Copyright
©The Author(s) 2016.
World J Clin Infect Dis. May 25, 2016; 6(2): 6-21
Published online May 25, 2016. doi: 10.5495/wjcid.v6.i2.6
Published online May 25, 2016. doi: 10.5495/wjcid.v6.i2.6
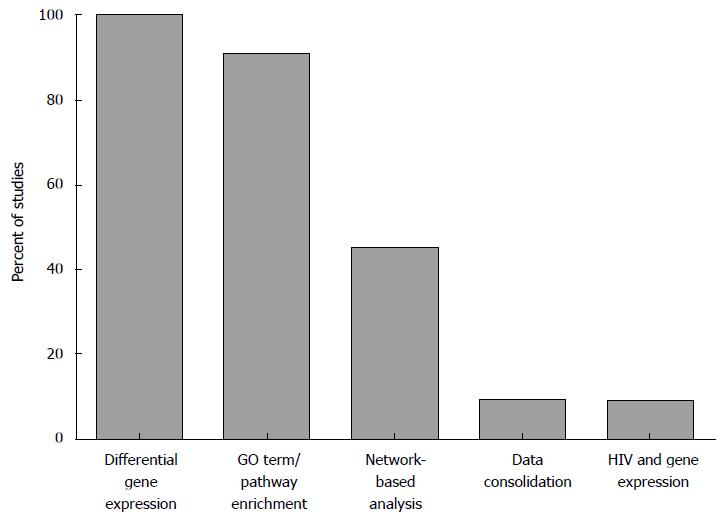
Figure 1 Summary of methods used across gene expression profiling studies in the field of human immunodeficiency virus latency and eradication.
Identification of DEGs and functional analysis of GO terms and pathways enriched for DEGs are the methods that are most commonly used across studies. Network-based analyses are used in a subset of studies; while methods that consolidate host gene expression with other data types (e.g., proteomics or HIV expression data) are scarce. DEGs: Differentially expressed genes; GO: Gene ontology; HIV: Human immunodeficiency virus.
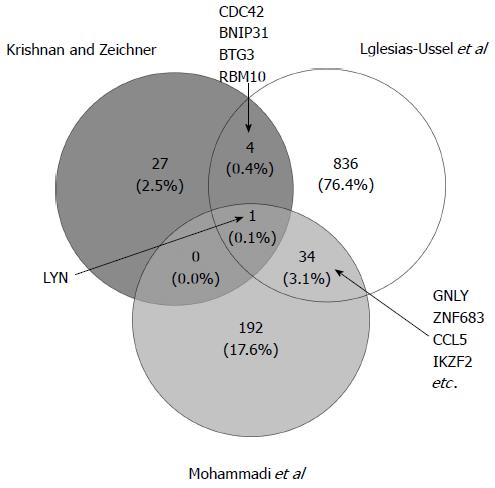
Figure 2 Venn diagram depicting differentially expressed genes across three latency models.
The overlapping genes were identified using the online tool Venny (http://bioinfogp.cnb.csic.es/tools/venny/index.html). Shown are the total number of differentially expressed genes and percent of total identified across all models[18,19,42]. For each overlap, gene symbols are listed. For the overlap between Iglesias-Ussel et al[19] and Mohammadi et al[42] studies, the four genes with the highest average absolute fold change are listed.
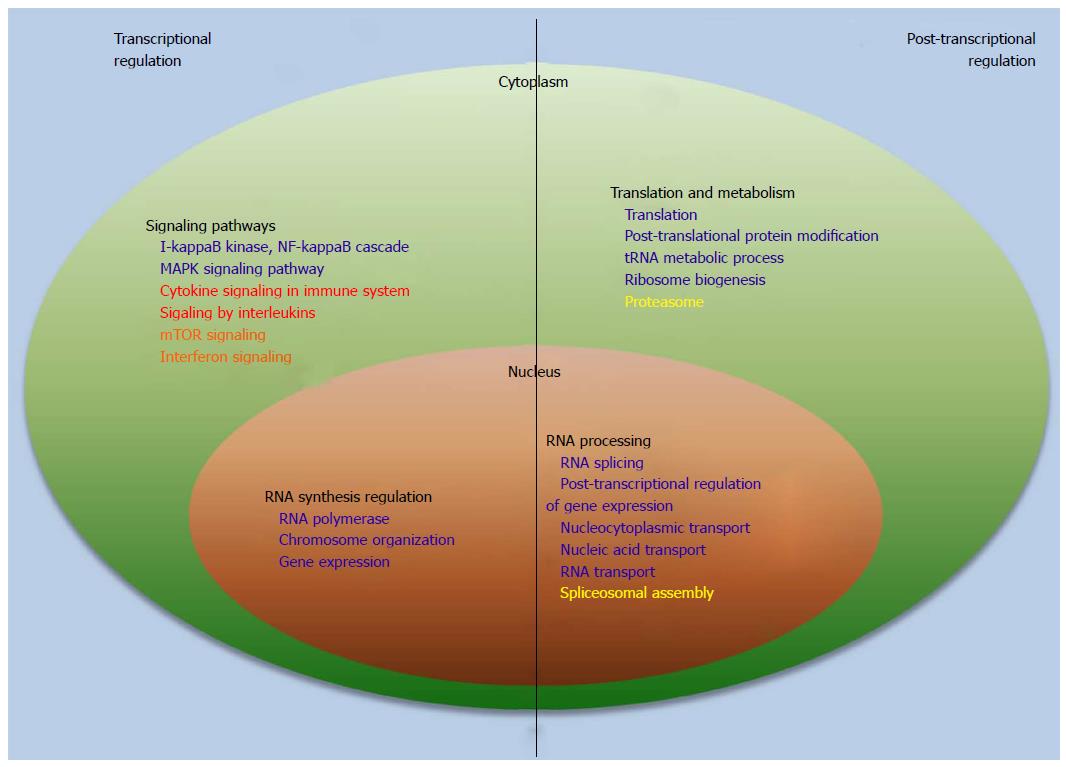
Figure 3 Transcriptional and post-transcriptional mechanisms of regulation of human immunodeficiency virus expression.
Pathway and GO term categories related to transcriptional and post-transcriptional regulation of HIV expression, identified in gene expression studies that compared latently infected and uninfected cells, are shown. Dark blue, Iglesias-Ussel et al[19]; Red, Mohammadi et al[42]; Brown, Evans et al[76]; Yellow, Krishnan and Zeichner[18]. GO: Gene ontology; HIV: Human immunodeficiency virus; mTOR: Mammalian target of rapamycin.
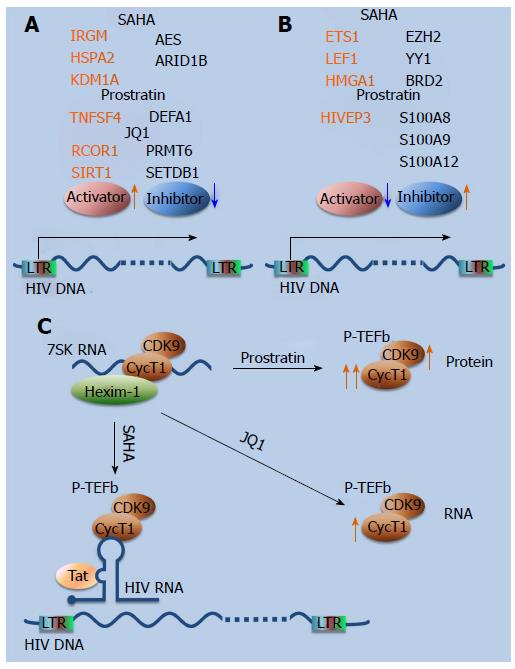
Figure 4 Main findings from gene expression studies using Latency reversing agents.
A: Novel mechanisms of HIV reactivation besides primary mechanisms of action of LRAs. These include upregulation (red arrow) of HIV activators (red oval) and downregulation (blue arrow) of repressors (blue oval). Examples for LRAs from 3 functional classes (HDACi, SAHA; PKC agonist, Prostratin; and bromodomain inhibitor, JQ1) are listed; B: Effects of LRAs on host genes that are inhibitory for HIV reactivation. These include upregulation (red arrow) of HIV repressors (blue oval) and downregulation (blue arrow) of activators (red oval). Examples for LRAs from 2 functional classes (HDACi, SAHA; and PKC agonist, Prostratin) are shown; C: LRAs of different classes act on components of p-TEFb complex via different mechanisms, contributing to HIV reactivation. SAHA induced dissociation of p-TEFb from the inactive 7SK RNA complex and facilitated its recruitment to the HIV LTR. Prostrain and JQ1 upregulated components of p-TEFb complex at the protein and RNA level, respectively (red arrows indicate upregulation). LRA: Latency reversing agent; HDACi: Histone deacetylase inhibitor; PKC: Protein kinase C; SAHA: Suberoylanilide hydroxamic acid; IGRM: Immunity-related GTPase family, M; HSPA2: Heat shock 70 kDA protein 2; KDM1A: Lysine (K)-specific demethylase; TNFSF4: Tumor necrosis factor (ligand) superfamily, member 4; RCOR1: REST coreceptor 1; SIRT1: Sirtuin 1; AES: Amino-terminal enhancer of split; ARID1B: AT rich interactive domain 1B, SWI1-like; DEFA1: Defensin alpha 1; PRMT6: Protein arginine methyltransferase 6; SETDB1: SET domain, bifurcated 1; ETS1: V-Ets avian erythroblastosis virus E26 oncogene homolog 1; LEF1: Lymphoid enhancer-binding factor 1; HMGA1: High mobility group AT-hook 1; HIVEP3: HIV type I enhancer binding protein 3; EZH2: Enhancer of zeste 2 polycomb repressive complex 2 subunit; YY1: YY1 transcription factor; BRD2: Bromodomain protein containing 2; S100A8: S100 Calcium Binding Protein A8; S100A9: S100 Calcium Binding Protein A9; S100A12: S100 Calcium Binding Protein A12; CDK9: Cyclin-dependent kinase 9; P-TEFb: Positive transcription elongation factor; CycT1: Cyclin T1; Hexim-1: Hexamethylene Bis-Acetamide Inducible 1; LTR: Long terminal repeat; Tat: Transactivator of transcription.
- Citation: White CH, Moesker B, Ciuffi A, Beliakova-Bethell N. Systems biology applications to study mechanisms of human immunodeficiency virus latency and reactivation. World J Clin Infect Dis 2016; 6(2): 6-21
- URL: https://www.wjgnet.com/2220-3176/full/v6/i2/6.htm
- DOI: https://dx.doi.org/10.5495/wjcid.v6.i2.6