Copyright
©The Author(s) 2025.
World J Exp Med. Mar 20, 2025; 15(1): 94022
Published online Mar 20, 2025. doi: 10.5493/wjem.v15.i1.94022
Published online Mar 20, 2025. doi: 10.5493/wjem.v15.i1.94022
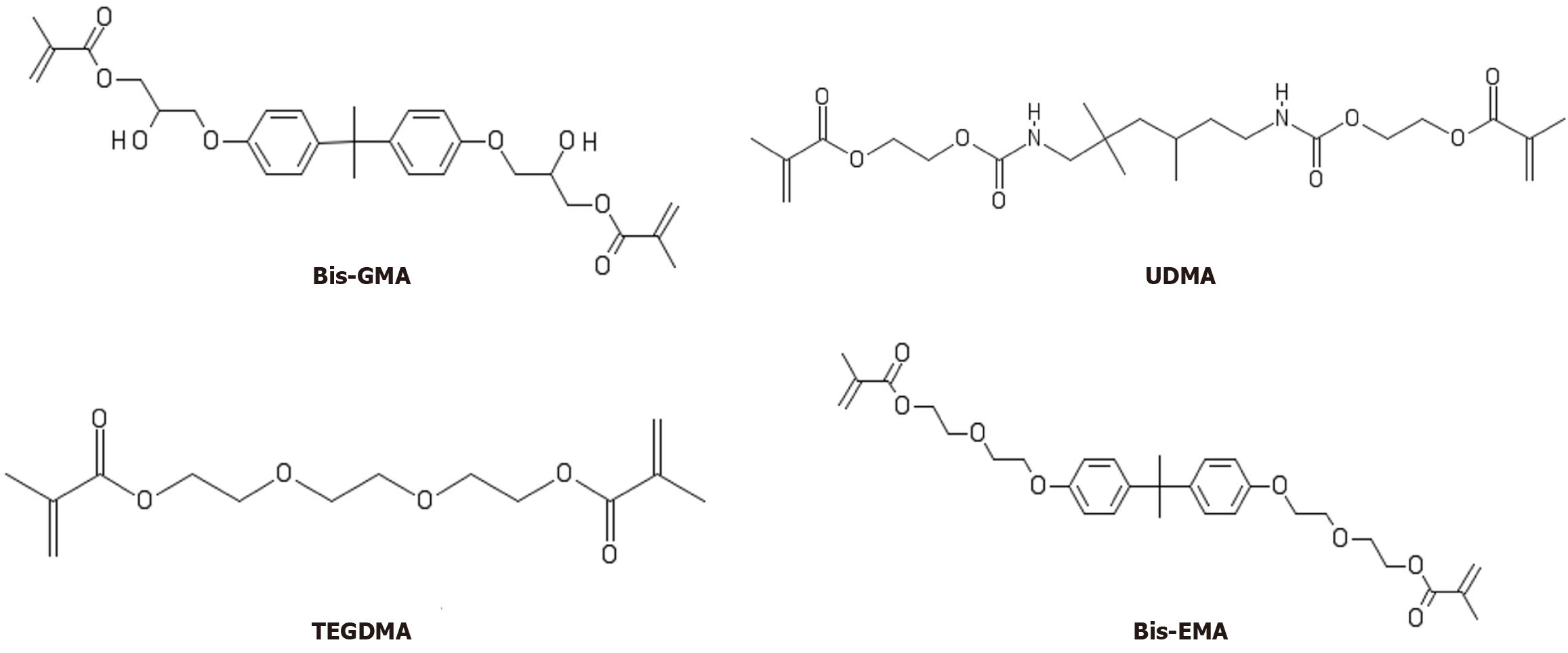
Figure 1 Chemical structure of the tested monomers: Bisphenol-A glycol dimethacrylate, bisphenol-A ethoxylated dimethacrylate, triethylene glycol dimethacrylate, and urethane dimethacrylate.
Bis-EMA: Bisphenol-A ethoxylated dimethacrylate; Bis-GMA: Bisphenol-A glycol dimethacrylate; UDMA: Urethane dimethacrylate; TEGDMA: Triethylene glycol dimethacrylate.
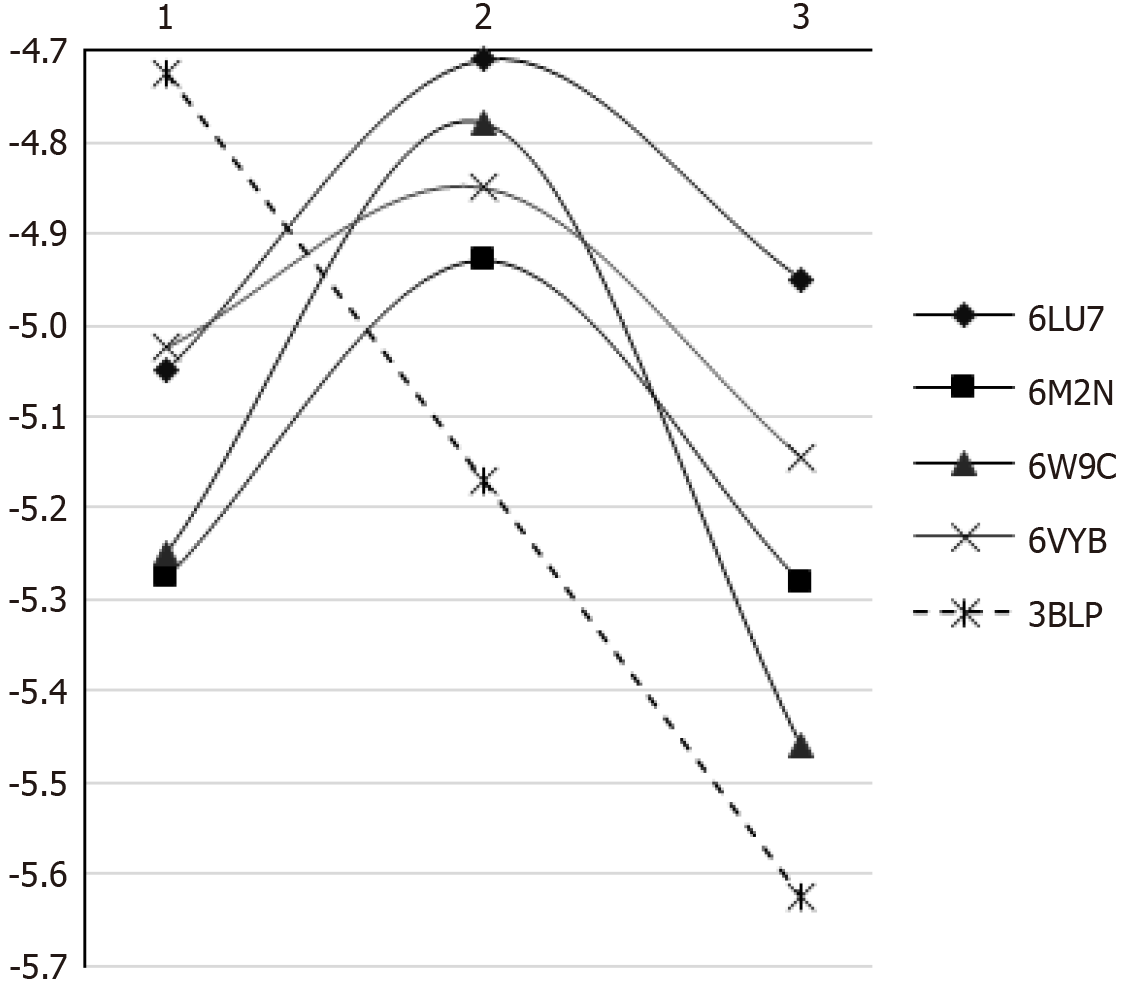
Figure 2 Binding energy between severe acute respiratory syndrome-coronavirus 2 proteins and resin composite chains.
1: Monomer; 2: Two-monomer chain; 3: Three-monomer chain.
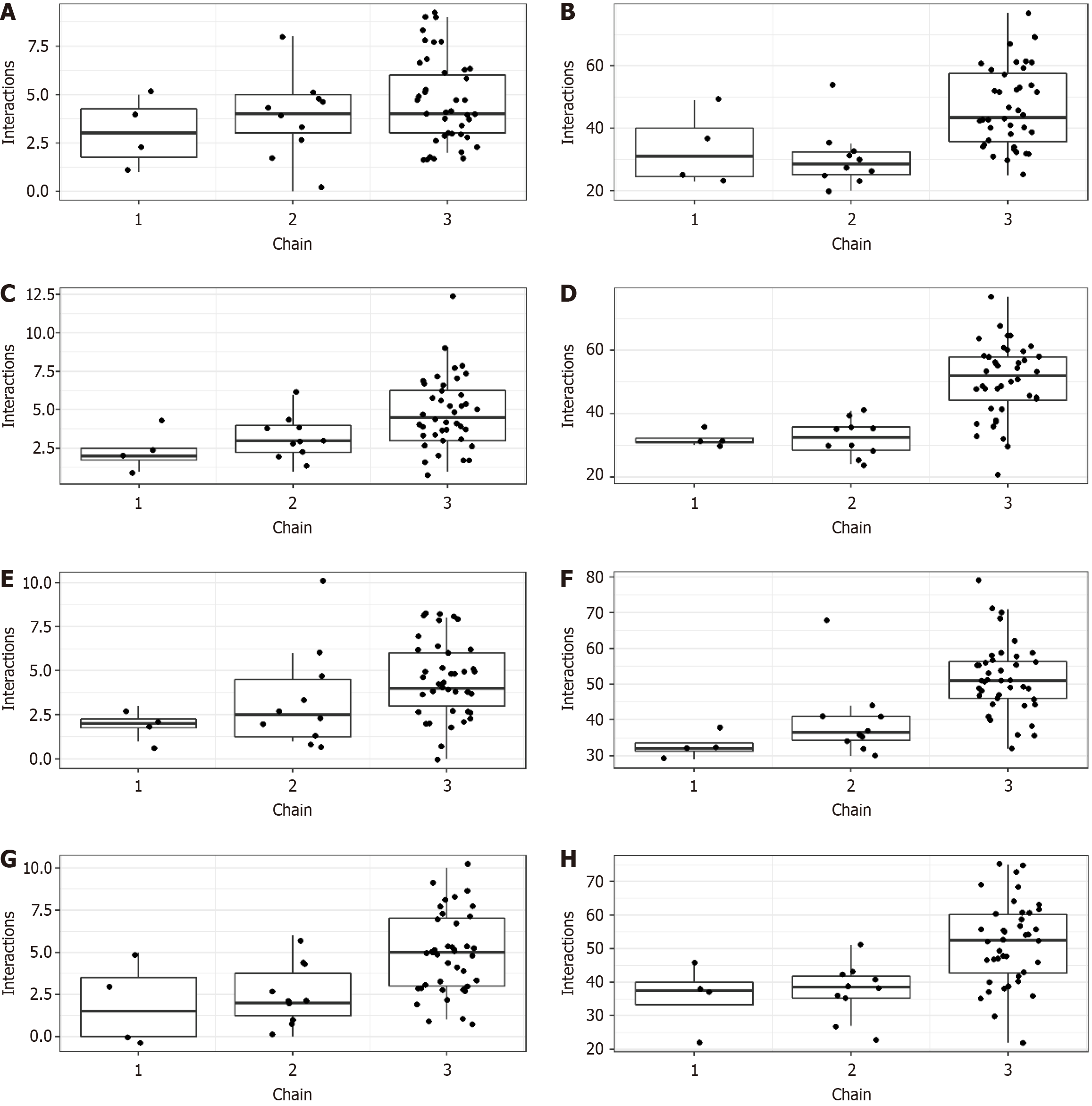
Figure 3 Interactions between severe acute respiratory syndrome-coronavirus 2 proteins and resin composite.
A: Hydrogen bond between 6LU7 and resin composite; B: Hydrophobic interactions between 6LU7 and resin composite; C: Hydrogen bond between 6M2N and resin composite; D: Hydrophobic interactions between 6M2N and resin composite; E: Hydrogen bond between 6VYB and resin composite; F: Hydrophobic interactions between 6VYB and resin composite; G: Hydrogen bond between 6W9C and resin composite; H: Hydrophobic interactions between 6W9C and resin composite.
Figure 4 Graphical interaction between unpolymerised chain (red), 2-monomer chain (yellow), 3-monomer polymerised chain (blue), and the severe acute respiratory syndrome-coronavirus 2 protein.
- Citation: Sette-de-Souza PH, Fernandes Costa MJ, Dutra Borges BC. SARS-CoV-2 proteins show great binding affinity to resin composite monomers and polymerized chains. World J Exp Med 2025; 15(1): 94022
- URL: https://www.wjgnet.com/2220-315x/full/v15/i1/94022.htm
- DOI: https://dx.doi.org/10.5493/wjem.v15.i1.94022