Published online Sep 29, 2022. doi: 10.5495/wjcid.v12.i2.50
Peer-review started: June 18, 2022
First decision: July 14, 2022
Revised: August 2, 2022
Accepted: September 21, 2022
Article in press: September 21, 2022
Published online: September 29, 2022
Processing time: 100 Days and 6.2 Hours
During the peak of the coronavirus diseases 2019 (COVID-19) pandemic, cli
To study the influence of SARS-CoV-2 anti-nucleocapsid-IgG seropositivity on clinical course and diseases severity in hospitalized COVID-19 patients.
We conducted a retrospective study of adults admitted to a tertiary care com
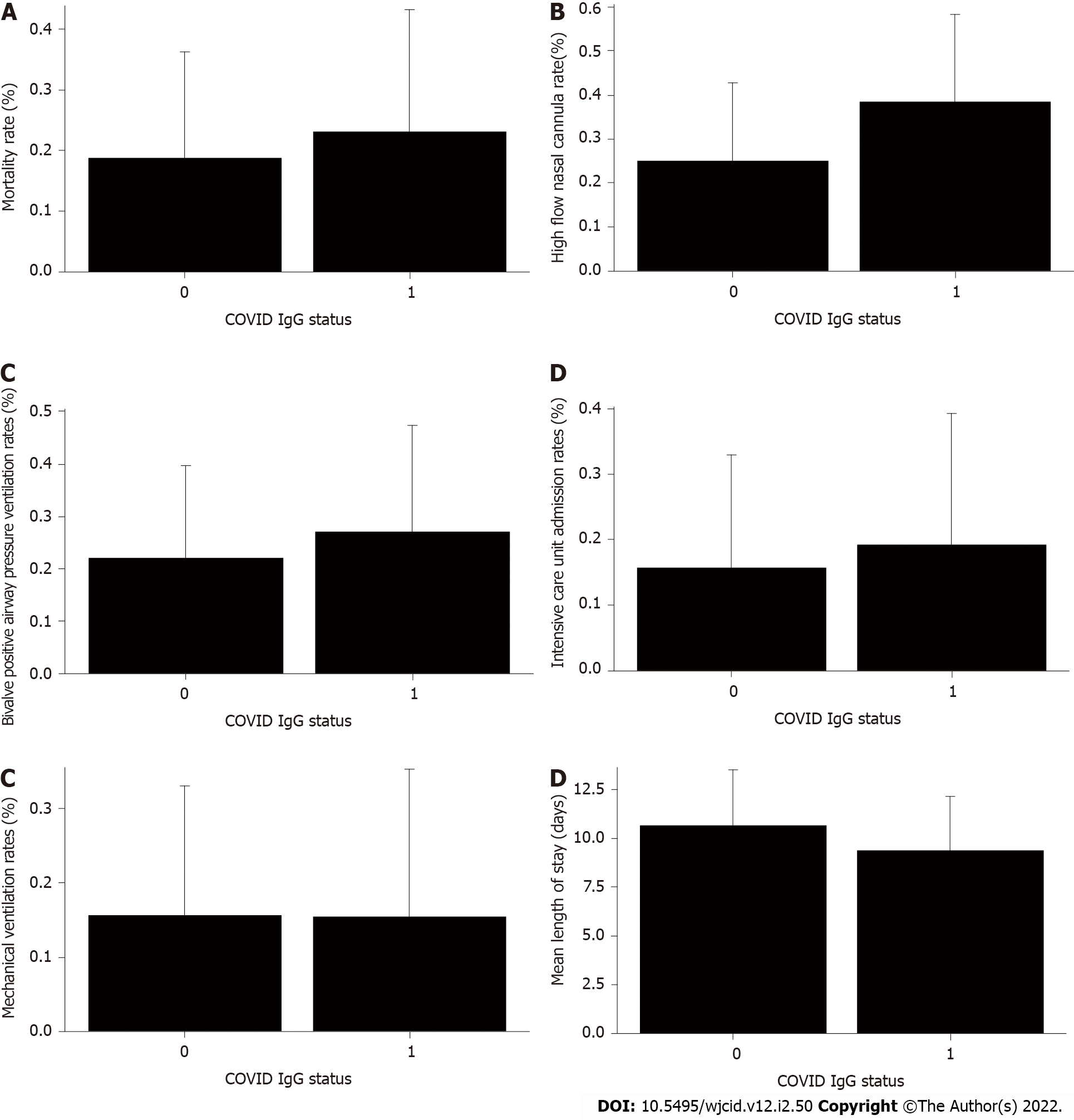
After a thorough examination of patient data, it was found that admission rates to the Intensive Care Unit, as well as the usage of BiPAP, HFNC and VENT support, in conjunction with patient outcomes, were not significantly different across IgG-N status. However, the LOS variable when assessed by IgG-N status was found to be significant (t value = 2.16, P value = 0.0349). IgG-N negative patients had higher than average LOS in comparison to IgG-N positive patients (15.12 vs 9.35 d). Even when removing the extreme value (an LOS of 158 d), IgG-N negative patients still had slightly higher than average stays (10.66 vs 9.35 d) but the relationship was no longer significant. For patient outcome/death, only age (numerical) was a significant predictor (F value = 4.66, P value = 0.0352). No other variables for any of the outcomes were significant predictors of clinical course or disease severity.
Our study demonstrated that IgG-N seroconversion had no significant association with clinical outcomes in hospitalized COVID-19 patients.
Core Tip: We intended to study an immunologic marker to predict the need for advanced oxygen supplement system and clinical outcome in order to support our hospital crisis management system during the peak of the pandemic. Our study demonstrated that presence of anti-nucleocapsid-IgG (IgG-N) against severe acute respiratory syndrome coronavirus 2 infection had no impact on the clinical outcome or disease severity in hospitalized coronavirus disease 2019 (COVID-19) patients. We did not find a correlation of statistical significance to use IgG-N as a biomarker to predict clinical outcome in COVID-19 patients admitted to a community hospital in North Dakota.
- Citation: Dzananovic B, Williamson M, Nwaigwe C, Routray C. Clinical significance of anti-nucleocapsid-IgG sero-positivity in SARS-CoV-2 infection in hospitalized patients in North Dakota. World J Clin Infect Dis 2022; 12(2): 50-60
- URL: https://www.wjgnet.com/2220-3176/full/v12/i2/50.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v12.i2.50
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) is a novel virus which belongs to the family of Coronaviridae, the causative agent for coronavirus diseases 2019 (COVID-19)[1]. SARS-CoV-2 emerged out of Wuhan in China and soon after, it spread to the entire world and thereby becoming a “Pandemic”[2-4]. After two years of rapid spread and the virus claiming over five million lives, healthcare system continues to scramble to protect patients from the atypical pneumonia-like illness caused by COVID-19. Diagnosis of SARS-CoV-2 infection is primarily dependent on reverse-transcription polymerase chain reaction (RT-PCR) testing of nasopharyngeal swab samples with more recent progress into rapid antigen testing[5,6]. Rapid community spread of the infection and dev
Taking a dive into the pathophysiology of COVID-19, a strong comprehension of the role of the hu
Practicing in a tertiary care community-based teaching hospital in North Dakota, United States, we have had experience with the pre-vaccine phase of COVID-19 pandemic. Noncompliance with public mask usage and rapid community transmission led to a sharp rise in COVID-19 illness and hospitalizations in North Dakota. In the midst of a healthcare crisis, we decided to investigate whether a qualitative IgG-N could be used as a molecular marker to determine the prognosis in the hospitalized patients. Basing our hypothesis on a theory that a rampaging community spread of SARS-CoV-2 infection led to a measurable IgG-N seroconversion of our population, thus impacting outcomes from hospitalization due to COVID-19, we retrospectively analyzed the data provided by the single-center community hospital from which we practiced.
All patients were admitted to the community hospital between December 1, 2020 and August 30, 2021. Fifty-nine patients were included in the study who were screened for IgG-N within 48 h of admission. We excluded all patients that had been admitted to the hospital with a non-COVID-19 diagnosis who incidentally tested positive for SARS-CoV-2 during screening. Patients with severe or critical COVID-19 illness as per the definition of National Institute of Health were included in the study. Those with mild and moderate illness were excluded from the study as most of them did not meet criteria for hospitalization. None of the included patients had been vaccinated against COVID-19. All the patients were confirmed positive for SARS-CoV-2 infection using RT-PCR from nasopharyngeal swab samples at admission. Both male and female patients aged 28 to96 were included in the study. All patients were checked for the presence of SARS-CoV-2-IgG-N within 48 h of admission by using Abbott SARS-CoV-2-IgG assay, that uses a two-step chemiluminescent microparticle immunoassay method with acridinium-labeled anti-human IgG, performed at North Dakota state laboratory. Admission blood samples identified 26 patients positive for IgG-N against SARS-CoV-2 and 33 negatives. In October 2021, we started data acquisition, reviewing the electronic medical record of included patients.
As this retrospective cohort study investigated the study population from patient admission to outcome, a thorough review of the electronic medical record was performed to capture data. This data was inclusive of the following: age, gender, body mass index (BMI), duration of symptoms prior to hospitalizations (DOS), length of hospital stay measured in days (LOS), admission to ICU, need for high flow nasal cannula (HFNC), bilevel positive airway pressure ventilation (BiPAP) or mechanical ventilation (VENT) for supplemental oxygen/support, as well as the final patient outcome - discharge or death. (Table 1)
| IgG-N positive | IgG-N negative | |
| Demographic information | ||
| Number | 26 (19 male, 7 female) | 32 (20 male, 12 female) |
| Mean age | 61.1 (17.6) | 60.0 (16.1) |
| Median age | 63.5 (25) | 63.5 (21) |
| Mean BMI | 33.7 (7.4) | 33.9 (7.7) |
| Median BMI | 32.8 (12) | 33.9 (10.1) |
| Mean DOS-d | 7.5 (5.6) | 6.5 (4.1) |
| Median DOS-d | 5.5 (6.0) | 7.0 (7.0) |
| Outcome | ||
| Mean LOS-d | 9.3 (5.6) | 10.7 (10.4) |
| Median LOS-d | 7.5 (7) | 6.5 (7) |
| Proportion of death | 0.23 | 0.19 |
| Proportion of ICU | 0.19 | 0.16 |
| Proportion of BiPAP | 0.27 | 0.22 |
| Proportion of VENT | 0.15 | 0.16 |
| Proportion of HFNC | 0.38 | 0.25 |
Formatting: Age, BMI and DOS were numerical variables. However, additional constructs split Age and BMI into two and three-group categories for some analyses. For example, BMI_2, patients with a BMI of < 29.9 were put in one group, and those > 30 in a second group. For BMI_3, patients with a BMI of < 25 were put into one group, those between BMI of 25-29.9 in a second group, and those with BMI > 30 in a third. For the variable labelled Age_2, patients < 75 were put in one group, and those 75+ in a second group. For Age_3, patients with an age of < 40 were put into the first group, those between 40-75 in a second, and those 75+ in a third group. It should be noted here that one patient had an extreme value for their LOS at-158 d. Models were run with both the patient included and excluded to determine the sensitivity of the models to this extreme value.
Correlation of outcomes: For each pair of outcomes (Death, ICU, BiPAP, VENT and HFNC), the phi coefficient (measure of association between binary variable, comparable to the Pearson coefficient for continuous normal variables) was calculated (Table 2).
| Death | ICU | BiPAP | VENT | HFNC | |
| Death | 1 | - | - | - | - |
| ICU | 0.67 | 1 | - | - | - |
| BiPAP | 0.71 | 0.70 | 1 | - | - |
| VENT | 0.60 | 0.94 | 0.65 | 1 | - |
| HFNC | 0.58 | 0.48 | 0.67 | 0.43 | 1 |
Outcomes by IgG status alone: For each variable, the outcome was modeled as a function of IgG-N status (positive or negative) using a generalized linear distribution. For LOS, it was determined that a negative binomial distribution had a better fit than a Poisson or Gaussian distribution, as evidence by a Pearson chi-square/df value closer to 1.0. The negative binomial distribution is also less sensitive to outliers. All other outcomes utilized a binary logistic regression model.
Outcomes by IgG-N status full model: For any single models that were significant, a multiple regression model was utilized, accounting for the consequential effect (if any) of the defined confounding variables of age, sex, BMI, and duration of symptoms.
LOS was modeled as a function of age using a negative binomial model. From there, age (categorical), BMI (numerical), and the interaction of BMI and age were each run with and without the extreme patient LOS-value noted in the previous section. The patient outcome/death was modeled as a function of LOS using a logistic model. Then, death was modeled as a function of age (numerical), and then age (categorical). The same was done for BMI. Finally, death was modeled as a function of sex. ICU, BiPAP, VENT, and HFNC were each modeled as a function of age (numerical), BMI (numerical), and sex separately.
Statistical analysis used SAS Studio V.3.8 (Cary, North Carolina, United States). The statistical review of the study was performed by a biomedical statistician.
We conducted a retrospective cohort study among fifty-nine adults aged between 28-96, admitted to the hospital with severe or critical COVID-19 illness between December 2020 and August 2021.
Unsurprisingly, most outcomes were strongly correlated. VENT and ICU rates were very strongly correlated (Phi Coeff = 0.94). All but one patient who went on mechanical ventilation was also admitted to the ICU. In contrast, VENT and HFNC rates were only moderately correlated (Phi Coeff = 0.43).
Patient outcome, ICU admissions, HFNC, BiPAP and VENT rates were not significantly different across IgG-N status (Figure 1A-E). However, LOS by IgG-N status was found to be significant (t value = 2.16, P value = 0.0349) when including the extreme value (LOS > 150 d). IgG-N negative patients had higher average LOS than IgG-N positive patients (15.12 vs 9.35 d). However, when removing the extreme value (LOS of 150 d), IgG-N negative patients still had slightly higher average LOS (10.66 vs 9.35 d), but the relationship was no longer significant (Figure 1F). Furthermore, median LOS was lower in IgG-N negative patients (6.5 vs 7.5 d).
Because LOS-days and IgG-N status was significant, at least when not removing the extreme value, the full model was considered which included age, BMI, and sex, and duration of symptoms. However, in the full model, IgG-N was not significant when controlling for the other variables. This remained true when using a model without the extreme value.
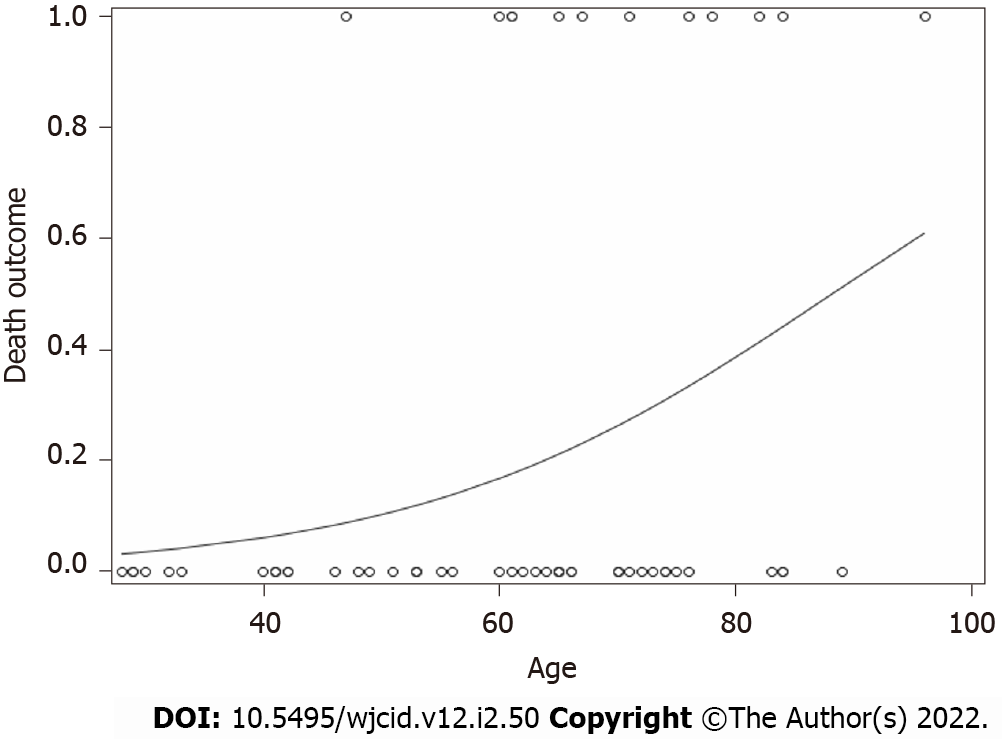
For death, only age (numerical) was a significant predictor (F value = 5.07, P value = 0.0283). As age increased, the probability of having an endpoint of one (Death) increased (Figure 2). No other variables for any of the outcomes were significant.
SARS-CoV-2 infection causes an atypical pneumonia like respiratory illness known as COVID-19 characterized by fever, dyspnea, anosmia and a worsening hypoxia[21,22]. Among those hospitalized with COVID-19, patients often required supplemental oxygen using HFNC, BiPAP and an increased admission into the ICU requiring mechanical ventilation depending on the severity of the respiratory failure and lung parenchymal involvement. The pathophysiology of COVID-19 is primarily an immune-mediated process with a variety of antibody signatures among which the IgG signatures were of interest to our study. A robust immune-mediated inflammatory cascade guides the pathophysiology of the COVID-19 illness[22-24]. Different proinflammatory cytokines such as IL-6 and TNF-α have been correlated with diseases severity[25].
There is some data available to understand the humoral response to SARS-CoV-2 infection and role of various IgG subtypes in the body’s line of defense. Much of it was inherited from the studies of SARS-CoV-1[26]. IgG antibodies directed towards the spike protein (IgG-S) and that of the nucleocapsid protein (IgG-N) are the two important components of humoral immunity against SARS-CoV-2 infection. SARS-CoV-2 uses the spike protein to bind to the target cell through its receptor-binding domain and therefore is the target site for neutralizing antibody, IgG-S[27]. The role of IgG-S in early viral clearance is crucial for favorable clinical course and survival[28-30]. IgG-S is considered the neutralizing antibody which may elicit a protection against SARS-CoV-2 by interfering with virion binding to host cell receptors, blocking cellular uptake and preventing endosomal processing of viral genome[13,27]. However, the kinetics of the antibody response becomes more complex to understand with current available literature, which is conflicting. In one interesting study, there is a link between the IgG-S response and COVID-19 severity, but the antibody response has to develop in a specific time window to improve viral clearance and disease outcomes. A faster antibody response was associated with better survival (within the first 14 d of infection) and deceased patients showed a slower antibody response although they reached higher IgG titers later in the disease trajectory[31]. Other studies have shown that, severely ill patients exhibit higher peak, faster and stronger antibody response compared to mildly symptomatic patients[13,32]. Severely ill COVID-19 patients have been found to produce a unique serologic signature with increased IgG-S with afucosylated Fc glycans. The Fc modification of IgG-S triggers activation of natural killers cells and enhances production of IL-6 and TNF-α by primary monocytes that results in more severe disease[33].
The role of IgG-N in the pathogenesis and clinical course of COVID-19 remains largely unknown. As to our current knowledge, severe COVID-19 is characterized by a series of inflammatory signatures including a cytokine storm, inflammatory alveolar infiltrates and formation of vascular microthrombi[33]. During the peak of the pandemic, clinicians took their chance to use different inflammatory markers such as C-reactive protein, platelet count, D-dimer and Ferritin, to name a few to monitor diseases progression and crisis planning. However, data to support the specificity of these inflammatory signatures as reliable prognostic markers for COVID-19 is limited[34,35]. As per one report by Batra et al[19], showed that titers of IgG-N at the time of admission can be a prognostic factor in the clinical course of the diseases and was associated with increased incidence of hypoxemia, admission into the ICU and extended length of stay in the hospital. In our study, we hypothesized that the presence of IgG-N at the time of admission into the hospital could be used as a marker of impending diseases severity and determine hospital course. We pursued a qualitative measurement of IgG-N on all our patients. Some key parameters such as the degree of hypoxemia, mean length of hospitalization, ICU admission, need for mechanical ventilation and patient outcome as in-hospital datasets were examined in our study group. We enrolled a total of 59 patients who were admitted with hypoxia secondary to COVID-19, out of which 26 (44%) patients had IgG-N antibody at the time of admission into the hospital. Our goal was to investigate the role of IgG-N as a marker to anticipate the clinical course in hospitalized patients. Based on our results, we concluded that IgG-N might not be a reliable predictor of COVID-19 diseases severity.
Our data indicate that age was a single independent predictor of death following hospitalization, which is in support of reports published earlier[36,37]. As age increased, the probability of death increased (Figure 2). Mortality rate was not significantly different in IgG-N positive group vs negative (Figure 1A). We did not find any statistical difference with the need to use HFNC between the two groups (Figure 1B). Many of our patient population had clinically progressive diseases with worsening respiratory failure requiring BiPAP or transfer to ICU to be intubated and placed on mechanical ventilation. After following the patient pool until discharge, we did not find any significant difference with the need to use BiPAP between the two groups with and without IgG-N at the time of admission (Figure 1C). The admission rate into the ICU and need for mechanical ventilation was not statistically significant either (Figures 1D and E). Although we saw an extended LOS among the IgG-N negative group, but after adjusting for the extreme outlier, the findings were no longer significant (Figure 1F). Furthermore, median LOS was actually lower in the IgG-N negative group, showing that the extreme value was skewing the LOS average.
Our study had several limitations. We did not measure the IgG-N antibody titers in our study and so we cannot imply if high vs low antibody titers have any direct impact on the disease severity and mortality in COVID-19. Since every individual patient was enrolled into the study only when they were symptomatic enough to meet criteria for hospitalization, especially hypoxic with oxygen saturation < 90%, it could be argued that they may be at different stage of the diseases course and different phase of the seroconversion. This could have confounded our findings since seroconversion and viral kinetics are time dependent phenomena. We did not standardize our patients based on their underlying comorbidities, which further could have influenced our results. More investigation using a larger sample size and different IgM/IgG subtypes is warranted to put more light in this area.
We have analyzed the presence or absence of IgG-N in patients admitted to the hospital with severe or critical COVID-19 illness and evaluated the effects of presence of IgG-N on clinical severity and outcome. Age happens to be the single independent risk factor for a worse outcome. Our analysis revealed no significant correlation between IgG-N status and degree of respiratory failure or mortality. The degree of respiratory failure was characterized by the utilization of high flow nasal canula, bilevel positive pressure ventilation and intubation with mechanical ventilation. IgG-N seroconversion had no significant effect on mean length of stay in the hospital. Further studies with large cohorts and risk-adjusted comorbidities are needed to demonstrate the more accurate role of IgG-N seroconversion on clinical outcome.
During the peak of the coronavirus disease 2019 (COVID-19) pandemic, hospitals and clinicians had to adapt quickly to the rapid spread of the infection in the community. In the absence of adequate literature, clinicians hypothesized and studied the utility of various protein markers to prognosticate their patients. We intended to study the correlation of anti-nucleocapsid-severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) antibody (IgG-N) presence with the clinical outcome in severely ill hospitalized COVID-19 patients.
We were interested in characterizing a correlation between presence or absence of IgG-N and clinical outcome in hospitalized COVID-19 patients. We wanted to test the ability of IgG-N in predicting the severity of illness, maximal oxygen support needed and final outcome in order to mobilize staff, manage intensive care unit (ICU) beds and ventilators as a part of the crisis management planning.
To study the effect of SARS-CoV-2 anti-nucleocapsid-IgG on COVID-19 diseases severity and outcome. We studied the effect of presence or absence of IgG-N on mean length of stay in the hospital, maximal oxygen support needed and mortality.
We conducted a retrospective cohort study on adults aged between 28-96, being admitted to the hospital with severe or critical COVID-19 illness. Blood sample was collected either at or within 48 h of admission to the hospital to check for SARS-CoV-2-IgG-N. A total of fifty nine patients were enrolled into the study. We utilized a binary logistic regression model to analyze the outcome data.
Our data demonstrated that the need for maximal oxygen support, mean length of stay in the hospital and mortality were not significantly different between the groups with or without IgG-N at the time of admission.
We concluded that IgG-N seroconversion had no significant correlation with the need for maximal oxygen support as well as mortality during the course of hospitalization. Length of stay in the hospital was not significantly different across the IgG-N status.
Our study demonstrated that presence of anti-nucleocapsid-IgG against SARS-CoV-2 infection had no impact on the clinical outcome or diseases severity in hospitalized COVID-19 patients. We did not find a correlation of statistical significance to use IgG-N as a biomarker to predict clinical outcome in COVID-19 patients admitted to a community hospital in North Dakota. Acknowledging the limitation of our study, we look forward to a future study with larger sample size and risk-adjusted comorbidities to investigate the association with better clarity.
We would like to acknowledge Ms. Shannon Yarbrough, for her contribution with medical library services.
Provenance and peer review: Unsolicited article; Externally peer reviewed.
Peer-review model: Single blind
Specialty type: Infectious diseases
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): D
Grade E (Poor): 0
P-Reviewer: Casaca W, Brazil; Freund O, Israel S-Editor: Liu JH L-Editor: A P-Editor: Liu JH
| 1. | Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, Si HR, Zhu Y, Li B, Huang CL, Chen HD, Chen J, Luo Y, Guo H, Jiang RD, Liu MQ, Chen Y, Shen XR, Wang X, Zheng XS, Zhao K, Chen QJ, Deng F, Liu LL, Yan B, Zhan FX, Wang YY, Xiao GF, Shi ZL. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579:270-273. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15248] [Cited by in RCA: 14123] [Article Influence: 2824.6] [Reference Citation Analysis (1)] |
| 2. | Lu H, Stratton CW, Tang YW. Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. J Med Virol. 2020;92:401-402. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1670] [Cited by in RCA: 1769] [Article Influence: 353.8] [Reference Citation Analysis (0)] |
| 3. | Zheng M, Gao Y, Wang G, Song G, Liu S, Sun D, Xu Y, Tian Z. Functional exhaustion of antiviral lymphocytes in COVID-19 patients. Cell Mol Immunol. 2020;17:533-535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1043] [Cited by in RCA: 1285] [Article Influence: 257.0] [Reference Citation Analysis (0)] |
| 4. | Zhang J, Litvinova M, Wang W, Wang Y, Deng X, Chen X, Li M, Zheng W, Yi L, Wu Q, Liang Y, Wang X, Yang J, Sun K, Longini IM Jr, Halloran ME, Wu P, Cowling BJ, Merler S, Viboud C, Vespignani A, Ajelli M, Yu H. Evolving epidemiology and transmission dynamics of coronavirus disease 2019 outside Hubei province, China: a descriptive and modelling study. Lancet Infect Dis. 2020;20:793-802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 458] [Cited by in RCA: 392] [Article Influence: 78.4] [Reference Citation Analysis (0)] |
| 5. | Liu R, Han H, Liu F, Lv Z, Wu K, Liu Y, Feng Y, Zhu C. Positive rate of RT-PCR detection of SARS-CoV-2 infection in 4880 cases from one hospital in Wuhan, China, from Jan to Feb 2020. Clin Chim Acta. 2020;505:172-175. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 404] [Cited by in RCA: 386] [Article Influence: 77.2] [Reference Citation Analysis (0)] |
| 6. | Muecksch F, Wise H, Batchelor B, Squires M, Semple E, Richardson C, McGuire J, Clearly S, Furrie E, Greig N, Hay G, Templeton K, Lorenzi JCC, Hatziioannou T, Jenks S, Bieniasz PD. Longitudinal Serological Analysis and Neutralizing Antibody Levels in Coronavirus Disease 2019 Convalescent Patients. J Infect Dis. 2021;223:389-398. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 209] [Cited by in RCA: 192] [Article Influence: 48.0] [Reference Citation Analysis (0)] |
| 7. | Ma H, Zeng W, He H, Zhao D, Jiang D, Zhou P, Cheng L, Li Y, Ma X, Jin T. Serum IgA, IgM, and IgG responses in COVID-19. Cell Mol Immunol. 2020;17:773-775. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 292] [Cited by in RCA: 351] [Article Influence: 70.2] [Reference Citation Analysis (0)] |
| 8. | Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O'Byrne A, Kouphou N, Galao RP, Betancor G, Wilson HD, Signell AW, Winstone H, Kerridge C, Huettner I, Jimenez-Guardeño JM, Lista MJ, Temperton N, Snell LB, Bisnauthsing K, Moore A, Green A, Martinez L, Stokes B, Honey J, Izquierdo-Barras A, Arbane G, Patel A, Tan MKI, O'Connell L, O'Hara G, MacMahon E, Douthwaite S, Nebbia G, Batra R, Martinez-Nunez R, Shankar-Hari M, Edgeworth JD, Neil SJD, Malim MH, Doores KJ. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598-1607. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1049] [Cited by in RCA: 933] [Article Influence: 186.6] [Reference Citation Analysis (0)] |
| 9. | To KK, Tsang OT, Leung WS, Tam AR, Wu TC, Lung DC, Yip CC, Cai JP, Chan JM, Chik TS, Lau DP, Choi CY, Chen LL, Chan WM, Chan KH, Ip JD, Ng AC, Poon RW, Luo CT, Cheng VC, Chan JF, Hung IF, Chen Z, Chen H, Yuen KY. Temporal profiles of viral load in posterior oropharyngeal saliva samples and serum antibody responses during infection by SARS-CoV-2: an observational cohort study. Lancet Infect Dis. 2020;20:565-574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2296] [Cited by in RCA: 2260] [Article Influence: 452.0] [Reference Citation Analysis (0)] |
| 10. | Moradi G, Mohamadi Bolbanabad A, Ahmadi S, Aghaei A, Bahrami F, Veysi A, Nasiri Kalmarzi R, Shokri A, Ghaderi E, Mohsenpour B, Mohammadi A. Persistence assessment of SARS-CoV-2-specific IgG antibody in recovered COVID-19 individuals and its association with clinical symptoms and disease severity: A prospective longitudinal cohort study. Int Immunopharmacol. 2021;98:107893. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 13] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 11. | Liu ZL, Liu Y, Wan LG, Xiang TX, Le AP, Liu P, Peiris M, Poon LLM, Zhang W. Antibody Profiles in Mild and Severe Cases of COVID-19. Clin Chem. 2020;66:1102-1104. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 46] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 12. | Xiao K, Yang H, Liu B, Pang X, Du J, Liu M, Liu Y, Jing X, Chen J, Deng S, Zhou Z, Yin L, Yan Y, Mou H, She Q. Antibodies Can Last for More Than 1 Year After SARS-CoV-2 Infection: A Follow-Up Study From Survivors of COVID-19. Front Med (Lausanne). 2021;8:684864. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 31] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 13. | Pang NY, Pang AS, Chow VT, Wang DY. Understanding neutralising antibodies against SARS-CoV-2 and their implications in clinical practice. Mil Med Res. 2021;8:47. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 14. | Chen W, Zhang J, Qin X, Wang W, Xu M, Wang LF, Xu C, Tang S, Liu P, Zhang L, Liu X, Zhang Y, Yi C, Hu Z, Yi Y. SARS-CoV-2 neutralizing antibody levels are correlated with severity of COVID-19 pneumonia. Biomed Pharmacother. 2020;130:110629. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 45] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 15. | Legros V, Denolly S, Vogrig M, Boson B, Siret E, Rigaill J, Pillet S, Grattard F, Gonzalo S, Verhoeven P, Allatif O, Berthelot P, Pélissier C, Thiery G, Botelho-Nevers E, Millet G, Morel J, Paul S, Walzer T, Cosset FL, Bourlet T, Pozzetto B. A longitudinal study of SARS-CoV-2-infected patients reveals a high correlation between neutralizing antibodies and COVID-19 severity. Cell Mol Immunol. 2021;18:318-327. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 246] [Article Influence: 61.5] [Reference Citation Analysis (0)] |
| 16. | Freund O, Tau L, Weiss TE, Zornitzki L, Frydman S, Jacob G, Bornstein G. Associations of vaccine status with characteristics and outcomes of hospitalized severe COVID-19 patients in the booster era. PLoS One. 2022;17:e0268050. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 19] [Article Influence: 6.3] [Reference Citation Analysis (1)] |
| 17. | Eroshenko N, Gill T, Keaveney MK, Church GM, Trevejo JM, Rajaniemi H. Implications of antibody-dependent enhancement of infection for SARS-CoV-2 countermeasures. Nat Biotechnol. 2020;38:789-791. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 90] [Cited by in RCA: 88] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 18. | Del Valle DM, Kim-Schulze S, Huang HH, Beckmann ND, Nirenberg S, Wang B, Lavin Y, Swartz TH, Madduri D, Stock A, Marron TU, Xie H, Patel M, Tuballes K, Van Oekelen O, Rahman A, Kovatch P, Aberg JA, Schadt E, Jagannath S, Mazumdar M, Charney AW, Firpo-Betancourt A, Mendu DR, Jhang J, Reich D, Sigel K, Cordon-Cardo C, Feldmann M, Parekh S, Merad M, Gnjatic S. An inflammatory cytokine signature predicts COVID-19 severity and survival. Nat Med. 2020;26:1636-1643. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1894] [Cited by in RCA: 1793] [Article Influence: 358.6] [Reference Citation Analysis (0)] |
| 19. | Batra M, Tian R, Zhang C, Clarence E, Sacher CS, Miranda JN, De La Fuente JRO, Mathew M, Green D, Patel S, Bastidas MVP, Haddadi S, Murthi M, Gonzalez MS, Kambali S, Santos KHM, Asif H, Modarresi F, Faghihi M, Mirsaeidi M. Role of IgG against N-protein of SARS-CoV2 in COVID19 clinical outcomes. Sci Rep. 2021;11:3455. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 59] [Cited by in RCA: 62] [Article Influence: 15.5] [Reference Citation Analysis (0)] |
| 20. | Tan W, Lu Y, Zhang J, Wang J, Dan Y, Tan Z, He X, Qian C, Sun Q, Hu Q, Liu H, Ye S, Xiang X, Zhou Y, Zhang W, Guo Y, Wang XH, He W, Wan X, Sun F, Wei Q, Chen C, Pan G, Xia J, Mao Q, Chen Y, Deng G. Viral Kinetics and Antibody Responses in Patients with COVID-19. Infectious Diseases. 2020;. [DOI] [Full Text] |
| 21. | Alimohamadi Y, Sepandi M, Taghdir M, Hosamirudsari H. Determine the most common clinical symptoms in COVID-19 patients: a systematic review and meta-analysis. J Prev Med Hyg. 2020;61:E304-E312. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 119] [Reference Citation Analysis (1)] |
| 22. | Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, Zhang L, Fan G, Xu J, Gu X, Cheng Z, Yu T, Xia J, Wei Y, Wu W, Xie X, Yin W, Li H, Liu M, Xiao Y, Gao H, Guo L, Xie J, Wang G, Jiang R, Gao Z, Jin Q, Wang J, Cao B. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. 2020;395:497-506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35178] [Cited by in RCA: 30106] [Article Influence: 6021.2] [Reference Citation Analysis (3)] |
| 23. | Tang F, Zhang X, Zhang B, Zhu B, Wang J. A nomogram prediction of outcome in patients with COVID-19 based on individual characteristics incorporating immune response-related indicators. J Med Virol. 2022;94:131-140. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 24. | Xu Z, Shi L, Wang Y, Zhang J, Huang L, Zhang C, Liu S, Zhao P, Liu H, Zhu L, Tai Y, Bai C, Gao T, Song J, Xia P, Dong J, Zhao J, Wang FS. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir Med. 2020;8:420-422. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5228] [Cited by in RCA: 5781] [Article Influence: 1156.2] [Reference Citation Analysis (2)] |
| 25. | Yuki K, Fujiogi M, Koutsogiannaki S. COVID-19 pathophysiology: A review. Clin Immunol. 2020;215:108427. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1218] [Cited by in RCA: 1191] [Article Influence: 238.2] [Reference Citation Analysis (0)] |
| 26. | Cevik M, Tate M, Lloyd O, Maraolo AE, Schafers J, Ho A. SARS-CoV-2, SARS-CoV, and MERS-CoV viral load dynamics, duration of viral shedding, and infectiousness: a systematic review and meta-analysis. Lancet Microbe. 2021;2:e13-e22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 937] [Cited by in RCA: 999] [Article Influence: 249.8] [Reference Citation Analysis (0)] |
| 27. | Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. 2022;23:3-20. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1831] [Cited by in RCA: 1913] [Article Influence: 637.7] [Reference Citation Analysis (0)] |
| 28. | Dispinseri S, Secchi M, Pirillo MF, Tolazzi M, Borghi M, Brigatti C, De Angelis ML, Baratella M, Bazzigaluppi E, Venturi G, Sironi F, Canitano A, Marzinotto I, Tresoldi C, Ciceri F, Piemonti L, Negri D, Cara A, Lampasona V, Scarlatti G. Neutralizing antibody responses to SARS-CoV-2 in symptomatic COVID-19 is persistent and critical for survival. Nat Commun. 2021;12:2670. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 270] [Cited by in RCA: 278] [Article Influence: 69.5] [Reference Citation Analysis (0)] |
| 29. | Yu J, Tostanoski LH, Peter L, Mercado NB, McMahan K, Mahrokhian SH, Nkolola JP, Liu J, Li Z, Chandrashekar A, Martinez DR, Loos C, Atyeo C, Fischinger S, Burke JS, Slein MD, Chen Y, Zuiani A, Lelis FJN, Travers M, Habibi S, Pessaint L, Van Ry A, Blade K, Brown R, Cook A, Finneyfrock B, Dodson A, Teow E, Velasco J, Zahn R, Wegmann F, Bondzie EA, Dagotto G, Gebre MS, He X, Jacob-Dolan C, Kirilova M, Kordana N, Lin Z, Maxfield LF, Nampanya F, Nityanandam R, Ventura JD, Wan H, Cai Y, Chen B, Schmidt AG, Wesemann DR, Baric RS, Alter G, Andersen H, Lewis MG, Barouch DH. DNA vaccine protection against SARS-CoV-2 in rhesus macaques. Science. 2020;369:806-811. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 925] [Cited by in RCA: 855] [Article Influence: 171.0] [Reference Citation Analysis (0)] |
| 30. | Folegatti PM, Ewer KJ, Aley PK, Angus B, Becker S, Belij-Rammerstorfer S, Bellamy D, Bibi S, Bittaye M, Clutterbuck EA, Dold C, Faust SN, Finn A, Flaxman AL, Hallis B, Heath P, Jenkin D, Lazarus R, Makinson R, Minassian AM, Pollock KM, Ramasamy M, Robinson H, Snape M, Tarrant R, Voysey M, Green C, Douglas AD, Hill AVS, Lambe T, Gilbert SC, Pollard AJ; Oxford COVID Vaccine Trial Group. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467-478. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1962] [Cited by in RCA: 1772] [Article Influence: 354.4] [Reference Citation Analysis (0)] |
| 31. | Lucas C, Klein J, Sundaram M, Liu F, Wong P, Silva J, Mao T, Oh JE, Tokuyama M, Lu P, Venkataraman A, Park A, Israelow B, Wyllie AL, Vogels CBF, Muenker MC, Casanovas-Massana A, Schulz WL, Zell J, Campbell M, Fournier JB, Grubaugh ND, Farhadian S, Wisnewski AV, Cruz CD, Omer S, Ko AI, Ring A, Iwasaki A. Kinetics of antibody responses dictate COVID-19 outcome. medRxiv. 2020;. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 37] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 32. | Lau EHY, Tsang OTY, Hui DSC, Kwan MYW, Chan WH, Chiu SS, Ko RLW, Chan KH, Cheng SMS, Perera RAPM, Cowling BJ, Poon LLM, Peiris M. Neutralizing antibody titres in SARS-CoV-2 infections. Nat Commun. 2021;12:63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 291] [Cited by in RCA: 262] [Article Influence: 65.5] [Reference Citation Analysis (0)] |
| 33. | Chakraborty S, Gonzalez J, Edwards K, Mallajosyula V, Buzzanco AS, Sherwood R, Buffone C, Kathale N, Providenza S, Xie MM, Andrews JR, Blish CA, Singh U, Dugan H, Wilson PC, Pham TD, Boyd SD, Nadeau KC, Pinsky BA, Zhang S, Memoli MJ, Taubenberger JK, Morales T, Schapiro JM, Tan GS, Jagannathan P, Wang TT. Proinflammatory IgG Fc structures in patients with severe COVID-19. Nat Immunol. 2021;22:67-73. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 226] [Cited by in RCA: 238] [Article Influence: 59.5] [Reference Citation Analysis (0)] |
| 34. | Ponti G, Maccaferri M, Ruini C, Tomasi A, Ozben T. Biomarkers associated with COVID-19 disease progression. Crit Rev Clin Lab Sci. 2020;57:389-399. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 467] [Cited by in RCA: 536] [Article Influence: 107.2] [Reference Citation Analysis (0)] |
| 35. | Xiao AT, Gao C, Zhang S. Profile of specific antibodies to SARS-CoV-2: The first report. J Infect. 2020;81:147-178. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 156] [Cited by in RCA: 163] [Article Influence: 32.6] [Reference Citation Analysis (0)] |
| 36. | Sun H, Ning R, Tao Y, Yu C, Deng X, Zhao C, Meng S, Tang F, Xu D. Risk Factors for Mortality in 244 Older Adults With COVID-19 in Wuhan, China: A Retrospective Study. J Am Geriatr Soc. 2020;68:E19-E23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 136] [Article Influence: 27.2] [Reference Citation Analysis (0)] |
| 37. | Collaborative TO, Williamson E, Walker AJ, Bhaskaran K, Bacon S, Bates C, Morton CE, Curtis HJ, Mehrkar A, Evans D, Inglesby P, Cockburn J, McDonald HI, MacKenna B, Tomlinson L, Douglas IJ, Rentsch CT, Mathur R, Wong A, Grieve R, Harrison D, Forbes H, Schultze A, Croker R, Parry J, Hester F, Harper S, Perera R, Evans S, Smeeth L, Goldacre B. OpenSAFELY: factors associated with COVID-19-related hospital death in the linked electronic health records of 17 million adult NHS patients. Published online, 2020. [DOI] [Full Text] |