillin/clavulanic acid precluded indirect pathogenicity (absence of S. pyogenes eradication) by protecting amoxicillin from degradation, and S. pyogenes was eradicated (ft > MIC of 100% in mixed inocula simulations)[75]. However the S. pneumoniae and the H. influenzaeβ-lactamase positive strains were selected regardless their susceptibility to amoxicillin/clavulanic acid (MIC = 2/1 μg/mL in both cases). This could be associated to a decrease in the amoxicillin ft > MIC value from approximately 41% in bacterial-free simulations to approximately 33% in simulations with the mixed inocula due to the partial degradation of amoxicillin by the β-lactamase produced by the BLPACR strain (resistant to amoxicillin/clavulanic acid)[75], being clavulanic acid concentrations not able to counter β-lactamase production completely. In the case of cefditoren both H. influenzae strains and S. pyogenes were eradicated (ft > MIC ≥ 58%) and S. pneumoniae was 2 log10 decreased (ft > MIC of approximately 26%)[75]. According to the results of this study, countering of co-pathogenicity seems to be gradual since β-lactamase inhibitors as clavulanic acid in tid regimens were able to counter it for very susceptible strains as S. pyogenes but not for those less susceptible as the S. pneumoniae strain, that was selected. This suggests that, at least in vitro, cefditoren, that is resistant to the action of β-lactamases, offers the advantage of countering co-pathogenicity.
CONCLUSION
PK/PD is a key instrument to improve antimicrobial stewardship since its analysis supports optimal selection, dosing and duration of therapy[3]. PK/PD should be aimed to the identification of the drug exposure measure that is closely associated not only with the ability to kill organisms but also to suppress the emergence of resistant subpopulations[5]. This last aim is especially important since the main ecological side-effect of antibiotics is the spread of resistance that contributes to increase morbidity, mortality and associated expenses. In an environment with a lack of robust pipeline of new antibiotics, PK/PD of available compounds should be considered part of each antibiotic treatment selection. In contrast to what occurs with other compounds, an extensive investigational program with cefditoren have explored PK/PD of this 3rd generation cephalosporin not only for therapeutic efficacy prediction but also for countering (from clonal and gene perspectives) spread of resistance traits among human-adapted respiratory pathogens. From the perspective of efficacy prediction the studies performed (including those investigating the effect of protein binding) showed the cefditoren pharmacodynamic coverage of the three main human-adapted respiratory pathogens responsible for community-acquired respiratory infections. This could be associated with the demonstrated efficacy of cefditoren in clinical trials of upper and lower respiratory tract infections reported in the literature[76,77]. From the perspective of the potential for countering spread of resistance, the pharmacodynamic studies performed with cefditoren showed its capability for countering intra-strain spread of resistance linked to ftsI gene mutations in H. influenzae, for countering the spread of H. influenzae resistant strains (BLNAR and BLPACR) in multi-strain H. influenzae niches or of S. pneumoniae strains with multiple resistance traits in multi-strain S. pneumoniae niches, and for overcoming indirect pathogenicity linked to β-lactamase production by H. influenzae that protects S. pyogenes in multibacterial niches.
The extensive information on cefditoren PK/PD gathered from published studies, apart from being an example of desirable investigations with old/new compounds, suggests an ecological potential for cefditoren (countering resistance spread among human-adapted commensals) in addition to its adequate pharmacodynamic coverage of respiratory pathogens (including those resistant to previous oral compounds) producing community-acquired infections.
Peer reviewer: Sang Hee Lee, Professor, Drug Resistance Proteomics Laboratory (NLRL), Department of Biological Sciences, Myongji University, 116 Myongjiro, 116 Myongjiro, Yongin 449-728, South Korea
S- Editor Wu X L- Editor A E- Editor WuX