Published online Sep 20, 2024. doi: 10.5493/wjem.v14.i3.94999
Revised: May 20, 2024
Accepted: June 21, 2024
Published online: September 20, 2024
Processing time: 152 Days and 17 Hours
A significant subset of individuals with epilepsy fails to respond to currently available antiepileptic drugs, resulting in heightened mortality rates, psychosocial challenges, and a diminished quality of life. Genetic factors, particularly within the SCN1A gene, and the pro-inflammatory cytokine response is important in intricating the drug resistance in idiopathic epilepsy cases. In this extended study, we determined the correlation of rs6732655A/T single nucleotide polymorphism to understand the causative association of SCN1A gene with epilepsy drug resis
To find the correlation of SCN1A gene rs6732655A/T polymorphism with the drug-resistant epilepsy and inflammatory response.
The study enrolled 100 age and sex-matched patients of both drug-resistant and drug-responsive epilepsy cases. We analysed the rs6732655A/T polymorphism to study its association and causative role in drug-resistant epilepsy cases using restriction fragment length polymorphism technique. The diagnostic performance of interleukin (IL)-1β, IL-6, and high mobility group box 1 (HMGB1) protein levels was evaluated in conjunction with genotypic outcome receiver operating characteristic analysis.
AT and AA genotypes of rs6732655 SCN1A gene polymorphism were associated with higher risk of drug resistance epilepsy. Serum biomarkers IL-6, IL1β and HMGB1 demonstrated diagnostic potential, with cutoff values of 4.63 pg/mL, 59.52 pg/mL and 7.99 ng/mL, respectively, offering valuable tools for epilepsy management. Moreover, specific genotypes (AA and AT) were found to be linked to the elevated levels of IL-1β and IL-6 and potentially reflecting increased oxidative stress and neuro-inflammation in drug-resistant cases supporting the previous reported outcome of high inflammatory markers response in drug resistance epilepsy.
SCN1A genotypes AA and AT are linked to higher drug-resistant epilepsy risk. These findings underscore the potential influence of inflammation and genetics on epilepsy treatment resistance.
Core Tip: Genetic factors, including SCN1A gene variants, and their pro-inflammatory cytokine response [interleukin (IL)-1β, IL-6, and high mobility group box 1 protein (HMGB1)], play crucial roles in drug-resistant epilepsy. This study investigates the correlation between SCN1A gene variants (rs6732655A/T) and drug resistance in epilepsy, confirming higher levels of IL-1β, IL-6, and HMGB1 in drug-resistant cases and suggesting specific genotypes (AA and AT) as potential biomarkers for oxidative stress and neuro-inflammation in drug-resistant idiopathic epilepsy.
- Citation: Viswas A, Dabla PK, Shrivastav D, Gupta S, Yadav M, Yadav S, Koner BC. SCN1A rs6732655A/T polymorphism: Diagnostic and therapeutic insights for drug-resistant epilepsy. World J Exp Med 2024; 14(3): 94999
- URL: https://www.wjgnet.com/2220-315x/full/v14/i3/94999.htm
- DOI: https://dx.doi.org/10.5493/wjem.v14.i3.94999
Epilepsy is a prevalent neurological disorder afflicting millions of people worldwide. Despite eminent progress in the
Numerous single nucleotide polymorphisms (SNPs) have been investigated in the context of drug-resistant epilepsy. These studies have primarily focused on genes related to multidrug resistance, such as multi-drug resistance 1 (MDR1) and multidrug resistance-associated protein 2 (MRP2), voltage-gated sodium channel subunits, including SCN1, SCN2, and SCN3, and metabolic enzymes like CYP2 and CYP3. The significance of voltage-gated sodium channels in this context is particularly noteworthy, given their relevance to AEDs that target sodium channels[4]. Recently, five novel de novo and inherited pathogenic/likely pathogenic SCN1A variants were identified in a Moroccan child with Dravet syndrome through exome analysis and confirmed by Sanger sequencing, including a novel pathogenic splice site variant (SCN1A c.965-2A>G). These findings enrich the mutations database for this major epilepsy gene and provide critical reference data to guide genetic counselling for SCN1A-related epilepsy disorders ranging from mild to severe phenotypes like Dravet syndrome[5,6]. Previous studies have investigated potential associations between genetic polymorphisms in the SCN1A gene and drug resistance in epilepsy, with some reporting significant findings. Margari et al[7] found that the AA genotype of the SCN1A rs6732655A/T polymorphism was associated with an increased risk of drug-resistant epilepsy.
In our previous study, we found an association between rs10167228A/T SNP in the SCN1A gene and drug resistance in epilepsy. There are several genetic variants of interest due to their potential implications in the development of drug resistance in the context of idiopathic generalized epilepsy (IGE)[8].
In this study, we focused on the rs6732655A/T SNP in the SCN1A gene and its interactions that may lead to a pro-inflammatory response. Furthermore, we recently reported the causative association of rs10167228A/T SNP of SCN1A gene as a risk factor for drug-resistant epilepsy. We also found that high mobility group box 1 protein (HMGB1), inter
In this study, we aimed to determine the correlation of specific genetic polymorphisms SNPrs6732655A/T within the SCN1A gene, which encodes a crucial sodium channel subunit and is intimately related to AEDs designed to act on these sodium channels. In addition to the genetic aspect, we also delve into the intricate web of neuro-inflammation, an emer
The study was performed out at the Department of Biochemistry and Department of Neurology, Govind Ballabh Pant Institute of postgraduate medical education and research, tertiary care super specialty hospital New Delhi, India. It invol
A total of 5 mL venous blood was collected under aseptic conditions. Blood samples were collected in plain vials and in ethylenediamine tetra acetic acid vials for biochemical parameters and inflammatory cytokine level analysis. The levels of serum IL-1β and IL-6 were estimated using a commercial enzyme-linked immunosorbent assay (ELISA) kit.
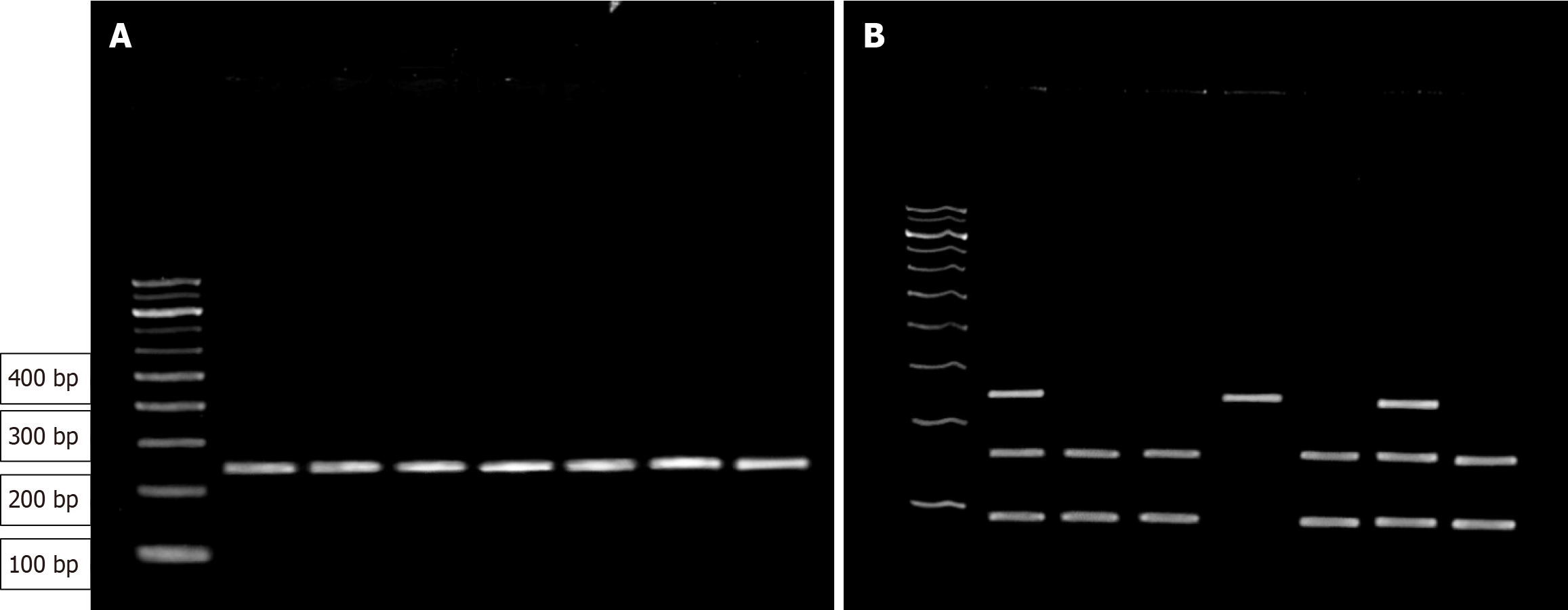
A total of 3 mL peripheral whole blood samples underwent DNA extraction via the column binding method utilizing the ReliaPrep™ Blood gDNA Miniprep System. A meticulous 1% agarose gel was prepared for electrophoresis, and the analysis of the rs6732655A/T polymorphism, the relevant region of the SCN1A gene was amplified via polymerase chain reaction (PCR) using specific primers (Table 1) and PCR conditions. PCR products were subjected to digestion with TaqI restriction endonuclease enzyme. This restriction fragment length polymorphism (RFLP)-PCR process exhibited specific outcomes for different alleles: The A allele remained uncut, yielding a single 250 bp band, while the T allele was cleaved into two smaller fragments (95 and 155 bp). Heterozygotes displayed a combination of both alleles, generating bands of 250 bp, 155 bp, and 95 bp. After PCR amplification, the products were resolved through electrophoresis in a 3% agarose gel (Figure 1).
| SNP | Primers | Sequence | Restriction enzyme |
| rs6732655A/T | Forward primer | 5’-CCAAATGGTGACACAGTGAA-3’ | RsaI |
| Reverse primer | 5’-GCCTTGATCACTTGTAGGACTTTT-3’ |
Data analysis was conducted using SPSS version 21 (IBM Corp., Armonk, NY, United States). Quantitative data were expressed using mean, standard deviation, frequency, and percentage for qualitative data. An independent t-test was used to compare two independent variables, and the normality of the data was assessed using the Kolmogorov-Smirnov test. Parametric and non-parametric variables were compared using Student t-tests, ANOVA, Fisher's exact test, Mann-Whitney U tests and Kruskal Wallis test. Receiver operating characteristic (ROC) curve analysis was employed for prediction analysis and used to measure the specificity and sensitivity. Statistical significance was set at P < 0.05 for all tests.
In our previous report, we found that the mean age in the drug-resistant IGE group (cases) and the drug-responsive IGE group (controls) was 27.30 ± 7.0 years (26 males and 24 females) and 22.74 ± 7.3 years (21 males and 29 females), respectively. The serum levels of IL-1β were 91.81 (65.09-126.94) pg/mL vs 35.23 (15.05-43.87) pg/mL; IL-6 was 7.15 (5.0-18.13) pg/mL vs 2.97 (2.44-4.03) pg/mL; and HMGB-1 was 11.29 (9.11-16.93) ng/mL vs 6.02 (4.44-7.87) ng/mL, all of which were significantly higher in drug-resistant cases compared to drug-responsive cases, respectively[8].
Our investigation for the rs6732655 polymorphism revealed that within the drug-resistant group, nine subjects (18%) had the AA genotype, 17 subjects (34%) possessed the AT genotype, and 24 subjects (48%) carried the TT genotype. In contrast, within the drug-responsive group, five subjects (10%) exhibited the AA genotype, 24 subjects (48%) had the AT genotype, and 21 subjects (42%) bore the TT genotype. Similarly, when comparing the distribution of rs6732655 genotypes across both study groups, our analysis demonstrated no statistically significant difference, with a P value of 0.281 (Table 2).
| SNP | Allele type | Drug resistant, n = 50 | Drug responsive, n = 50 | χ2 | P value |
| rs6732655 | AA | 9 (18) | 5 (10) | 2.538 | 0.281 |
| AT | 17 (34) | 24 (48) | |||
| TT | 24 (48) | 21 (42) |
Risk analysis was performed to examine the relationship between distinct rs6732655 genotypes and drug resistance. The AT genotype exhibited a higher risk association with an odds ratio of 1.58 [95% confidence interval (CI): 0.456-5.444]; (P value = 0.47). Conversely, a lower risk association was observed with the AA genotype, characterized by an odds ratio of 0.62 (95%CI: 0.264-1.456, P = 0.27) (Table 3).
| SNP | Genotype | OR | 95%CI | P value |
| rs6732655 | AA | 0.62 | 0.264-1.456 | 0.27 |
| AT | 1.58 | 0.456-5.444 | 0.47 | |
| TT | Reference | Reference | Reference |
Serum levels of IL-1β, IL-6, and HMGB1 in rs6732655 genotype: Analysis of the serum levels of IL-1β, IL-6, and HMGB1 in both drug-resistant and drug-responsive cases was performed. In the drug-resistant group, the median serum IL-1β levels for the AA, AT, and TT genotypes were 150.05 pg/mL (IQR: 114.35-194 pg/mL), 89.35 pg/mL (IQR: 81.55-105.22 pg/mL), and 71.36 pg/mL (IQR: 25.80-126.67 pg/mL), respectively. The level of serum IL-1β among the different genotypes was significantly different (P = 0.003). Similarly, the median serum IL-1β levels for the AA, AT, and TT genotypes within the drug-responsive group were 56.71 pg/mL (IQR: 48.97-57.77 pg/mL), 33.21 pg/mL (IQR: 15.18-40.68 pg/mL), and 30.00 pg/mL (IQR: 14.34-42.25 pg/mL), respectively. Similar to the IL-1β serum levels in drug-resistant patients, the levels of IL-1β were also significantly different among the different genotypes of the drug responsive cases (P = 0.004).
In the drug-resistant group, the median IL-6 levels serum were significantly different between the three genotypes (P ≤ 0.01). The IL-6 values for the AT, AA, and TT genotypes were 29.06 pg/mL (IQR: 14.73-51.75 pg/mL), 6.89 pg/mL (IQR: 5.06-11.47 pg/mL), and 5.61 pg/mL (IQR: 3.17-11.49 pg/mL), respectively. In the three genotypes of the drug-responsive group, the IL-6 levels for AA, AT, and TT were 5.02 pg/mL (IQR: 4.48-8.27 pg/mL), 2.95 pg/mL (IQR: 2.17-3.77 pg/mL), respectively.
Furthermore, within the drug-resistant group, the median serum HMGB1 levels for the AA, AT, and TT genotypes were 17.21 ng/mL (IQR: 15.56-26.23 ng/mL), 10.72 ng/mL (IQR: 9.85-11.92 ng/mL), and 9.39 ng/mL (IQR: 7.89-19.92 ng/mL) respectively, with significant differences observed (P = 0.005). However, there was no significant difference in the median HMGB1 serum levels of the AA, AT, and TT genotypes [AA-7.98 ng/mL (IQR: 7.14-8.68), AT-5.86 ng/mL (IQR: 4.11-6.91 ng/mL), TT-5.92 ng/mL (IQR: 3.94-7.87 ng/mL; P = 0.06] (Table 4).
| Parameter | SNP | Allele type | Drug resistant, n = 50 | P value | Drug responsive, n = 50 | P value |
| IL-1β in pg/mL | rs6732655 | AA | 150.05 (114.35-194) | 0.003a | 56.71 (48.97-57.77) | 0.004a |
| AT | 89.35 (81.55-105.22) | 33.21 (15.18-40.68) | ||||
| TT | 71.36 (25.80-126.67) | 30.00 (14.34-42.25) | ||||
| IL-6 in pg/mL | rs6732655 | AA | 29.06 (14.73-51.75) | 0.001a | 5.02 (4.48-8.27) | 0.005a |
| AT | 6.89 (5.06-11.47) | 2.95 (2.17-3.77) | ||||
| TT | 5.61 (3.17-11.49) | 2.82 (2.49-3.85) | ||||
| HMGB-1 in ng/mL | rs6732655 | AA | 17.21 (15.56-26.23) | 0.005a | 7.98 (7.14-8.68) | 0.067 |
| AT | 10.72 (9.85-11.92) | 5.86 (4.11-6.91) | ||||
| TT | 9.39 (7.89-19.92) | 5.92 (3.94-7.87) |
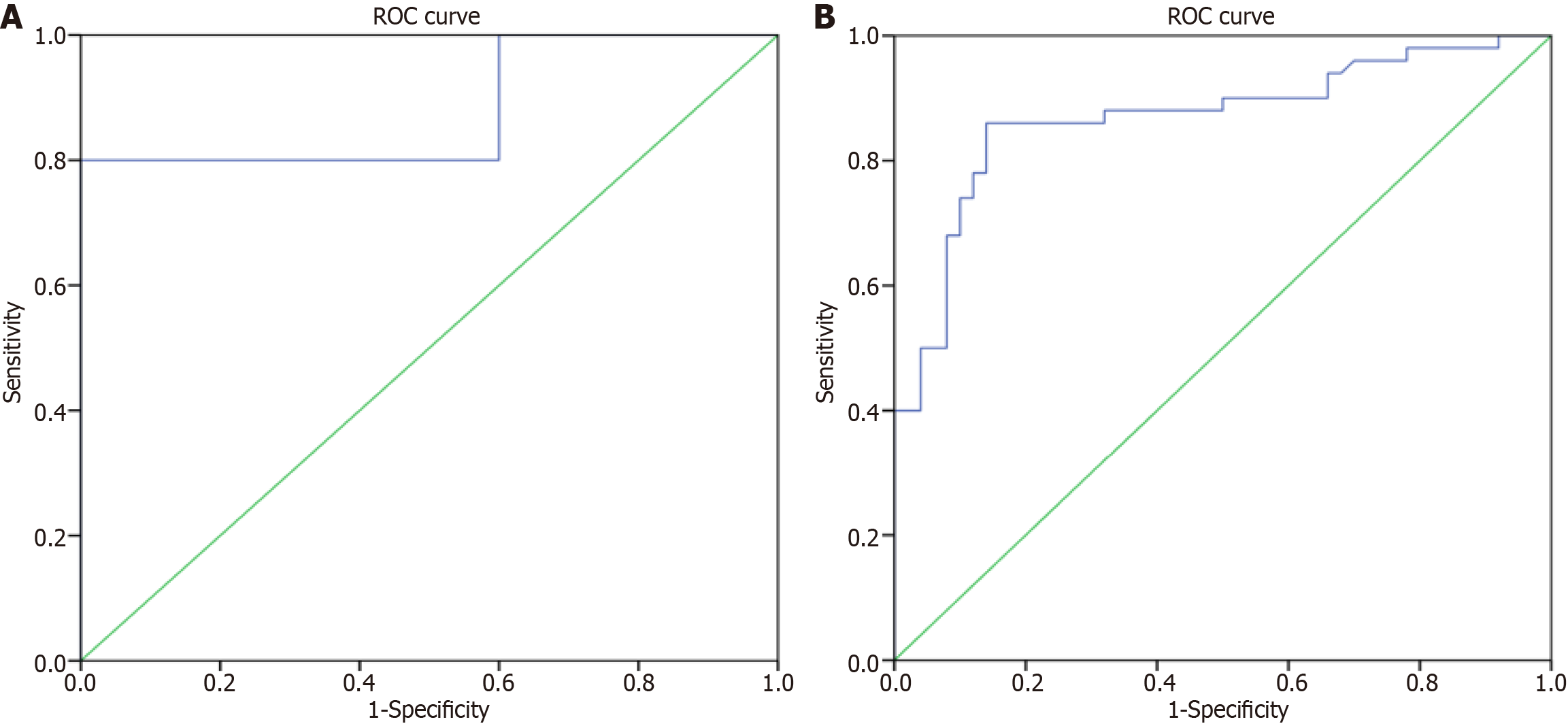
ROC analysis was conducted to evaluate the diagnostic performance of serum IL-1β, IL-6, and HMGB1 levels in distinguishing between drug-resistant and drug-responsive groups. The cutoff value for serum IL-1β levels was determined to be 59.52 pg/mL. Additionally, ROC analysis was performed to assess the diagnostic performance of serum IL-6 levels, with a cutoff value of 4.63 pg/mL. Furthermore, serum HMGB1 levels were also subjected to ROC analysis, revealing a cutoff value of 7.99 ng/mL. These analyses aimed to provide insights into the ability of these serum biomarkers to discriminate between drug-resistant and drug-responsive groups, offering potential diagnostic utility for epilepsy management (Table 5).
| Factor | AUC | Asymptotic 95%CI | Optimum cutoff | Sensitivity | Specificity | P value | |
| Lower bound | Upper bound | ||||||
| IL-1β | 0.880 | 0.808 | 0.952 | 59.52 | 80.0% | 100% | < 0.001 |
| IL-6 | 0.869 | 0.795 | 0.943 | 4.63 | 86.0% | 86.0% | < 0.001 |
Despite the availability of diverse AEDs, over one-third of epilepsy patients remain resistant to treatment, experiencing elevated mortality, psychosocial challenges, and reduced quality of life, often succumbing to SUDEP. Identifying SNPs associated with drug-resistant epilepsy is complex due to its multifaceted nature. Prominent SNPs studied include those in multidrug resistance genes (MDR1, MRP2), voltage-gated sodium channel subunits (SCN1, SCN2, SCN3), and meta
In our previously study, we observed elevated levels of inflammatory markers and oxidative stress in the drug-resistant cases, the comparison of the rs10167228 genotype between drug-resistant and drug-responsive groups did not reveal a significant association[8]. In this study, assessment of specific genotype associations with drug resistance compared to drug-sensitive individuals revealed that the drug-resistant population had 48% TT genotype, 34% AT genotype, and 18% AA genotype (Table 3). Furthermore, we found that the rs6732655 polymorphism is associated with a 1.5-fold higher risk in individuals with the AT genotype (Table 4). Similarly, various studies have shown a correlation between the severity of epileptic seizures and increased levels of cytokines in the blood plasma (HMGB-1, TLR4, IL-1β, IL-1R1, and TNF-α) of patients with drug-resistant epilepsy[9,10]. Increased levels of cytokines have also been observed in cerebrospinal fluid (IL-1β)[11]. Abe et al[12] found a link between the SCN1A gene's functional polymorphism rs3812718 (c.603-91G>A) and pharmacological response in terms of carbamazepine (CBZ) and phenytoin effective dosage. A population-specific link with CBZ and multiple drug resistant epilepsy has been described for the SCN1A IVS5-91GA intronic polymorphism of the SCN1A gene. Margari et al[7] suggested that intronic polymorphisms rs6730344, rs6732655, and rs10167228 within the SCN1A gene may represent potential factors that increase the risk of drug resistance. Additionally, the presence of the AA and AT genotypes in the rs1962842 intronic polymorphism has also been identified as a contributing risk factor in the drug-resistant patient group. As a result, these SCN1A gene polymorphisms could have a significant impact on how individuals with drug-resistant epilepsy respond to AEDs, which holds important implications for clinical management[7]. Namazi et al[13] explored the potential impact of SCN1A gene polymorphisms on the plasma concentrations of CBZ and its active metabolite carbamazepine 10, 11-epoxide (CBZE) in epileptic patients. However, the results did not reveal a significant association between SCN1A gene polymorphisms and CBZ and CBZE levels. This suggests that other factors may play a more significant role in inter-individual variations in response to CBZ.
Further, in our study, the ROC curve analysis revealed a statistically significant area under curve (AUC) value of 0.86 (P < 0.001), indicating high diagnostic accuracy with a sensitivity and specificity of 86% at an optimal cutoff value of 4.63 pg/mL for serum IL-6. Similarly, for serum IL-1β, ROC analysis yielded an AUC of 0.880 (P < 0.001), with a sensitivity of 80% and specificity of 100% at an optimal cutoff value of 59.52 pg/mL. Therefore, 4.63 pg/mL and 59.52 pg/mL can serve as valuable cutoff values for discriminating drug-resistant cases from drug-responsive subjects based on serum IL-6 and IL-1β, respectively (Figure 2).
Similarly, several authors reported that in drug-resistant temporal lobe epileptic tissues, increased NF-κB expression and annexin V-positive neurons particularly correlated with IL-6 levels[14]. HMGB1 levels were shown to increase approximately 3–4 h after a drug-resistant epilepsy episode, highlighting the potential of HMGB1 as a promising marker[15]. Zhu et al[16] suggested that HMGB1 has a predictive value > 9.625 ng/mL for epilepsy prognosis in children.
Further, we observed the associations between different genotypes of the rs6732655 gene polymorphism. Significant links were observed between different genotypes of the rs6732655 gene polymorphism and serum levels of IL-1β, IL-6, and HMGB1. Those with AA and AT genotypes demonstrated substantially higher levels of IL-1β and IL-6, indicative of potentially elevated oxidative stress and neuroinflammation compared to individuals with TT genotypes. HMGB1 levels, however, exhibited variation in drug-sensitive groups only among AA genotypes. As reported in our recent study, individuals with AA and AT genotypes exhibited significantly higher IL-1β and IL-6 levels, possibly attributed to increased oxidative stress and neuroinflammation compared to those with TT genotypes.
Our study investigates genetic polymorphisms associated alterations in the SCN1A gene and associated raised serum inflammatory markers in drug-resistant epilepsy cases. We found the positive associations between SCN1A gene poly
The study highlights a potential association between inflammation and drug resistance in IGE, particularly in epilepsy patients resistant to treatment. Notably, specific genetic variants, such as SCN1A rs6732655A/T genotypes (AT and AA), may predispose individuals to heightened drug resistance risk. These findings emphasize the need for further exploration into the intricate interplay between genetic factors and inflammatory processes in shaping drug response in IGE, offering promising avenues for advancing personalized treatment strategies and therapeutic interventions.
Thanks to the Multidisciplinary Research Unit (MRU), Maulana Azad Medical College, New Delhi for providing molecu
| 1. | Sirven JI. Epilepsy: A Spectrum Disorder. Cold Spring Harb Perspect Med. 2015;5:a022848. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 118] [Cited by in RCA: 110] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 2. | Rheims S, Sperling MR, Ryvlin P. Drug-resistant epilepsy and mortality-Why and when do neuromodulation and epilepsy surgery reduce overall mortality. Epilepsia. 2022;63:3020-3036. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 19] [Reference Citation Analysis (0)] |
| 3. | Smolarz B, Makowska M, Romanowicz H. Pharmacogenetics of Drug-Resistant Epilepsy (Review of Literature). Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 23] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 4. | Kim YO, Kim MK, Woo YJ, Lee MC, Kim JH, Park KW, Kim EY, Roh YI, Kim CJ. Single nucleotide polymorphisms in the multidrug resistance 1 gene in Korean epileptics. Seizure. 2006;15:67-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 5. | Zeng B, Zhang H, Lu Q, Fu Q, Yan Y, Lu W, Ma P, Feng C, Qin J, Luo L, Yang B, Zou Y, Liu Y. Identification of five novel SCN1A variants. Front Behav Neurosci. 2023;17:1272748. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 6. | El Mouhi H, Amllal N, Abbassi M, Nedbour A, Jalte M, Lyahyai J, Chafai Elalaoui S, Bouguenouch L, Chaouki S. Identification of novel and de novo variant in the SCN1A gene confirms Dravet syndrome in Moroccan child: a case report. Mol Biol Rep. 2024;51:233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 7. | Margari L, Legrottaglie AR, Vincenti A, Coppola G, Operto FF, Buttiglione M, Cassano A, Bartolomeo N, Mariggiò MA. Association between SCN1A gene polymorphisms and drug resistant epilepsy in pediatric patients. Seizure. 2018;55:30-35. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 28] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 8. | Viswas A, Dabla PK, Gupta S, Yadav M, Tanwar A, Upreti K, Koner BC. SCN1A Genetic Alterations and Oxidative Stress in Idiopathic Generalized Epilepsy Patients: A Causative Analysis in Refractory Cases. Ind J Clin Biochem. 2023;. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Reference Citation Analysis (0)] |
| 9. | Lipatova LV, Serebryanaya NB, Sivakova NA. The role of neuroinflammation in the pathogenesis of epilepsy. RJTAO. 2018;10:38-45. [DOI] [Full Text] |
| 10. | Han Y, Yang L, Liu X, Feng Y, Pang Z, Lin Y. HMGB1/CXCL12-Mediated Immunity and Th17 Cells Might Underlie Highly Suspected Autoimmune Epilepsy in Elderly Individuals. Neuropsychiatr Dis Treat. 2020;16:1285-1293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 11. | Borger V, Hamed M, Taube J, Aydin G, Ilic I, Schneider M, Schuss P, Güresir E, Becker A, Helmstaedter C, Elger CE, Vatter H. Resective temporal lobe surgery in refractory temporal lobe epilepsy: prognostic factors of postoperative seizure outcome. J Neurosurg. 2021;135:760-769. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 12. | Abe T, Seo T, Ishitsu T, Nakagawa T, Hori M, Nakagawa K. Association between SCN1A polymorphism and carbamazepine-resistant epilepsy. Br J Clin Pharmacol. 2008;66:304-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 81] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 13. | Namazi S, Azarpira N, Javidnia K, Emami M, Rahjoo R, Berahmand R, Borhani-Haghighi A. SCN1A and SCN1B gene polymorphisms and their association with plasma concentrations of carbamazepine and carbamazepine 10, 11 epoxide in Iranian epileptic patients. Iran J Basic Med Sci. 2015;18:1215-1220. [PubMed] |
| 14. | Lorigados Pedre L, Morales Chacón LM, Pavón Fuentes N, Robinson Agramonte MLA, Serrano Sánchez T, Cruz-Xenes RM, Díaz Hung ML, Estupiñán Díaz B, Báez Martín MM, Orozco-Suárez S. Follow-Up of Peripheral IL-1β and IL-6 and Relation with Apoptotic Death in Drug-Resistant Temporal Lobe Epilepsy Patients Submitted to Surgery. Behav Sci (Basel). 2018;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 15. | Engel J Jr, Pitkänen A. Biomarkers for epileptogenesis and its treatment. Neuropharmacology. 2020;167:107735. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 81] [Article Influence: 16.2] [Reference Citation Analysis (0)] |
| 16. | Zhu M, Chen J, Guo H, Ding L, Zhang Y, Xu Y. High Mobility Group Protein B1 (HMGB1) and Interleukin-1β as Prognostic Biomarkers of Epilepsy in Children. J Child Neurol. 2018;33:909-917. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |