Published online May 20, 2016. doi: 10.5493/wjem.v6.i2.37
Peer-review started: August 31, 2015
First decision: October 27, 2015
Revised: March 2, 2016
Accepted: March 17, 2016
Article in press: March 18, 2016
Published online: May 20, 2016
Processing time: 263 Days and 14.5 Hours
Safe and effective gene therapy approaches require targeted tissue-specific transfer of a therapeutic transgene. Besides traditional approaches, such as transcriptional and transductional targeting, microRNA-dependent post-transcriptional suppression of transgene expression has been emerging as powerful new technology to increase the specificity of vector-mediated transgene expression. MicroRNAs are small non-coding RNAs and often expressed in a tissue-, lineage-, activation- or differentiation-specific pattern. They typically regulate gene expression by binding to imperfectly complementary sequences in the 3’ untranslated region (UTR) of the mRNA. To control exogenous transgene expression, tandem repeats of artificial microRNA target sites are usually incorporated into the 3’ UTR of the transgene expression cassette, leading to subsequent degradation of transgene mRNA in cells expressing the corresponding microRNA. This targeting strategy, first shown for lentiviral vectors in antigen presenting cells, has now been used for tissue-specific expression of vector-encoded therapeutic transgenes, to reduce immune response against the transgene, to control virus tropism for oncolytic virotherapy, to increase safety of live attenuated virus vaccines and to identify and select cell subsets for pluripotent stem cell therapies, respectively. This review provides an introduction into the technical mechanism underlying microRNA-regulation, highlights new developments in this field and gives an overview of applications of microRNA-regulated viral vectors for cardiac, suicide gene cancer and hematopoietic stem cell therapy, as well as for treatment of neurological and eye diseases.
Core tip: Post-transcriptional microRNA-induced suppression of gene expression is a simple new, highly efficient technology to restrict transgene expression to a specific tissue. It is based on the insertion of a target sequence for a cell-specifically expressed microRNA, typically into the 3’ untranslated region of a transgene expression cassette. MicroRNA-induced regulation can result in an up to 100-fold reduction of transgene expression in tissues where expression is not desired. This targeting strategy can be used in combination with other targeting strategies to further improve vector specificity for gene therapeutic approaches.
- Citation: Geisler A, Fechner H. MicroRNA-regulated viral vectors for gene therapy. World J Exp Med 2016; 6(2): 37-54
- URL: https://www.wjgnet.com/2220-315X/full/v6/i2/37.htm
- DOI: https://dx.doi.org/10.5493/wjem.v6.i2.37
Tissue-specific targeting of viral vectors is a key requirement for safe and efficient gene therapy. However, many viral vectors transduce a broad range of cell types. Improvement of specificity can be achieved by direct vector application into a defined location within the organ. But in general, transgene delivery by this means remains near the injection site and the application can lead to tissue injury, making this procedure unsuitable for many applications[1,2]. Other targeting strategies focus on improvement of target specificity after systemic vector application. In this regard, several approaches of transductional targeting have been successfully employed, including switching the virus serotype and capsid engineering via directed evolution, creation of vectors with chimeric or mosaic structures and insertion of antibodies or bi-specific fusion proteins containing the targeting ligands[3-5]. However, being based on the various natural entry mechanisms of infection of many viruses, this strategy has limitations. Transcriptional target-specific expression of the transgene using tissue-specific promoters represents another frequently used approach to improve vector specificity. Although this targeting strategy has been used successfully in many applications, it is limited by the small number of promoters whose activity is exclusively restricted to the target tissue and sufficiently strong to induce transgene expression at therapeutic levels[6,7]. Based on discovery of the RNA interference mechanism enabling post-transcriptional suppression of gene expression[8], microRNA-dependent suppression of transgene expression has been emerging as a promising new approach to improve vector targeting. Since its initial application for reduction of off-target transgene expression in antigen presenting cells (APC)[9], microRNA-dependent control of transgene expression has been used to restrict transgene expression in gene therapeutic approaches, to control oncolytic viruses, for live attenuated virus vaccine development and basic research. In this review we will provide an overview of this technology, its application in gene therapy and discuss perspectives for its further development.
MicroRNAs are small endogenous non-coding RNA sequences of approximately 21 bp that post-transcriptionally regulate gene expression of about more than 60% of human protein-coding genes[10,11]. They are involved in most of cellular processes, including development, differentiation, proliferation and apoptosis[12-16]. MicroRNAs are highly conserved between species and expressed specifically and at certain levels dependent on tissue, lineage or differentiation state[17]. More than 2500 unique mature human microRNAs have been identified so far (http://www.mirbase.org/). Individual microRNA species can vary widely in copy number ranging from less than 10 to more than 30000 copies per cell[18-20]. Beside the tissue-specific expression profile, several microRNAs are dysregulated in cancer[21,22], infectious diseases[23] or diseases of the heart[24] and liver[25], which gives them potential as targets for new therapies.
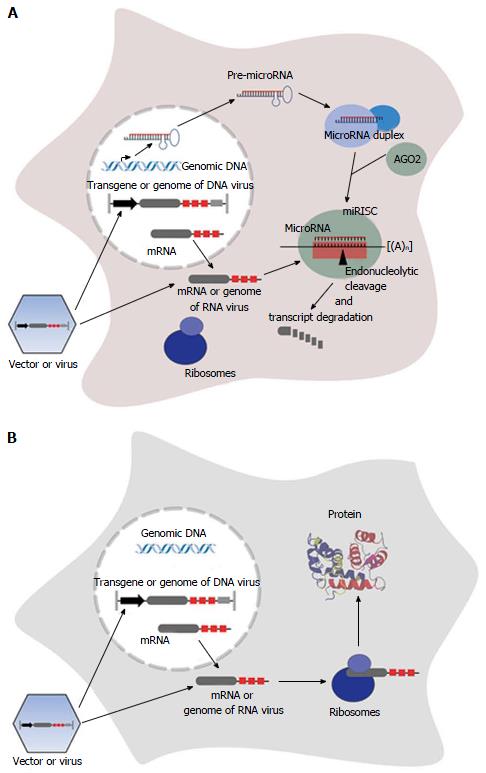
MicroRNAs are usually processed from a precursor molecule (pri-microRNA) that folds into hairpin structures with imperfectly base-paired stems. Pri-microRNAs are further processed by nuclear and cytoplasmatic cleavage proteins, resulting in a short RNA duplex (see references for greater detail[10,26]). One strand of the duplex, the guide strand (microRNA), is selected based on the relative free energies of the microRNA duplex ends[27,28] and is loaded into a multi enzyme complex, the RNA-induced silencing complex (RISC). The less common product is defined as the passenger strand (microRNA*), and is assumed to be degraded[26]. Alternatively both strands of the RNA duplex, namely the 5’ strand (miR-5p) and the 3’ strand (miR-3p) become mature functional microRNAs[29-33]. The mature microRNA is associated with an Argonaute (AGO) family protein, that constitutes the core of the RISC, and functions by base-paired binding to the corresponding target site located in the mRNA, resulting in repression of protein synthesis[10]. However, some microRNAs are able to activate mRNA translation[34-38]. Complete complementarity of microRNA and its target site leads to endonucleolytical central cleavage of the microRNA/mRNA-duplex by AGO2[26,39-43], using a mechanism similar to RNA-interference mediated by siRNAs. In plants, this mechanism is the predominant one[44], while the prevalent mechanism in animals involves binding with incomplete complementarity[12], resulting in inhibition of translation and/or initiation of mRNA degradation[10,39,45-50]. In contrast to plants, whose microRNA target sites are mostly located in protein-coding regions, target sites in animals are often found as repeats in the 3’ UTR of the mRNA[51-54].
Some important rules relating to the interaction between microRNA and its target site were determined by experimental and bioinformatic analysis. The mRNA targeting specificity of a microRNA is determined by perfect match between the seed sequence, a conserved sequence which is usually situated at positions 2-7 or 2-8 of the 5’-end of the microRNA, with a corresponding sequence in the mRNA[12,51-54]. An adenine opposite microRNA position 1 and an adenine or uracil opposite microRNA position 9 are not essential, but increase the efficacy of binding[54]. MicroRNAs exhibiting the same seed sequence belong to the same microRNA family and can regulate the same mRNA targets[51]. A single microRNA species can regulate the production of hundreds of proteins, most likely by recognizing the same seed-matched sequence in the mRNA[55]. A second common characteristic of endogenous microRNA-mRNA interaction comprises nucleotide mismatches in microRNA positions 9-12, thereby most likely preventing an AGO2-mediated cleavage of the target mRNA[10,56]. A third characteristic is that matches within the seed region alone are not always sufficient to induce gene repression; stabilization of microRNA binding may need additional complementarity in the 3’ part of the microRNA. In particular if seed matching is suboptimal, nucleotides at microRNA positions 13-16 become important[51,57]. Thus the 3’ portion can help to compensate for a single nucleotide mismatch in the seed region, as experimentally confirmed for let7 sites in Caenorhabditis elegans lin-4[58] and for miR-196 site in mammalian Hoxb8[43] mRNA. Additional factors can influence binding stability and thereby the efficacy of microRNA-mediated gene regulation[54,57,59,60]. Thus a more effective repression can result from an AU-rich nucleotide composition near the target site[57]. Additionally, more than 15 nucleotides between miR-TS and the stop codon may reduce competition between proteins involved in translation and microRNA-mediated silencing, respectively[57].
Endogenously expressed microRNAs can also be used to specifically modulate the expression of an exogenously applied nucleic acid as a therapeutic cDNA. Therefore artificial microRNA target sites, referred to as miR-TS in this review, serve as targets for a specific microRNA, resulting in post-transcriptional silencing of the transcript[9] (Figure 1). In contrast to transcriptional targeting using tissue-specific promoters that positively regulate transgene expression, expression is negatively controlled by tissue-specifically expressed microRNAs. It was shown that insertion of miR-TS completely complementary to the microRNA in an arrangement of multiple tandem repeats of miR-TS results in strong repression of transgene expression in cells with corresponding microRNA expression[9,17,61]. In this case the transcript is endonucleolytically cleaved, similar to degradation by siRNAs, and the microRNA-RISC is rapidly recycled[39]. Usually the microRNA guide strand mediates transgene repression, thus a corresponding sequence representing the miRTS is inserted into the transgene expression cassette. However, recently Kim et al[62] discovered that expression of a therapeutic transgene can also be regulated by the passenger strand of a microRNA (miR-122) linked to the transgene, thereby eliminating the risk of affecting expression of endogenous microRNA guide strand-regulated genes.
There are several factors influencing the efficacy of microRNA-regulated transgene expression systems. First the cell type- and tissue-specific expression of a chosen microRNA candidate is an important factor that must be taken into account for microRNA-regulated transgene expression systems. Many suitable microRNAs fulfilling this requirement have already been defined (please see reference[63,64]). Among them miR-122, a microRNA, almost exclusively expressed in liver tissue, represents the best candidate for a specific hepatocyte-de-targeting[20,65,66]. Indeed, successful miR-122-mediated suppression of transgene expression in liver was found in several studies[67-69]. However, specific microRNA expression may not be sufficient to achieve a stringent microRNA-dependent transgene repression.
Second, the expression level of a microRNA is crucial for microRNA-mediated regulation. High-throughput assays revealed that only the most abundant microRNAs within a cell mediate significant target suppression[70]. Consequently a fraction of microRNAs that is highly and specifically expressed in certain tissues, possibly involved in defining and maintaining tissue identity[71], are the most potent to de-target transgene expression. In general more highly expressed microRNAs repress their targets to a greater degree[67]. However, the relationship between microRNA expression and its repression activity is nonlinear; in one case it was shown that a 10-fold increase of microRNA concentration results only in a 10% increase of repression[72]. In addition, microRNA expression level is not the only determinant of repressive activity and repression capacity can differ among microRNAs. Kozomara et al[72] showed that although some microRNAs exhibit similar expression levels, they mediate very different repression levels. Conversely some microRNAs repress their targets to similar degrees despite of large difference in expression levels. Another key factor for microRNA-mediated repression of a miRTS-containing transcript is the need for a certain minimal microRNA expression threshold, quantified as 100 copies/pg small RNA by Brown et al[17]. The value is not only important because it defines the minimal amount which is necessary to suppress transgene repression but it also indicates that under a certain level, microRNAs are unable to induce repression. This consideration becomes important if the chosen microRNA is expressed in both the tissue where transgene expression is intended as well as in the tissue where it is not. Therefore microRNAs expressed under the minimal expression limit may not affect transgene expression in the targeted tissue.
Third, the configuration of the miR-TS is another important factor affecting repression efficacy. Corresponding to the different mechanism of microRNA-induced gene silencing, complete complementary miR-TS allow per se higher suppression of the transgene than imperfectly complementary sequences[33,61,73-75]. As perfectly complementary target sequences are endonucleolytically cleaved between microRNA positions 10/11, the microRNA is rapidly recycled[39,69,76-78]. Consequently complete complementarity reduces the risk of microRNA saturation and thus induction of undesirable side effects because the bioavailability of the cognate microRNA to regulate its natural targets is maintained[79].
In addition, the number of miR-TS repeats affects microRNA-mediated suppression of transgene expression. In general an increased number of miR-TS enhances microRNA-dependent transgene repression. But repression efficacy does not linearly correlate with an increasing number of miRTS, and above a certain number of miR-TS, only a relatively modest increase of repression of transgene expression is observed[67,73,80]. Thus maximal suppression of transgene expression of viral vectors, even under optimal conditions (highly and specifically expressed microRNA, use of tandem miRTS repeats), rarely exceeds 100-fold[9,61,67,68,81]. The reason for this limitation is currently not fully understood, but different factors may be involved. It seems that the rate-limiting steps in microRNA activity are the level and dynamics of association of the target mRNA with the microRNA-loaded RISC, rather than level of target transgene mRNA[17,72,82], which is directly dependent of vector and expression systems. We and others have shown that 3-4 identical repeats of miRTS are sufficient for efficient transgene repression (Table 1). An increase to 6 or even 12 copies is possible, but may only have a marginal effect on repression efficacy[67,80,83]. Moreover, an increase of miR-TS repeats can induce saturation of the respective microRNA, as more target sites means more substrate for the microRNA to bind, resulting in potentially undesirable off-target effects[76]. As shown by several studies, different miRTS can be inserted in series in order to de-target various cell types with different microRNA expression profiles[17,61,69,84]. On the other hand, a combination of miRTS corresponding to different microRNAs for a given cell type or tissue can enhance microRNA-mediated transgene suppression, especially if the microRNAs are only expressed at moderate levels[85]. Moreover, employing cooperative microRNAs may reduce the risk saturating the function of one microRNA[67]. An enhancement of microRNA-mediated suppression of transgene expression can also be achieved by combination of microRNA regulation with other regulation systems. Thus, Bennett et al[86] developed an adenoviral vector (AdV) system utilizing both microRNA-mediated regulation and the Cre recombinase (Cre)-loxP recombination system to further reduce hepatic transgene expression and to achieve relatively pure spleen-/dendritic cell-specific expression thereby improving AdV-based vaccination against transgene products. Although insertion of miR122TS into the transgene expression cassette of AdV suppressed liver expression by about 100-fold, significant transgene expression levels were still found in the liver[86,87]. Therefore they engineered an AdV carrying the transgene flanked by loxP sites and containing miR-122TS, while another AdV expressed Cre equipped with miR-TS for miR142-3p, which is highly expressed in the spleen. Intravenous co-administration of the AdVs resulted in an approximately 3800-fold reduction of hepatic transgene expression and a slightly reduced, but substantial, transgene expression in the marginal zone of the spleen, which was sufficient to exhibit therapeutic value[86]. In most studies, small 4 to 6 nucleotide long spacer sequences have been used to separate miR-TS repeats[9,61,67,68]. Introduction of spacers may, in general, reduce steric hindrance of enzyme complexes binding to microRNA/miR-TS duplexes and thus facilitate better repression. Nevertheless spacer-free multimeric miR-TS were also successfully applied for transgene repression[69]. Furthermore it is thought that, compared to longer spacers, insertion of shorter spacers might reduce the risk of formation of secondary structures around the miR-TS that might disturb base-paring of microRNA with the miR-TS[88,89]. Thus whether and to what degree spacer sequences affect efficacy of designed microRNA-regulated suppression systems remains to be elucidated. Besides the functional importance of applied miR-TS, copy number of miR-TS, as well as spacing between miR-TS, are relevant to viral vector construction, as it directly enlarges the transgene expression cassette inserted into the vector genome. As miRTS are small in size (about 22 bp), insertion of tandem repeats of miR-TS, including spacer sequences, generally does not constitute a capacity problem for most viral vectors. However, for some viral vectors with low packaging capacity, such as self-complementary adeno-associated virus (AAV) vectors[90], keeping down the total length of miR-TS could be an essential requirement. In this regard, our group investigated whether shortening of miR-TS corresponding to miR122 affects the miR-122-mediated repression function. Indeed, we found that a deletion of up to 5 nucleotides from the 5’ end of the miR-122TS was well tolerated and did not influence transgene suppression[67]. However, more studies are necessary to confirm our results for other miRTS.
| Target cell/tissue for therapy | miR-TS for | Number miRTS | Vector | MicroRNA-regulated transgene | De-targeted cell/tissue | Ref. |
| Heart | ||||||
| Heart | miR-122 | 3, 5 | AAV9 | EGFP, lacZ, luciferase | Liver | [67,68] |
| Heart | miR-122 | 3 | AAV9 | Human S100A1 | Liver, skeletal muscle | [61] |
| miR-206 | 3 | |||||
| Cancer | ||||||
| PyMT mouse breast cancer | miR-1 | 4 | AAV9- ESGLSQS | Herpes simplex virus thymidinkinase (and ganciclovir), luciferase | Heart | [129] |
| Metastatic hepatocellular carcinoma model | miR-122 | 4 | AAV8 | Herpes simplex virus thymidinkinase (and ganciclovir), luciferase | Hepatocytes/liver | [128] |
| Glioblastoma cells in vitro | miR-128 | 2 | Lentiviral | Herpes simplex virus thymidinkinase (and ganciclovir) | Neuronal differentiated cells | [127] |
| Subcutaneous fibrosarcoma xenografts | miR-122 | 4 | Adenoviral | Luciferase | Hepatocytes/liver | [181] |
| Intradermal melanoma tumor | miR-122 | 4 | Adenoviral | Herpes simplex virus thymidinkinase | Hepatocytes/liver | [87] |
| Human glioma xenografts | miR-31 miR-127 miR-143 | 3 3 3 | Baculoviral | Herpes simplex virus thymidinkinase (and ganciclovir) | Astrocytes/brain | [85] |
| Hepatocellular carcinoma model | miR-181 | 2 | Adenoviral | Human telomerase reverse transcriptase RNA-targeting ribozyme | Hematopoietic stem cell- and progenitor-derived blood cells | [130] |
| miR-122 | 3 | Hepatocytes/liver | [131] | |||
| HSC and iPSC | ||||||
| Tumor-infiltrating monocytes/macrophages | miR-126 miR-130a | 2 2 | Lentiviral | Interferon-α | Hematopoietic stem and progenitor cells | [134] |
| Differentiated myeloid cells | miR-126 | 2 | Lentiviral | Gp91phox | Hematopoietic stem and progenitor cells | [135] |
| Differentiated hematopoietic cells | miR-126 miR-130a | 4 4 | Lentiviral | Galactocerebrosidase | Hematopoietic stem and progenitor cells | [133] |
| Lymphoid progenitors, myeloid cells | miR150 | 4 | Lentiviral | GFP | Differentiated T and B lymphocytes | [132] |
| Post-thymic T cells | miR181a | 4 | Lentiviral | GFP, inhibition of negative thymic selection | Developing T cells | [182] |
| Myeloid cells Immature dendritic cells | miR223 miR155 | 4 4 | Lentiviral | EGFP, NGFR | Granulocytes and monocytes, mature dendritic cells | [17] |
| Human fetal fibroblasts | miR-302a, d miR-142-3p | 4 4 | Lentiviral | EGFP, mCherry | Human ES, neural progenitors | [137] |
| miR-155 | 4 | |||||
| miR-223 | 4 | |||||
| ES, iPSC | miR-292-3p | 4 | Lentiviral | GFP, differentiation of ES to neural progenitors | Pluripotent cells | [138] |
| ES, iPSC | let-7a | 4 | Lentiviral | GFP | Non pluripotent cells | [136] |
| CNS and eye | ||||||
| CNS | miR-1 | 3 | AAV9 | lacZ, EGFP | Heart, liver | [69] |
| miR-122 | 3 | |||||
| CNS/astrocytes | miR-124 | 4 | Lentiviral | lacZ, glial glutamate transporter | Neurons | [141] |
| CNS/cortical excitatory neurons | miR-128 miR-221 | 4, 8, 12 4, 8 | Lentiviral | EGFP | Cortical inhibitory neurons | [83] |
| Retina/eye | miR-124 miR-204 | 4 4 | AAV5 AAV8 | EGFP | Photoreceptors/retinal pigment epithelium | [143] |
| Photoreceptor/eye | miR-181c | 4 | AAV2 (quadY-F+T-V) | GFP | Ganglion cells and inner retina | [144] |
| Immune system and other applications | ||||||
| Skeletal muscle | miR-142-3p | 2, 4, 8 | AAV1 | Ovalbumin human sarcoglycan | Antigen presenting cells | [152,153] |
| Hepatocytes/liver | miR-142-3p | 4 | Lentiviral | Factor IX, VIII, induction immunologic tolerance | Antigen presenting cells | [9,145,147,183] |
| Dendritic cells/spleen | miR-142-3p miR-122 | 4 4 | Adenoviral | Luciferase, lacZ, combination with Cre-loxP system | Hepatocytes/liver | [86] |
| Neural stem cells | miR124a | 4 | Lentiviral | GFP, identification of neurons | Neurons | [184] |
| Adipose tissue | miR-122 | 3, 8 | AAV8 | Human leptin | Hepatocytes/liver | [185] |
| Intramuscular delivery | miR-122 miR-206 | 6 6 | AAV8 | 201Ig immunoadhesin, luciferase | Liver, skeletal muscle | [186] |
| Human fibroblasts HFL1 | miR-124 | 4 | Lentiviral, replication deficient | Neural conversion genes Ascl1, Brn2 and Myt1L | Human induced neurons | [179] |
| Skeletal muscle | miR-208a | 2 | AAV9 | Human calpain3 | Heart | [81] |
Although a systematic investigation defining common standards for insertion of miR-TS into a transgene expression system is currently not available, location of the miR-TS in the mRNA is obviously important for repression efficacy. Based on knowledge of microRNA target recognition and microRNA-induced cleavage of cellular targets, it becomes obvious that secondary structures of the mRNA itself or structures formed by miR-TS insertion can also affect transgene suppression as accessibility to the target mRNA is hindered[78,91]. Since the 3’ UTR of mRNAs almost always lack secondary RNA structure[92], miR-TS can be placed there, usually near the stop codon, but insertion within the 5’ UTR or the open reading frame is also possible[93-96]. Other factors influencing repression capacity of a given microRNA include increased nuclear localization of a microRNA, its stability[97] and the stability of the microRNA/miR-TS duplex[70,72]. Concerning the latter, analysis of the free energy of multiple duplexes revealed a weak negative correlation of duplex stability and microRNA-mediated regression. Therefore lower stability seems to favor transient interaction with target mRNAs and efficient down-regulation of multiple targets[39,72].
In summary, optimal microRNA-mediated repression of the vector-encoded transgene needs high expression of the microRNA, insertion of 3-4 copies of miR-TS with complete complementarity, uniqueness with regard to regulation by other microRNA candidates and insertion within a site with low secondary structure. All these aspects need to be pre-evaluated for optimal microRNA-mediated regulation of transgene expression of viral vectors.
Although this technology displays many advantages compared to other methods, it is not without concerns. Given that endogenous microRNAs are involved in target regulation, miR-TS containing transgene mRNAs are in competition with cellular target mRNAs. By the use of complete complementary miR-TS, enabling rapid microRNA recycling, and defining an optimal target site number, the risk of microRNA saturation can be minimized. We and others showed that endogenous microRNA profile or microRNA-regulated genes were not altered after administration of optimized miRTS-bearing vectors[61,67,69,81].
Several promising results using microRNA-dependent regulation have been obtained for different disease models and vector systems (Table 1).
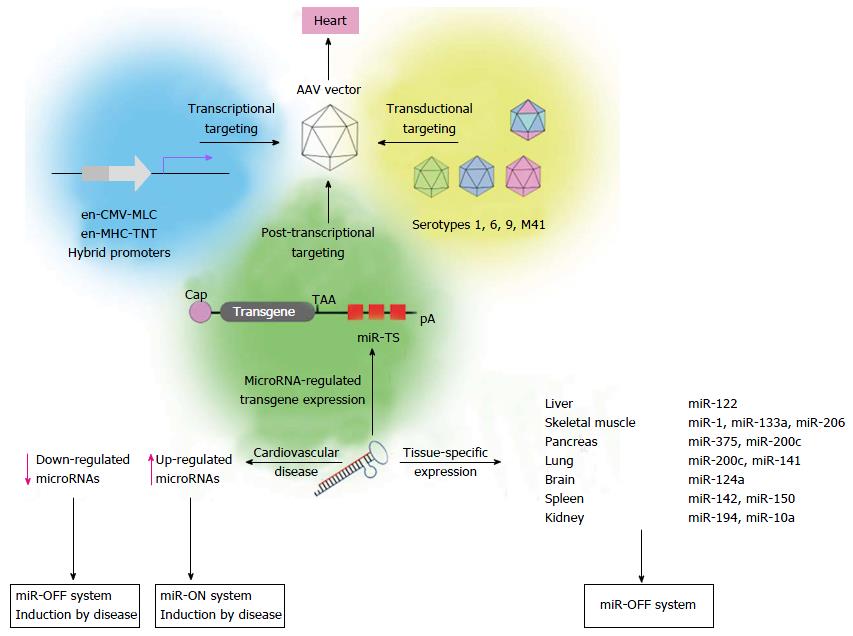
AAV vectors have been established as the leading vector type for myocardial gene transfer in preclinical and clinical applications[98]. Their main advantage is their ability to transduce cardiomyocytes in vivo at high efficiency. In addition, AAV vectors allow long lasting expression of encoded transgenes and do not trigger a strong immune response or inflammation[99-102]. Importantly AAV vector-driven expression is performed without integration of the vector genome into the host genome[103] which, in consequence, reduces the risk of long term, irreversible side effects[104-106]. As conventional AAV vectors containing capsid proteins of serotype 2 (AAV2) revealed a natural cardiac tropism, their transduction efficiency was limited. Therefore discovery of naturally occurring AAV serotypes carrying variations in the amino acid sequence of the capsid protein[107,108] enabled the development of pseudotyped AAV vectors and led to substantial improvement of cardiac gene transfer. AAV9 vectors are the most cardiotropic serotype upon systemic vector administration for transgene delivery into the heart of rodents[109-115], whereas AAV6 vectors seems to be superior to other serotypes in large animals[116]. In humans, a clinical phase 2b trial showed promising results upon intracoronary administration of an AAV1 vector expressing the calcium regulatory protein SERCA2a in patients with advanced heart failure[117]. However, improved cardiac transduction by serotype switch did not abrogate the intrinsic property of AAV resulting in sustained transduction of a broad range of tissues, in particular liver and skeletal muscle.
Transcriptional targeting represents another approach to increase cardiac specificity of AAV vectors. To restrict AAV vector-mediated transgene expression to the heart upon intravenous injection, a variety of cardiac-specific promoters have been investigated. Promoters of the heavy chain (MHC)[118-120] and the light chain (MLC)[121,122] of cardiac protein myosin are most potent to transcriptionally mediate heart-specific transgene expression. However, endogenous cardiac-specific promoters are too large to be packaged into AAV vectors and their core elements alone provide only weak transgene expression. Smaller hybrid promoters consisting of a cardiac core element and another strong enhancer element increased cardiac expression[109,122] but transgene expression was also detected in other tissues, especially in the liver, skeletal muscle and pancreas[61,122]. At this point it seems unlikely that promoter-only approaches will sufficiently target the vectors, therefore microRNA-dependent regulation of AAV vector-mediated transgene expression has shifted into focus to further improve cardiac specificity of AAV vectors (Figure 2). Two studies carried out by Qiao et al[68] and Geisler et al[67] addressed microRNA-mediated suppression of transgene expression in liver after systemic AAV9 vector application. Among three microRNA candidates (miR-122, miR-192 and miR-148a) that have been described to be selectively expressed in liver[65,123], we found that miR-122 was most abundantly expressed in murine liver, whereas expression in murine heart was approximately 5 orders of magnitude lower. Systemic application of AAV9 vectors containing miR122TS in the 3’ UTR of an EGFP reporter confirmed effectiveness of miR-122-mediated suppression in the liver, whereas cardiac expression remained unaffected. Interestingly, microRNA-mediated transgene suppression in the liver was by far more efficient than transcriptional control by the en-CMV-MLC0.26 cardiac hybrid promoter[67]. The other study carried out by Qiao et al[68] confirmed improved specificity of miR-122-regulated AAV9 vectors. Similar to our study, they reported that microRNA-mediated increase of cardiac specific expression was even more efficient than transcriptional control of transgene expression by an en-MHC-TNT hybrid promoter.
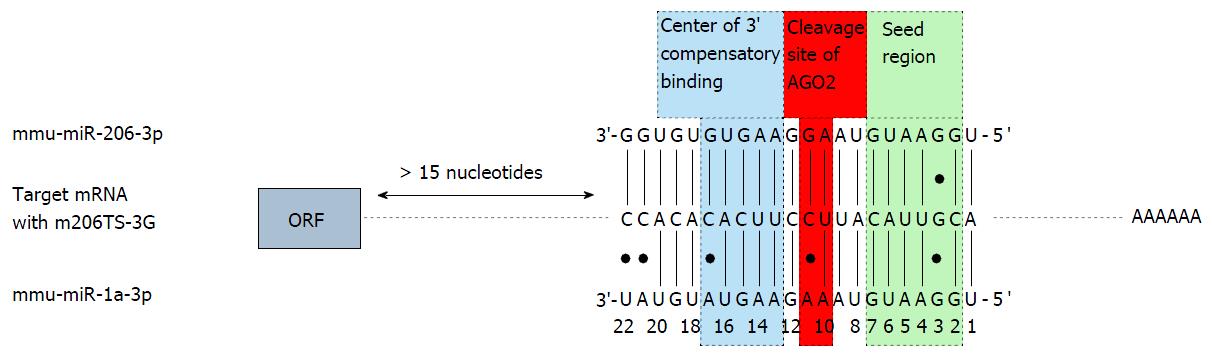
Abundant transgene expression upon systemic AAV9 vector application can also be found in the skeletal muscle. Therefore transgene suppression in this tissue is necessary to further enhance the cardio-specificity of AAV vector-mediated gene transfer. According to the guidelines for microRNA selection (see above), only miR206 fulfills the major requirements of microRNA-mediated repression: It is highly expressed in the skeletal muscle and absent in the heart. However, we found that a transgene containing miR-206TS was strongly suppressed in both the skeletal muscle and heart of mice following systemic application of AAV9 vector. The reason for this is binding of the miR-1, which shows high sequence homology to miR-206 and is highly expressed in heart tissue. By introducing single nucleotide substitutions into the seed region of the microRNA/miR-206TS duplex, we generated mutated miR-206TS that became resistant to miR-1 regulation but remained fully sensitive to miR-206[61] (Figure 3). This was a result of compensatory effects induced by perfect complementarity of the 3’ portion of miR206 to mutated miR206TS. In vivo transgene expression of AAV9 vectors bearing mutated miR206TS and miR122TS was strongly suppressed in both skeletal muscle and the liver, whereas expression in heart remained unaffected[61]. Interestingly, contrary to the generally applicable concept of miR-TS recognition and repression by microRNAs, we observed that insertion of single nucleotide mutations within miR206TS can even enhance miR206-mediated transgene repression. Although the underlying mechanism remains to be elucidated, we observed similar results for miR122TS repression but not for other modified miR-TS (data not published).
Regulation of transgene expression by endogenous microRNAs has been exploited for vector-mediated suicide gene therapy with Herpes Simplex Virus thymidine kinase (HSVtk). Expression of the HSV-tk mediates phosphorylation of the prodrug ganciclovir (GCV), thus inhibiting DNA replication in rapidly dividing cancer cells. As phosphorylated GCV is also toxic in normal cells[124], tumor cell-specific expression of HSV-tk is required. AdVs are widely used in cancer gene therapy. However, they exhibit strong hepatotropism after systemic application and can induce severe hepatotoxicity after HSV-tk delivery[125,126]. Repression of HSV-tk containing miR-122TS induced by miR122 distinctly reduced hepatotoxicity upon local delivery by AdV, without altering the antitumor effects, as miR-122 is weakly expressed in transduced melanoma tumor cells but highly expressed in hepatocytes[87]. In another approach, a lentiviral vector (LV) expressing HSVtk in combination with ganciclovir showed improved tumor specificity in vitro by miR128-mediated transgene regulation, a microRNA that is differentially expressed between glioblastoma cells and normal brain tissue[127]. Other studies confirmed the reduction of side effects by microRNA-dependent suppression of suicide genes in cancer gene therapy approaches. Wu et al[85] found that in addition to transcriptional control by a glioma-specific promoter, insertion of miRTS for the miR31, miR127 and miR143 into baculoviral vectors led to repression of HSV-tk in normal neural cells. As these microRNAs are more highly expressed in these cells than in glioma cells, this approach led to reduced HSVtk/GCV induced cytotoxicity. More recently, Della Peruta et al[128] improved tumor specificity of systemically applied AAV8 vectors expressing HSVtk linked with miR-122TS in combination with a liver-specific promoter in a syngeneic metastatic murine hepatocellular carcinoma (HCC) model. Administration of the vector and GCV treatment resulted in a reduction of cancer growth and number of metastases without liver toxicity. Moreover Trepel et al[129] used miR-TS of the miR1 to inhibit cytotoxicity in heart after HSV-tk delivery with an AAV9 vector capsid variant. Animals treated with the vector were protected from cardiac HSV-tk expression and drug-induced development of dilative cardiomyopathy, whereas delivery of the suicide gene to tumors significantly inhibited tumor growth after GCV treatment. Another study recently carried out by Won et al[130] improved specificity of cancer gene therapy by developing microRNA-regulated trans-splicing ribozyme that targets human telomerase reverse transcriptase (hTERT) RNA in cancer cells. The group I intron-based ribozyme targets and cleaves its substrate RNA and trans-splices an exon attached at its 3’ end (e.g., a therapeutic RNA) onto the cleaved target RNA, resulting in expression of the therapeutic RNA and repression of substrate RNA. As the ribozyme specifically targets hTERT RNA positive cancer cells, but also hematopoietic stem cell-derived blood cells, the ribozyme was modified by inserting target sites for the blood cell-specific miR181a downstream of its 3’ exon. Analysis of AdV-mediated expression of the hTERT-targeting trans-splicing ribozyme harboring miR181aTS with HSV-tk gene as a 3’ exon under control of a liver-specific promoter and GCV treatment resulted in specific anticancer effects. Moreover systemic vector application in an orthotopic multifocal HCC mouse model demonstrated a regression of liver tumor nodules and tumor volume, with minimal hepatotoxicity[130]. In a similar approach, AdV-mediated administration of the hTERT-targeting trans-splicing ribozyme containing miR122TS resulted in efficient anti-cancer effects and reduced hepatotoxicity[131].
For hematopoietic stem cell (HSC) therapy, lineage- and differentiation stage-specific microRNA expression can be used to specifically express a transgene in a discrete subset of progeny. Although integrating vectors, such as LVs, mediate long-term expression in progenitor cells, expression needs to be often restricted to their differentiated progeny. Therefore Brown et al[17] showed that miR223 de-targets transgene expression in the myeloid progeny of HSC (granulocytes and monocytes), thus restricting expression to the lymphoid progeny. Similarly, transgenes were equipped with miR-TS for miR150 and miR-155, thus repressing their expression in mature T and B cells and mature dendritic cells, but not in lymphoid progenitors or immature dendritic cells, respectively[17,132]. On the other hand, microRNA-regulation technology also worked if the expression of a toxic transgene needs to be avoided in hematopoietic pluripotent stem/progenitors cells (HSPC). In this regard it has been shown that miR-126 and miR-130a, both expressed in the HSPC compartment, but not in mature blood cells, suppressed LV-mediated transgene expression of galactocerebrosidase containing the corresponding miR-TS in HSC, allowing long-term hematopoiesis after vector transduction and a stable bone marrow graft in mice. Moreover a robust expression of galactocerebrosidase in mature hematopoietic cells enabled successful treatment of globoid cell leukodystrophy in a mouse model[133]. In a similar approach Escobar et al[134] targeted interferon-α expression to tumor-infiltrating monocytes/macrophages using a combination of transcriptional and miR-126-/miR130a-mediated control during post-transplant hematopoiesis to limit HSC exposure to the transgene thereby inhibiting breast cancer progression. Furthermore in an approach using therapeutic LV encoding gp91phox linked with miR126TS under the control of a myeloid-specific promoter, high levels of myeloid-specific transgene expression was achieved for X-linked chronic granulomatous disease therapy, entirely sparing the miR-126 positive CD34+ HSC compartment. As ectopic gp91phox expression in HSC could cause production of reactive oxygen species that may damage DNA, alter cell growth or induce apoptosis, the transcriptionally and post-transcriptionally regulated LV reduced offtarget expression and effectively corrected the X-phenotype in gp91phox-deficient mice[135].
MicroRNA-regulated reporter expression systems can also be used to track the differentiation of stem cells and have been widely disseminated in the field of pluripotent stem cell technology (induced pluripotent stem cells, embryonic stem cells)[17,136-138]. A big challenge in this field is the in vitro generation of a homogenous cell population of a pre-defined lineage which can be successfully transplanted. Therefore cell type- and differentiation-specific microRNAs are helpful to identify and select cells with desired differentiation fates. Upon differentiation, cells can be selected either by combining miRTS with a positive or negative regulator such as neomycin or thymidine kinase[17,136] or by combining with a reporter, such as GFP, and sorting by fluorescent activated cell sorting (FACS)[138], respectively.
Gene therapy holds promise for treating of genetic as well as metabolic disorders of the central nervous system (CNS) and in particular AAVs have received growing interest as viral vector systems for gene delivery in the last few years. Besides the well-known generally favorable properties of AAV vectors, several AAV serotypes are able to cross the blood-brain barrier and thus effect an efficient transduction of cells of the CNS with systemic vector application[139]. Based on the broad range of tissue targeting by AAV9 vectors, Xie et al[69] exploited miR-TS technology to restrict AAV9 vectors to the CNS. Therefore they constructed AAV9 vectors with miR-TS completely complementary to hepatocyte-specific miR-122 and muscle-specific miR-1 in order to repress reporter gene expression in liver, heart and the skeletal muscle. In vivo comparison of vectors, each bearing one or three miR122TS or/and miR-1TS, found that the number of miR-TS repeats inversely correlated with reporter expression levels in tissues where the corresponding microRNA is expressed. Moreover systemic application of the microRNA-regulated AAV9 vector resulted in distinct suppression of the transgene in the liver and muscle, whereas it was not altered in the brain. Most vector systems transducing the CNS have a strong neurotropism. However, for several neurodegenerative disorders, certain cell types of the CNS, such as astrocytes, need to be specifically targeted, as they exhibit important physiological functions and may display altered functions. Among CNS-specific microRNAs, miR-124 is specifically expressed in neurons but absent in astrocytes[140]. Accordingly, Colin et al[141] demonstrated that insertion of miR-124TS into a LV is sufficient to repress transgene expression in neuronal cells in vitro and in vivo, whereas expression remained unaffected in astrocytes. Moreover, exploiting microRNA regulation enables more than targeting transgene expression to neurons or supporting cells. Thus Sayeg et al[83] showed that a combinatory regulation of a locally applied LV by miR128 and miR221 targeted transgene expression between neuronal subtypes in the murine neocortex as expression was selectively inhibited in excitatory neurons but not in inhibitory cells.
Inherent retinal degeneration of the eye results from impaired function of the retinal pigment epithelium (RPE) or photoreceptor (PR) cells and leads to severe visual deficits and blindness[142]. In an approach to restrict AAV-vector-mediated EGFP reporter gene expression to either RPE or PRs, Karali et al[143] subretinally applied miRTSbearing AAV5 vectors into the eye of mice and pigs. AAV vectors containing miR-204TS were selectively repressed in RPE, which was in agreement with the miR204 expression in RPE. In contrast the EGFP reporter was repressed in PRs but not in RPE if the vector was equipped with miR-TS for miR-124, a microRNA that is expressed in PRs but absent in RPE. For advanced retinal gene therapy, Kay et al[144] recently developed an AAV2 vector capsid mutant with a strongly increased transduction efficacy for PRs upon intravitreal vector delivery. Furthermore, transgene expression was restricted to PRs only by use of a combinatorial approach of a PR-specific promoter and incorporation of multiple target sites for miR-181c, a microRNA which is expressed in retinal ganglion cells and the inner retina.
The concept of microRNA-mediated negative regulation of transgene expression was first shown by Brown et al[9] in order to reduce off-target transgene expression in APC upon systemic injection of LV. By combining a hepatocytic-specific promoter and target sites for the hematopoietic-lineage specific miR-142-3p, a reduced transgene expression in APCs allowed a long-term expression of clotting factor F.IX for stable correction of hemophilia B mice upon systemic LV administration[145,146]. Furthermore it was shown that this combined approach induced an immunological tolerance for the transgene[147-149] resulting from hepatocyte-directed expression[147,150]. Further studies confirmed these results for other vector systems. Intramuscular application of AAV vectors leads to appearance of immune responses against the transgene and rapid decrease of transgene expression, possibly as a result of vector transduction of APC[151]. Thus Majowicz et al[152] and Boisgerault et al[153] inserted target sites for miR1423p into the transgene expression cassette of AAV1 vectors. Upon intramuscular application they observed prolonged expression of the transgene in the skeletal muscle, which correlated with reduced immunogenicity to the transgene.
Besides the development of microRNA-regulated viral vectors in order to de-target transgene expression in certain tissues, lineages or differentiation states or to identify cell subsets, modification of tropism of replication competent oncolytic viruses (OV) has gained growing interest. Although OVs preferentially replicate in tumor cells, application to cancer gene therapy is limited by their replication and corresponding induction of toxicity in normal tissues. By insertion of miRTS for tissue-specific microRNAs into the viral genome, microRNAs have been used to reduce replication in normal cells, while maintaining their oncolytic potential in tumor cells. This was initially shown by Kelly et al[154], who engineered a coxsackie A21 OV with miRTS for the skeletal muscle-specific miR206 and muscle-specific miR133a, thus reducing myotoxicity while maintaining oncolytic potential. MicroRNA-regulation of OVs to increase tumor specificity has been successfully applied for several RNA[84,155-158] and DNA[80,93,159-167] viruses in the meantime. For detailed information the reader is referred to Ruiz et al[168].
Another aspect that needs to be addressed is the enhancement of safety of live attenuated virus vaccines by microRNA-regulation[169]. Insertion of miRTS into the viral genome leads to reduced primary replication in the viral target organ, whereas weaker replication in other tissues induces protective immunity of the host. This was successfully shown for poliovirus[170], influenza virus[171], flaviviruses[172-175] and alphavirus[176].
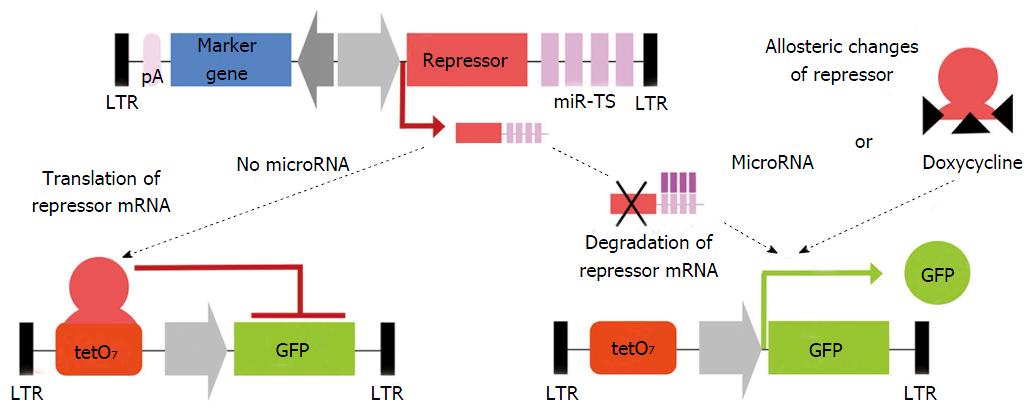
Activity of a given microRNA can be easily identified by coupling corresponding miR-TS to a reporter gene such as lacZ, GFP or luciferase and analysis of reporter gene expression. This allows dynamic measurement of microRNA expression over time on a single cell basis. Because reporter gene expression is observed as microRNA-mediated shut down of expression, the absence of signal might be not distinguishable from false negative results of failed transduction or transgene silencing. By LV-mediated co-expression of the microRNA-regulated sensor reporter and a second non-microRNA-regulated reporter, both under the control of a bidirectional promoter, microRNA activity can be measured independently from vector dose, transduction level and promoter activity[9,17,133]. This principle has been applied to construct a microRNA reporter library for analysis of microRNA activity in FACS-sorted cells. This technique has many advantages, as it allows the measurement of microRNA bioactivity in heterogeneous cell populations such as HSC[133] or neural stem cells[177]. However, for microRNAs with expression levels close to the detection threshold of the reporter system, its reliability is limited and other methods are preferred. Thus recently a miR-ON system (Figure 4) was generated to identify small subpopulations of microRNA-expressing cells and to select them directly. This system includes transduction of a repressor containing miR-TS, together with a reporter under its regulation. Reporter activity is observed only if the upstream repressor mRNA is degraded and cells expressing corresponding microRNA become selectable by reporter gene expression[178].
Furthermore microRNA-mediated regulation of transgene expression can help to improve direct cell conversion as shown for conversion of human fibroblasts into functional neurons (human induced neurons) by a self-regulating vector. Insertion of miR-TS for the neuron-specific miR-124 linked to the neural reprogramming genes Ascl1, Brn2 and Myt1L of an integration deficient LV facilitated a down-regulation of conversion gene expression once the cell has reached a stable neuronal fate, thereby allowing for a more complete functional maturation of the cells in culture[179].
For additional applications in further fields, the reader is referred to Table 1.
In this review we have described the features of microRNA-regulated transgene expression, including its technical mechanisms and its application to transgene delivery by viral vectors. The variety of examples showed how microRNA regulation can be diversely exploited in different contexts as vectors, routes of application or fields of research in general. Compared to other targeting strategies, including transcriptional and transductional targeting, microRNA-regulation displays many advantages. Insertion of approximately 100 bp sequence of miR-TS can be easily accomplished by conventional cloning techniques. In general the small size of miRTS prevents interference with the packaging capacity for therapeutic genes or foreign genes, thus making it applicable for both viral vectors and viruses. This system has now become a major technique for specific application of therapeutic transgenes, for increased tumor specificity of oncolytic viruses and enhanced safety of recombinant live attenuated virus vaccines. Nevertheless each miR-TS needs to be carefully evaluated with regard to its optimal effectiveness. Moreover, as many viral vectors have a broad tissue tropism, which in general requires the use of several target sites to restrict a transgene to a specific tissue or cell type, target site optimization is required. The miR-ON system may be an alternative to overcome this disadvantage. Importantly, it may also be a platform to develop microRNA-regulated shRNA and artificial microRNA expression systems, which are currently not available.
Based on the results of the overall studies investigating potential side effects induced by microRNA-mediated transgene regulation, no major side effects have been observed to date, suggesting the mechanism is safe for therapeutic use. But there is a need to consider specific guidelines concerning the complementary between target sites and its corresponding microRNA, the number of target sites and the strength of transgene expression. In summary, microRNA regulation has been emerged as a powerful new technology to improve vector-mediated transgene expression in vitro and in vivo. It has the potential to be translated into clinical applications, thereby improving efficacy and safety of gene- and virotherapeutic approaches to the degree that their use can become more common and be applied to additional indications.
We thank Erik Wade for critical reading of the manuscript and helpful comments.
P- Reviewer: Guo ZS S- Editor: Kong JX L- Editor: A E- Editor: Li D
| 1. | Toivonen R, Koskenvuo J, Merentie M, Söderström M, Ylä-Herttuala S, Savontaus M. Intracardiac injection of a capsid-modified Ad5/35 results in decreased heart toxicity when compared to standard Ad5. Virol J. 2012;9:296. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 2. | Gupta S, Maitra R, Young D, Gupta A, Sen S. Silencing the myotrophin gene by RNA interference leads to the regression of cardiac hypertrophy. Am J Physiol Heart Circ Physiol. 2009;297:H627-H636. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 13] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 3. | Mizuguchi H, Hayakawa T. Targeted adenovirus vectors. Hum Gene Ther. 2004;15:1034-1044. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 129] [Cited by in RCA: 128] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 4. | Yamamoto M, Curiel DT. Current issues and future directions of oncolytic adenoviruses. Mol Ther. 2010;18:243-250. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 141] [Cited by in RCA: 158] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 5. | Liu Y, Siriwon N, Rohrs JA, Wang P. Generation of Targeted Adeno-Associated Virus (AAV) Vectors for Human Gene Therapy. Curr Pharm Des. 2015;21:3248-3256. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 13] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 6. | Qiao J, Doubrovin M, Sauter BV, Huang Y, Guo ZS, Balatoni J, Akhurst T, Blasberg RG, Tjuvajev JG, Chen SH. Tumor-specific transcriptional targeting of suicide gene therapy. Gene Ther. 2002;9:168-175. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 86] [Cited by in RCA: 87] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 7. | Moulin V, Morgan ME, Eleveld-Trancikova D, Haanen JB, Wielders E, Looman MW, Janssen RA, Figdor CG, Jansen BJ, Adema GJ. Targeting dendritic cells with antigen via dendritic cell-associated promoters. Cancer Gene Ther. 2012;19:303-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 8. | Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806-811. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10522] [Cited by in RCA: 10144] [Article Influence: 375.7] [Reference Citation Analysis (1)] |
| 9. | Brown BD, Venneri MA, Zingale A, Sergi Sergi L, Naldini L. Endogenous microRNA regulation suppresses transgene expression in hematopoietic lineages and enables stable gene transfer. Nat Med. 2006;12:585-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 390] [Cited by in RCA: 364] [Article Influence: 19.2] [Reference Citation Analysis (0)] |
| 10. | Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351-379. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2112] [Cited by in RCA: 2444] [Article Influence: 162.9] [Reference Citation Analysis (0)] |
| 11. | Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5833] [Cited by in RCA: 6541] [Article Influence: 384.8] [Reference Citation Analysis (0)] |
| 12. | Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215-233. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14460] [Cited by in RCA: 16085] [Article Influence: 1005.3] [Reference Citation Analysis (2)] |
| 13. | Brennecke J, Hipfner DR, Stark A, Russell RB, Cohen SM. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25-36. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1568] [Cited by in RCA: 1572] [Article Influence: 71.5] [Reference Citation Analysis (0)] |
| 14. | Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83-86. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2463] [Cited by in RCA: 2548] [Article Influence: 115.8] [Reference Citation Analysis (0)] |
| 15. | Harfe BD. MicroRNAs in vertebrate development. Curr Opin Genet Dev. 2005;15:410-415. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 268] [Cited by in RCA: 274] [Article Influence: 13.7] [Reference Citation Analysis (0)] |
| 16. | Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005;15:200-205. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 241] [Cited by in RCA: 244] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 17. | Brown BD, Gentner B, Cantore A, Colleoni S, Amendola M, Zingale A, Baccarini A, Lazzari G, Galli C, Naldini L. Endogenous microRNA can be broadly exploited to regulate transgene expression according to tissue, lineage and differentiation state. Nat Biotechnol. 2007;25:1457-1467. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 461] [Cited by in RCA: 443] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 18. | Chen C, Ridzon DA, Broomer AJ, Zhou Z, Lee DH, Nguyen JT, Barbisin M, Xu NL, Mahuvakar VR, Andersen MR. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005;33:e179. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3676] [Cited by in RCA: 3907] [Article Influence: 195.4] [Reference Citation Analysis (0)] |
| 19. | Liang Y, Ridzon D, Wong L, Chen C. Characterization of microRNA expression profiles in normal human tissues. BMC Genomics. 2007;8:166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 761] [Cited by in RCA: 841] [Article Influence: 46.7] [Reference Citation Analysis (0)] |
| 20. | Chang J, Nicolas E, Marks D, Sander C, Lerro A, Buendia MA, Xu C, Mason WS, Moloshok T, Bort R. miR-122, a mammalian liver-specific microRNA, is processed from hcr mRNA and may downregulate the high affinity cationic amino acid transporter CAT-1. RNA Biol. 2004;1:106-113. [PubMed] |
| 21. | Lan H, Lu H, Wang X, Jin H. MicroRNAs as potential biomarkers in cancer: opportunities and challenges. Biomed Res Int. 2015;2015:125094. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 150] [Cited by in RCA: 225] [Article Influence: 22.5] [Reference Citation Analysis (0)] |
| 22. | Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857-866. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5705] [Cited by in RCA: 6030] [Article Influence: 317.4] [Reference Citation Analysis (0)] |
| 23. | Zeng FR, Tang LJ, He Y, Garcia RC. An update on the role of miRNA-155 in pathogenic microbial infections. Microbes Infect. 2015;17:613-621. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 39] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 24. | Joladarashi D, Thandavarayan RA, Babu SS, Krishnamurthy P. Small engine, big power: microRNAs as regulators of cardiac diseases and regeneration. Int J Mol Sci. 2014;15:15891-15911. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 35] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 25. | D’Anzeo M, Faloppi L, Scartozzi M, Giampieri R, Bianconi M, Del Prete M, Silvestris N, Cascinu S. The role of micro-RNAs in hepatocellular carcinoma: from molecular biology to treatment. Molecules. 2014;19:6393-6406. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 26. | Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet. 2008;9:102-114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3661] [Cited by in RCA: 3994] [Article Influence: 234.9] [Reference Citation Analysis (0)] |
| 27. | Khvorova A, Reynolds A, Jayasena SD. Functional siRNAs and miRNAs exhibit strand bias. Cell. 2003;115:209-216. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1801] [Cited by in RCA: 1798] [Article Influence: 81.7] [Reference Citation Analysis (0)] |
| 28. | Schwarz DS, Hutvágner G, Du T, Xu Z, Aronin N, Zamore PD. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1969] [Cited by in RCA: 1956] [Article Influence: 88.9] [Reference Citation Analysis (0)] |
| 29. | Yang X, Du WW, Li H, Liu F, Khorshidi A, Rutnam ZJ, Yang BB. Both mature miR-17-5p and passenger strand miR-17-3p target TIMP3 and induce prostate tumor growth and invasion. Nucleic Acids Res. 2013;41:9688-9704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 157] [Cited by in RCA: 157] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 30. | Chang KW, Kao SY, Wu YH, Tsai MM, Tu HF, Liu CJ, Lui MT, Lin SC. Passenger strand miRNA miR-31* regulates the phenotypes of oral cancer cells by targeting RhoA. Oral Oncol. 2013;49:27-33. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 52] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 31. | Okamura K, Phillips MD, Tyler DM, Duan H, Chou YT, Lai EC. The regulatory activity of microRNA* species has substantial influence on microRNA and 3’ UTR evolution. Nat Struct Mol Biol. 2008;15:354-363. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 418] [Cited by in RCA: 397] [Article Influence: 23.4] [Reference Citation Analysis (0)] |
| 32. | Biasiolo M, Sales G, Lionetti M, Agnelli L, Todoerti K, Bisognin A, Coppe A, Romualdi C, Neri A, Bortoluzzi S. Impact of host genes and strand selection on miRNA and miRNA* expression. PLoS One. 2011;6:e23854. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 33. | Yang JS, Phillips MD, Betel D, Mu P, Ventura A, Siepel AC, Chen KC, Lai EC. Widespread regulatory activity of vertebrate microRNA* species. RNA. 2011;17:312-326. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 267] [Article Influence: 19.1] [Reference Citation Analysis (0)] |
| 34. | Vasudevan S, Tong Y, Steitz JA. Cell-cycle control of microRNA-mediated translation regulation. Cell Cycle. 2008;7:1545-1549. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 131] [Cited by in RCA: 126] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 35. | Vasudevan S, Tong Y, Steitz JA. Switching from repression to activation: microRNAs can up-regulate translation. Science. 2007;318:1931-1934. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1948] [Cited by in RCA: 2051] [Article Influence: 113.9] [Reference Citation Analysis (0)] |
| 36. | Ørom UA, Nielsen FC, Lund AH. MicroRNA-10a binds the 5’UTR of ribosomal protein mRNAs and enhances their translation. Mol Cell. 2008;30:460-471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 918] [Cited by in RCA: 1019] [Article Influence: 59.9] [Reference Citation Analysis (0)] |
| 37. | Henke JI, Goergen D, Zheng J, Song Y, Schüttler CG, Fehr C, Jünemann C, Niepmann M. microRNA-122 stimulates translation of hepatitis C virus RNA. EMBO J. 2008;27:3300-3310. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 492] [Cited by in RCA: 527] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 38. | Niepmann M. Activation of hepatitis C virus translation by a liver-specific microRNA. Cell Cycle. 2009;8:1473-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 62] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 39. | Hutvágner G, Zamore PD. A microRNA in a multiple-turnover RNAi enzyme complex. Science. 2002;297:2056-2060. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1474] [Cited by in RCA: 1472] [Article Influence: 64.0] [Reference Citation Analysis (0)] |
| 40. | Llave C, Xie Z, Kasschau KD, Carrington JC. Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science. 2002;297:2053-2056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1203] [Cited by in RCA: 1156] [Article Influence: 50.3] [Reference Citation Analysis (0)] |
| 41. | Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185-197. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1365] [Cited by in RCA: 1447] [Article Influence: 68.9] [Reference Citation Analysis (0)] |
| 42. | Song JJ, Smith SK, Hannon GJ, Joshua-Tor L. Crystal structure of Argonaute and its implications for RISC slicer activity. Science. 2004;305:1434-1437. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 999] [Cited by in RCA: 1032] [Article Influence: 49.1] [Reference Citation Analysis (0)] |
| 43. | Yekta S, Shih IH, Bartel DP. MicroRNA-directed cleavage of HOXB8 mRNA. Science. 2004;304:594-596. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1267] [Cited by in RCA: 1289] [Article Influence: 61.4] [Reference Citation Analysis (0)] |
| 44. | Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAS and their regulatory roles in plants. Annu Rev Plant Biol. 2006;57:19-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2045] [Cited by in RCA: 1750] [Article Influence: 92.1] [Reference Citation Analysis (0)] |
| 45. | Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843-854. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8672] [Cited by in RCA: 8876] [Article Influence: 277.4] [Reference Citation Analysis (0)] |
| 46. | Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553-563. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 997] [Cited by in RCA: 1020] [Article Influence: 51.0] [Reference Citation Analysis (0)] |
| 47. | Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, Bartel DP, Linsley PS, Johnson JM. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769-773. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3585] [Cited by in RCA: 3742] [Article Influence: 187.1] [Reference Citation Analysis (0)] |
| 48. | Rehwinkel J, Behm-Ansmant I, Gatfield D, Izaurralde E. A crucial role for GW182 and the DCP1: DCP2 decapping complex in miRNA-mediated gene silencing. RNA. 2005;11:1640-1647. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 359] [Cited by in RCA: 357] [Article Influence: 17.9] [Reference Citation Analysis (0)] |
| 49. | Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, Inoue K, Enright AJ, Schier AF. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75-79. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1197] [Cited by in RCA: 1200] [Article Influence: 63.2] [Reference Citation Analysis (0)] |
| 50. | Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855-862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2890] [Cited by in RCA: 2952] [Article Influence: 92.3] [Reference Citation Analysis (0)] |
| 51. | Brennecke J, Stark A, Russell RB, Cohen SM. Principles of microRNA-target recognition. PLoS Biol. 2005;3:e85. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1694] [Cited by in RCA: 1733] [Article Influence: 91.2] [Reference Citation Analysis (0)] |
| 52. | Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504-511. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1178] [Cited by in RCA: 1223] [Article Influence: 58.2] [Reference Citation Analysis (0)] |
| 53. | Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15-20. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8847] [Cited by in RCA: 9285] [Article Influence: 464.3] [Reference Citation Analysis (0)] |
| 54. | Nielsen CB, Shomron N, Sandberg R, Hornstein E, Kitzman J, Burge CB. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA. 2007;13:1894-1910. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 286] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 55. | Selbach M, Schwanhäusser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2577] [Cited by in RCA: 2720] [Article Influence: 160.0] [Reference Citation Analysis (0)] |
| 56. | Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes Dev. 2001;15:188-200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2287] [Cited by in RCA: 2292] [Article Influence: 95.5] [Reference Citation Analysis (0)] |
| 57. | Grimson A, Farh KK, Johnston WK, Garrett-Engele P, Lim LP, Bartel DP. MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol Cell. 2007;27:91-105. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2851] [Cited by in RCA: 2957] [Article Influence: 164.3] [Reference Citation Analysis (0)] |
| 58. | Vella MC, Choi EY, Lin SY, Reinert K, Slack FJ. The C. elegans microRNA let-7 binds to imperfect let-7 complementary sites from the lin-41 3’UTR. Genes Dev. 2004;18:132-137. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 357] [Cited by in RCA: 362] [Article Influence: 17.2] [Reference Citation Analysis (0)] |
| 59. | Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 235] [Cited by in RCA: 252] [Article Influence: 14.0] [Reference Citation Analysis (0)] |
| 60. | Kertesz M, Iovino N, Unnerstall U, Gaul U, Segal E. The role of site accessibility in microRNA target recognition. Nat Genet. 2007;39:1278-1284. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1820] [Cited by in RCA: 1890] [Article Influence: 105.0] [Reference Citation Analysis (0)] |
| 61. | Geisler A, Schön C, Größl T, Pinkert S, Stein EA, Kurreck J, Vetter R, Fechner H. Application of mutated miR-206 target sites enables skeletal muscle-specific silencing of transgene expression of cardiotropic AAV9 vectors. Mol Ther. 2013;21:924-933. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 28] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 62. | Kim SJ, Lee CH, Lee SW. Targeting the MicroRNA Passenger Strand for Regulating Therapeutic Transgenes. Nucleic Acid Ther. 2015;25:209-218. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 63. | Sakurai F, Katayama K, Mizuguchi H. MicroRNA-regulated transgene expression systems for gene therapy and virotherapy. Front Biosci (Landmark Ed). 2011;16:2389-2401. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 17] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 64. | Gentner B, Naldini L. Exploiting microRNA regulation for genetic engineering. Tissue Antigens. 2012;80:393-403. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 65. | Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401-1414. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3103] [Cited by in RCA: 2974] [Article Influence: 165.2] [Reference Citation Analysis (1)] |
| 66. | Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735-739. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2502] [Cited by in RCA: 2596] [Article Influence: 112.9] [Reference Citation Analysis (0)] |
| 67. | Geisler A, Jungmann A, Kurreck J, Poller W, Katus HA, Vetter R, Fechner H, Müller OJ. microRNA122-regulated transgene expression increases specificity of cardiac gene transfer upon intravenous delivery of AAV9 vectors. Gene Ther. 2011;18:199-209. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 109] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 68. | Qiao C, Yuan Z, Li J, He B, Zheng H, Mayer C, Li J, Xiao X. Liver-specific microRNA-122 target sequences incorporated in AAV vectors efficiently inhibits transgene expression in the liver. Gene Ther. 2011;18:403-410. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 130] [Cited by in RCA: 123] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 69. | Xie J, Xie Q, Zhang H, Ameres SL, Hung JH, Su Q, He R, Mu X, Seher Ahmed S, Park S. MicroRNA-regulated, systemically delivered rAAV9: a step closer to CNS-restricted transgene expression. Mol Ther. 2011;19:526-535. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 132] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 70. | Mullokandov G, Baccarini A, Ruzo A, Jayaprakash AD, Tung N, Israelow B, Evans MJ, Sachidanandam R, Brown BD. High-throughput assessment of microRNA activity and function using microRNA sensor and decoy libraries. Nat Methods. 2012;9:840-846. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 299] [Cited by in RCA: 325] [Article Influence: 25.0] [Reference Citation Analysis (0)] |
| 71. | Yi R, Fuchs E. MicroRNAs and their roles in mammalian stem cells. J Cell Sci. 2011;124:1775-1783. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 83] [Cited by in RCA: 74] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 72. | Kozomara A, Hunt S, Ninova M, Griffiths-Jones S, Ronshaugen M. Target repression induced by endogenous microRNAs: large differences, small effects. PLoS One. 2014;9:e104286. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 26] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 73. | Doench JG, Petersen CP, Sharp PA. siRNAs can function as miRNAs. Genes Dev. 2003;17:438-442. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 909] [Cited by in RCA: 908] [Article Influence: 41.3] [Reference Citation Analysis (0)] |
| 74. | Wang B, Love TM, Call ME, Doench JG, Novina CD. Recapitulation of short RNA-directed translational gene silencing in vitro. Mol Cell. 2006;22:553-560. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 137] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 75. | Baccarini A, Chauhan H, Gardner TJ, Jayaprakash AD, Sachidanandam R, Brown BD. Kinetic analysis reveals the fate of a microRNA following target regulation in mammalian cells. Curr Biol. 2011;21:369-376. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 202] [Cited by in RCA: 191] [Article Influence: 13.6] [Reference Citation Analysis (0)] |
| 76. | Ebert MS, Neilson JR, Sharp PA. MicroRNA sponges: competitive inhibitors of small RNAs in mammalian cells. Nat Methods. 2007;4:721-726. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1744] [Cited by in RCA: 1653] [Article Influence: 91.8] [Reference Citation Analysis (0)] |
| 77. | Elbashir SM, Martinez J, Patkaniowska A, Lendeckel W, Tuschl T. Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. EMBO J. 2001;20:6877-6888. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1037] [Cited by in RCA: 1033] [Article Influence: 43.0] [Reference Citation Analysis (0)] |
| 78. | Ameres SL, Martinez J, Schroeder R. Molecular basis for target RNA recognition and cleavage by human RISC. Cell. 2007;130:101-112. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 404] [Cited by in RCA: 395] [Article Influence: 21.9] [Reference Citation Analysis (0)] |
| 79. | Brown BD, Naldini L. Exploiting and antagonizing microRNA regulation for therapeutic and experimental applications. Nat Rev Genet. 2009;10:578-585. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 319] [Cited by in RCA: 296] [Article Influence: 18.5] [Reference Citation Analysis (0)] |
| 80. | Leja J, Nilsson B, Yu D, Gustafson E, Akerström G, Oberg K, Giandomenico V, Essand M. Double-detargeted oncolytic adenovirus shows replication arrest in liver cells and retains neuroendocrine cell killing ability. PLoS One. 2010;5:e8916. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 37] [Cited by in RCA: 38] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 81. | Roudaut C, Le Roy F, Suel L, Poupiot J, Charton K, Bartoli M, Richard I. Restriction of calpain3 expression to the skeletal muscle prevents cardiac toxicity and corrects pathology in a murine model of limb-girdle muscular dystrophy. Circulation. 2013;128:1094-1104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 42] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 82. | Wee LM, Flores-Jasso CF, Salomon WE, Zamore PD. Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell. 2012;151:1055-1067. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 307] [Cited by in RCA: 295] [Article Influence: 22.7] [Reference Citation Analysis (0)] |
| 83. | Sayeg MK, Weinberg BH, Cha SS, Goodloe M, Wong WW, Han X. Rationally Designed MicroRNA-Based Genetic Classifiers Target Specific Neurons in the Brain. ACS Synth Biol. 2015;4:788-795. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 22] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 84. | Baertsch MA, Leber MF, Bossow S, Singh M, Engeland CE, Albert J, Grossardt C, Jäger D, von Kalle C, Ungerechts G. MicroRNA-mediated multi-tissue detargeting of oncolytic measles virus. Cancer Gene Ther. 2014;21:373-380. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 49] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 85. | Wu C, Lin J, Hong M, Choudhury Y, Balani P, Leung D, Dang LH, Zhao Y, Zeng J, Wang S. Combinatorial control of suicide gene expression by tissue-specific promoter and microRNA regulation for cancer therapy. Mol Ther. 2009;17:2058-2066. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 86. | Bennett D, Sakurai F, Shimizu K, Matsui H, Tomita K, Suzuki T, Katayama K, Kawabata K, Mizuguchi H. Further reduction in adenovirus vector-mediated liver transduction without largely affecting transgene expression in target organ by exploiting microrna-mediated regulation and the Cre-loxP recombination system. Mol Pharm. 2012;9:3452-3463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 9] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 87. | Suzuki T, Sakurai F, Nakamura S, Kouyama E, Kawabata K, Kondoh M, Yagi K, Mizuguchi H. miR-122a-regulated expression of a suicide gene prevents hepatotoxicity without altering antitumor effects in suicide gene therapy. Mol Ther. 2008;16:1719-1726. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 53] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 88. | Otaegi G, Pollock A, Sun T. An Optimized Sponge for microRNA miR-9 Affects Spinal Motor Neuron Development in vivo. Front Neurosci. 2011;5:146. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 38] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 89. | Tay FC, Lim JK, Zhu H, Hin LC, Wang S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv Drug Deliv Rev. 2015;81:117-127. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 89] [Cited by in RCA: 108] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 90. | McCarty DM, Monahan PE, Samulski RJ. Self-complementary recombinant adeno-associated virus (scAAV) vectors promote efficient transduction independently of DNA synthesis. Gene Ther. 2001;8:1248-1254. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 510] [Cited by in RCA: 538] [Article Influence: 22.4] [Reference Citation Analysis (0)] |
| 91. | Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNA that targets Hand2 during cardiogenesis. Nature. 2005;436:214-220. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1242] [Cited by in RCA: 1240] [Article Influence: 62.0] [Reference Citation Analysis (0)] |
| 92. | Forman JJ, Coller HA. The code within the code: microRNAs target coding regions. Cell Cycle. 2010;9:1533-1541. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 133] [Cited by in RCA: 119] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 93. | Ylösmäki E, Hakkarainen T, Hemminki A, Visakorpi T, Andino R, Saksela K. Generation of a conditionally replicating adenovirus based on targeted destruction of E1A mRNA by a cell type-specific MicroRNA. J Virol. 2008;82:11009-11015. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 97] [Cited by in RCA: 97] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 94. | Lytle JR, Yario TA, Steitz JA. Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5’ UTR as in the 3’ UTR. Proc Natl Acad Sci USA. 2007;104:9667-9672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 820] [Cited by in RCA: 901] [Article Influence: 50.1] [Reference Citation Analysis (0)] |
| 95. | Kloosterman WP, Wienholds E, Ketting RF, Plasterk RH. Substrate requirements for let-7 function in the developing zebrafish embryo. Nucleic Acids Res. 2004;32:6284-6291. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 182] [Cited by in RCA: 183] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 96. | Easow G, Teleman AA, Cohen SM. Isolation of microRNA targets by miRNP immunopurification. RNA. 2007;13:1198-1204. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 240] [Article Influence: 13.3] [Reference Citation Analysis (0)] |
| 97. | Bail S, Swerdel M, Liu H, Jiao X, Goff LA, Hart RP, Kiledjian M. Differential regulation of microRNA stability. RNA. 2010;16:1032-1039. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 248] [Cited by in RCA: 235] [Article Influence: 15.7] [Reference Citation Analysis (0)] |
| 98. | Zacchigna S, Zentilin L, Giacca M. Adeno-associated virus vectors as therapeutic and investigational tools in the cardiovascular system. Circ Res. 2014;114:1827-1846. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 88] [Cited by in RCA: 115] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 99. | McCaffrey AP, Fawcett P, Nakai H, McCaffrey RL, Ehrhardt A, Pham TT, Pandey K, Xu H, Feuss S, Storm TA. The host response to adenovirus, helper-dependent adenovirus, and adeno-associated virus in mouse liver. Mol Ther. 2008;16:931-941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 75] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 100. | Jooss K, Yang Y, Fisher KJ, Wilson JM. Transduction of dendritic cells by DNA viral vectors directs the immune response to transgene products in muscle fibers. J Virol. 1998;72:4212-4223. [PubMed] |
| 101. | Fisher KJ, Jooss K, Alston J, Yang Y, Haecker SE, High K, Pathak R, Raper SE, Wilson JM. Recombinant adeno-associated virus for muscle directed gene therapy. Nat Med. 1997;3:306-312. [PubMed] |
| 102. | Röger C, Pozzuto T, Klopfleisch R, Kurreck J, Pinkert S, Fechner H. Expression of an engineered soluble coxsackievirus and adenovirus receptor by a dimeric AAV9 vector inhibits adenovirus infection in mice. Gene Ther. 2015;22:458-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 103. | McCarty DM, Young SM, Samulski RJ. Integration of adeno-associated virus (AAV) and recombinant AAV vectors. Annu Rev Genet. 2004;38:819-845. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 296] [Cited by in RCA: 328] [Article Influence: 16.4] [Reference Citation Analysis (0)] |
| 104. | Pacak CA, Byrne BJ. AAV vectors for cardiac gene transfer: experimental tools and clinical opportunities. Mol Ther. 2011;19:1582-1590. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 58] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 105. | Hajjar RJ, Zsebo K, Deckelbaum L, Thompson C, Rudy J, Yaroshinsky A, Ly H, Kawase Y, Wagner K, Borow K. Design of a phase 1/2 trial of intracoronary administration of AAV1/SERCA2a in patients with heart failure. J Card Fail. 2008;14:355-367. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 183] [Cited by in RCA: 156] [Article Influence: 9.2] [Reference Citation Analysis (0)] |
| 106. | Ortolano S, Spuch C, Navarro C. Present and future of adeno associated virus based gene therapy approaches. Recent Pat Endocr Metab Immune Drug Discov. 2012;6:47-66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 26] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 107. | Gao G, Vandenberghe LH, Wilson JM. New recombinant serotypes of AAV vectors. Curr Gene Ther. 2005;5:285-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 373] [Article Influence: 18.7] [Reference Citation Analysis (0)] |
| 108. | Gao GP, Alvira MR, Wang L, Calcedo R, Johnston J, Wilson JM. Novel adeno-associated viruses from rhesus monkeys as vectors for human gene therapy. Proc Natl Acad Sci USA. 2002;99:11854-11859. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1153] [Cited by in RCA: 1187] [Article Influence: 51.6] [Reference Citation Analysis (0)] |
| 109. | Pleger ST, Most P, Boucher M, Soltys S, Chuprun JK, Pleger W, Gao E, Dasgupta A, Rengo G, Remppis A. Stable myocardial-specific AAV6-S100A1 gene therapy results in chronic functional heart failure rescue. Circulation. 2007;115:2506-2515. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 136] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 110. | Zincarelli C, Soltys S, Rengo G, Rabinowitz JE. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol Ther. 2008;16:1073-1080. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 883] [Cited by in RCA: 1112] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 111. | Bish LT, Morine K, Sleeper MM, Sanmiguel J, Wu D, Gao G, Wilson JM, Sweeney HL. Adeno-associated virus (AAV) serotype 9 provides global cardiac gene transfer superior to AAV1, AAV6, AAV7, and AAV8 in the mouse and rat. Hum Gene Ther. 2008;19:1359-1368. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 214] [Cited by in RCA: 233] [Article Influence: 14.6] [Reference Citation Analysis (0)] |
| 112. | Pacak CA, Mah CS, Thattaliyath BD, Conlon TJ, Lewis MA, Cloutier DE, Zolotukhin I, Tarantal AF, Byrne BJ. Recombinant adeno-associated virus serotype 9 leads to preferential cardiac transduction in vivo. Circ Res. 2006;99:e3-e9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 288] [Cited by in RCA: 310] [Article Influence: 16.3] [Reference Citation Analysis (0)] |
| 113. | Inagaki K, Fuess S, Storm TA, Gibson GA, Mctiernan CF, Kay MA, Nakai H. Robust systemic transduction with AAV9 vectors in mice: efficient global cardiac gene transfer superior to that of AAV8. Mol Ther. 2006;14:45-53. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 447] [Cited by in RCA: 490] [Article Influence: 25.8] [Reference Citation Analysis (0)] |
| 114. | Fechner H, Sipo I, Westermann D, Pinkert S, Wang X, Suckau L, Kurreck J, Zeichhardt H, Müller O, Vetter R. Cardiac-targeted RNA interference mediated by an AAV9 vector improves cardiac function in coxsackievirus B3 cardiomyopathy. J Mol Med (Berl). 2008;86:987-997. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 115. | Suckau L, Fechner H, Chemaly E, Krohn S, Hadri L, Kockskämper J, Westermann D, Bisping E, Ly H, Wang X. Long-term cardiac-targeted RNA interference for the treatment of heart failure restores cardiac function and reduces pathological hypertrophy. Circulation. 2009;119:1241-1252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 180] [Cited by in RCA: 164] [Article Influence: 10.3] [Reference Citation Analysis (0)] |
| 116. | Bish LT, Sleeper MM, Brainard B, Cole S, Russell N, Withnall E, Arndt J, Reynolds C, Davison E, Sanmiguel J. Percutaneous transendocardial delivery of self-complementary adeno-associated virus 6 achieves global cardiac gene transfer in canines. Mol Ther. 2008;16:1953-1959. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 69] [Cited by in RCA: 61] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 117. | Greenberg B, Yaroshinsky A, Zsebo KM, Butler J, Felker GM, Voors AA, Rudy JJ, Wagner K, Hajjar RJ. Design of a phase 2b trial of intracoronary administration of AAV1/SERCA2a in patients with advanced heart failure: the CUPID 2 trial (calcium up-regulation by percutaneous administration of gene therapy in cardiac disease phase 2b). JACC Heart Fail. 2014;2:84-92. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 99] [Cited by in RCA: 110] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 118. | Pacak CA, Sakai Y, Thattaliyath BD, Mah CS, Byrne BJ. Tissue specific promoters improve specificity of AAV9 mediated transgene expression following intra-vascular gene delivery in neonatal mice. Genet Vaccines Ther. 2008;6:13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 66] [Cited by in RCA: 82] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 119. | Salva MZ, Himeda CL, Tai PW, Nishiuchi E, Gregorevic P, Allen JM, Finn EE, Nguyen QG, Blankinship MJ, Meuse L. Design of tissue-specific regulatory cassettes for high-level rAAV-mediated expression in skeletal and cardiac muscle. Mol Ther. 2007;15:320-329. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 137] [Cited by in RCA: 173] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 120. | Aikawa R, Huggins GS, Snyder RO. Cardiomyocyte-specific gene expression following recombinant adeno-associated viral vector transduction. J Biol Chem. 2002;277:18979-18985. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 50] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 121. | Phillips MI, Tang Y, Schmidt-Ott K, Qian K, Kagiyama S. Vigilant vector: heart-specific promoter in an adeno-associated virus vector for cardioprotection. Hypertension. 2002;39:651-655. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 65] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 122. | Müller OJ, Leuchs B, Pleger ST, Grimm D, Franz WM, Katus HA, Kleinschmidt JA. Improved cardiac gene transfer by transcriptional and transductional targeting of adeno-associated viral vectors. Cardiovasc Res. 2006;70:70-78. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 123] [Cited by in RCA: 121] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 123. | Sempere LF, Freemantle S, Pitha-Rowe I, Moss E, Dmitrovsky E, Ambros V. Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 2004;5:R13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1115] [Cited by in RCA: 1226] [Article Influence: 58.4] [Reference Citation Analysis (0)] |
| 124. | Maron A, Havaux N, Le Roux A, Knoops B, Perricaudet M, Octave JN. Differential toxicity of ganciclovir for rat neurons and astrocytes in primary culture following adenovirus-mediated transfer of the HSVtk gene. Gene Ther. 1997;4:25-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 12] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 125. | Mizuguchi H, Hayakawa T. Enhanced antitumor effect and reduced vector dissemination with fiber-modified adenovirus vectors expressing herpes simplex virus thymidine kinase. Cancer Gene Ther. 2002;9:236-242. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 35] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 126. | Brand K, Arnold W, Bartels T, Lieber A, Kay MA, Strauss M, Dörken B. Liver-associated toxicity of the HSV-tk/GCV approach and adenoviral vectors. Cancer Gene Ther. 1997;4:9-16. [PubMed] |
| 127. | Skalsky RL, Cullen BR. Reduced expression of brain-enriched microRNAs in glioblastomas permits targeted regulation of a cell death gene. PLoS One. 2011;6:e24248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 141] [Cited by in RCA: 146] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 128. | Della Peruta M, Badar A, Rosales C, Chokshi S, Kia A, Nathwani D, Galante E, Yan R, Arstad E, Davidoff AM. Preferential targeting of disseminated liver tumors using a recombinant adeno-associated viral vector. Hum Gene Ther. 2015;26:94-103. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 28] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 129. | Trepel M, Körbelin J, Spies E, Heckmann MB, Hunger A, Fehse B, Katus HA, Kleinschmidt JA, Müller OJ, Michelfelder S. Treatment of multifocal breast cancer by systemic delivery of dual-targeted adeno-associated viral vectors. Gene Ther. 2015;22:840-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 16] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 130. | Won YS, Jeong JS, Kim SJ, Ju MH, Lee SW. Targeted anticancer effect through microRNA-181a regulated tumor-specific hTERT replacement. Cancer Lett. 2015;356:918-928. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 131. | Kim J, Won R, Ban G, Ju MH, Cho KS, Young Han S, Jeong JS, Lee SW. Targeted Regression of Hepatocellular Carcinoma by Cancer-Specific RNA Replacement through MicroRNA Regulation. Sci Rep. 2015;5:12315. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 18] [Cited by in RCA: 20] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 132. | Lachmann N, Jagielska J, Heckl D, Brennig S, Pfaff N, Maetzig T, Modlich U, Cantz T, Gentner B, Schambach A. MicroRNA-150-regulated vectors allow lymphocyte-sparing transgene expression in hematopoietic gene therapy. Gene Ther. 2012;19:915-924. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 133. | Gentner B, Visigalli I, Hiramatsu H, Lechman E, Ungari S, Giustacchini A, Schira G, Amendola M, Quattrini A, Martino S. Identification of hematopoietic stem cell-specific miRNAs enables gene therapy of globoid cell leukodystrophy. Sci Transl Med. 2010;2:58ra84. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 179] [Cited by in RCA: 170] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 134. | Escobar G, Moi D, Ranghetti A, Ozkal-Baydin P, Squadrito ML, Kajaste-Rudnitski A, Bondanza A, Gentner B, De Palma M, Mazzieri R. Genetic engineering of hematopoiesis for targeted IFN-α delivery inhibits breast cancer progression. Sci Transl Med. 2014;6:217ra3. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 76] [Cited by in RCA: 97] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 135. | Chiriaco M, Farinelli G, Capo V, Zonari E, Scaramuzza S, Di Matteo G, Sergi LS, Migliavacca M, Hernandez RJ, Bombelli F. Dual-regulated lentiviral vector for gene therapy of X-linked chronic granulomatosis. Mol Ther. 2014;22:1472-1483. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 53] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 136. | Di Stefano B, Maffioletti SM, Gentner B, Ungaro F, Schira G, Naldini L, Broccoli V. A microRNA-based system for selecting and maintaining the pluripotent state in human induced pluripotent stem cells. Stem Cells. 2011;29:1684-1695. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 24] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 137. | Kamata M, Liang M, Liu S, Nagaoka Y, Chen IS. Live cell monitoring of hiPSC generation and differentiation using differential expression of endogenous microRNAs. PLoS One. 2010;5:e11834. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 27] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 138. | Sachdeva R, Jönsson ME, Nelander J, Kirkeby A, Guibentif C, Gentner B, Naldini L, Björklund A, Parmar M, Jakobsson J. Tracking differentiating neural progenitors in pluripotent cultures using microRNA-regulated lentiviral vectors. Proc Natl Acad Sci USA. 2010;107:11602-11607. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 34] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 139. | Murlidharan G, Samulski RJ, Asokan A. Biology of adeno-associated viral vectors in the central nervous system. Front Mol Neurosci. 2014;7:76. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 113] [Cited by in RCA: 130] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 140. | Smirnova L, Gräfe A, Seiler A, Schumacher S, Nitsch R, Wulczyn FG. Regulation of miRNA expression during neural cell specification. Eur J Neurosci. 2005;21:1469-1477. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 502] [Cited by in RCA: 534] [Article Influence: 26.7] [Reference Citation Analysis (0)] |
| 141. | Colin A, Faideau M, Dufour N, Auregan G, Hassig R, Andrieu T, Brouillet E, Hantraye P, Bonvento G, Déglon N. Engineered lentiviral vector targeting astrocytes in vivo. Glia. 2009;57:667-679. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 120] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 142. | Perrault I, Rozet JM, Gerber S, Ghazi I, Leowski C, Ducroq D, Souied E, Dufier JL, Munnich A, Kaplan J. Leber congenital amaurosis. Mol Genet Metab. 1999;68:200-208. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 107] [Cited by in RCA: 112] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 143. | Karali M, Manfredi A, Puppo A, Marrocco E, Gargiulo A, Allocca M, Corte MD, Rossi S, Giunti M, Bacci ML. MicroRNA-restricted transgene expression in the retina. PLoS One. 2011;6:e22166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 48] [Cited by in RCA: 55] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 144. | Kay CN, Ryals RC, Aslanidi GV, Min SH, Ruan Q, Sun J, Dyka FM, Kasuga D, Ayala AE, Van Vliet K. Targeting photoreceptors via intravitreal delivery using novel, capsid-mutated AAV vectors. PLoS One. 2013;8:e62097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 119] [Cited by in RCA: 132] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 145. | Brown BD, Cantore A, Annoni A, Sergi LS, Lombardo A, Della Valle P, D’Angelo A, Naldini L. A microRNA-regulated lentiviral vector mediates stable correction of hemophilia B mice. Blood. 2007;110:4144-4152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 197] [Cited by in RCA: 191] [Article Influence: 10.6] [Reference Citation Analysis (0)] |
| 146. | Follenzi A, Battaglia M, Lombardo A, Annoni A, Roncarolo MG, Naldini L. Targeting lentiviral vector expression to hepatocytes limits transgene-specific immune response and establishes long-term expression of human antihemophilic factor IX in mice. Blood. 2004;103:3700-3709. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 162] [Cited by in RCA: 163] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 147. | Annoni A, Brown BD, Cantore A, Sergi LS, Naldini L, Roncarolo MG. In vivo delivery of a microRNA-regulated transgene induces antigen-specific regulatory T cells and promotes immunologic tolerance. Blood. 2009;114:5152-5161. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 103] [Cited by in RCA: 112] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 148. | Goudy KS, Annoni A, Naldini L, Roncarolo MG. Manipulating Immune Tolerance with Micro-RNA Regulated Gene Therapy. Front Microbiol. 2011;2:221. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 14] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 149. | Mátrai J, Cantore A, Bartholomae CC, Annoni A, Wang W, Acosta-Sanchez A, Samara-Kuko E, De Waele L, Ma L, Genovese P. Hepatocyte-targeted expression by integrase-defective lentiviral vectors induces antigen-specific tolerance in mice with low genotoxic risk. Hepatology. 2011;53:1696-1707. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 117] [Cited by in RCA: 120] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 150. | Kron MW, Espenlaub S, Engler T, Schirmbeck R, Kochanek S, Kreppel F. miRNA-mediated silencing in hepatocytes can increase adaptive immune responses to adenovirus vector-delivered transgenic antigens. Mol Ther. 2011;19:1547-1557. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 151. | Lu Y, Song S. Distinct immune responses to transgene products from rAAV1 and rAAV8 vectors. Proc Natl Acad Sci USA. 2009;106:17158-17162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 53] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 152. | Majowicz A, Maczuga P, Kwikkers KL, van der Marel S, van Logtenstein R, Petry H, van Deventer SJ, Konstantinova P, Ferreira V. Mir-142-3p target sequences reduce transgene-directed immunogenicity following intramuscular adeno-associated virus 1 vector-mediated gene delivery. J Gene Med. 2013;15:219-232. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 50] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 153. | Boisgerault F, Gross DA, Ferrand M, Poupiot J, Darocha S, Richard I, Galy A. Prolonged gene expression in muscle is achieved without active immune tolerance using microrRNA 142.3p-regulated rAAV gene transfer. Hum Gene Ther. 2013;24:393-405. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 46] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 154. | Kelly EJ, Hadac EM, Greiner S, Russell SJ. Engineering microRNA responsiveness to decrease virus pathogenicity. Nat Med. 2008;14:1278-1283. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 161] [Cited by in RCA: 174] [Article Influence: 10.2] [Reference Citation Analysis (0)] |
| 155. | Edge RE, Falls TJ, Brown CW, Lichty BD, Atkins H, Bell JC. A let-7 MicroRNA-sensitive vesicular stomatitis virus demonstrates tumor-specific replication. Mol Ther. 2008;16:1437-1443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 100] [Cited by in RCA: 101] [Article Influence: 5.9] [Reference Citation Analysis (0)] |
| 156. | Kelly EJ, Nace R, Barber GN, Russell SJ. Attenuation of vesicular stomatitis virus encephalitis through microRNA targeting. J Virol. 2010;84:1550-1562. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 87] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 157. | Leber MF, Bossow S, Leonard VH, Zaoui K, Grossardt C, Frenzke M, Miest T, Sawall S, Cattaneo R, von Kalle C. MicroRNA-sensitive oncolytic measles viruses for cancer-specific vector tropism. Mol Ther. 2011;19:1097-1106. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 80] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 158. | Ratnik K, Viru L, Merits A. Control of the rescue and replication of Semliki Forest virus recombinants by the insertion of miRNA target sequences. PLoS One. 2013;8:e75802. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 5] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 159. | Callegari E, Elamin BK, D’Abundo L, Falzoni S, Donvito G, Moshiri F, Milazzo M, Altavilla G, Giacomelli L, Fornari F. Anti-tumor activity of a miR-199-dependent oncolytic adenovirus. PLoS One. 2013;8:e73964. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 44] [Cited by in RCA: 48] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 160. | Ylösmäki E, Lavilla-Alonso S, Jäämaa S, Vähä-Koskela M, af Hällström T, Hemminki A, Arola J, Mäkisalo H, Saksela K. MicroRNA-mediated suppression of oncolytic adenovirus replication in human liver. PLoS One. 2013;8:e54506. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 161. | Sugio K, Sakurai F, Katayama K, Tashiro K, Matsui H, Kawabata K, Kawase A, Iwaki M, Hayakawa T, Fujiwara T. Enhanced safety profiles of the telomerase-specific replication-competent adenovirus by incorporation of normal cell-specific microRNA-targeted sequences. Clin Cancer Res. 2011;17:2807-2818. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 33] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 162. | Jin H, Lv S, Yang J, Wang X, Hu H, Su C, Zhou C, Li J, Huang Y, Li L. Use of microRNA Let-7 to control the replication specificity of oncolytic adenovirus in hepatocellular carcinoma cells. PLoS One. 2011;6:e21307. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 44] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 163. | Lee CY, Rennie PS, Jia WW. MicroRNA regulation of oncolytic herpes simplex virus-1 for selective killing of prostate cancer cells. Clin Cancer Res. 2009;15:5126-5135. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 164. | Fu X, Rivera A, Tao L, De Geest B, Zhang X. Construction of an oncolytic herpes simplex virus that precisely targets hepatocellular carcinoma cells. Mol Ther. 2012;20:339-346. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 49] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 165. | Hikichi M, Kidokoro M, Haraguchi T, Iba H, Shida H, Tahara H, Nakamura T. MicroRNA regulation of glycoprotein B5R in oncolytic vaccinia virus reduces viral pathogenicity without impairing its antitumor efficacy. Mol Ther. 2011;19:1107-1115. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 40] [Article Influence: 2.9] [Reference Citation Analysis (0)] |
| 166. | Cawood R, Chen HH, Carroll F, Bazan-Peregrino M, van Rooijen N, Seymour LW. Use of tissue-specific microRNA to control pathology of wild-type adenovirus without attenuation of its ability to kill cancer cells. PLoS Pathog. 2009;5:e1000440. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 99] [Cited by in RCA: 106] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 167. | Bofill-De Ros X, Gironella M, Fillat C. miR-148a- and miR-216a-regulated oncolytic adenoviruses targeting pancreatic tumors attenuate tissue damage without perturbation of miRNA activity. Mol Ther. 2014;22:1665-1677. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 31] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 168. | Ruiz AJ, Russell SJ. MicroRNAs and oncolytic viruses. Curr Opin Virol. 2015;13:40-48. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 53] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 169. | Lauring AS, Jones JO, Andino R. Rationalizing the development of live attenuated virus vaccines. Nat Biotechnol. 2010;28:573-579. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 269] [Cited by in RCA: 224] [Article Influence: 14.9] [Reference Citation Analysis (0)] |
| 170. | Barnes D, Kunitomi M, Vignuzzi M, Saksela K, Andino R. Harnessing endogenous miRNAs to control virus tissue tropism as a strategy for developing attenuated virus vaccines. Cell Host Microbe. 2008;4:239-248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 142] [Cited by in RCA: 139] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 171. | Perez JT, Pham AM, Lorini MH, Chua MA, Steel J, tenOever BR. MicroRNA-mediated species-specific attenuation of influenza A virus. Nat Biotechnol. 2009;27:572-576. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 121] [Cited by in RCA: 121] [Article Influence: 7.6] [Reference Citation Analysis (0)] |
| 172. | Yen LC, Lin YL, Sung HH, Liao JT, Tsao CH, Su CM, Lin CK, Liao CL. Neurovirulent flavivirus can be attenuated in mice by incorporation of neuron-specific microRNA recognition elements into viral genome. Vaccine. 2013;31:5915-5922. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 21] [Cited by in RCA: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 173. | Heiss BL, Maximova OA, Thach DC, Speicher JM, Pletnev AG. MicroRNA targeting of neurotropic flavivirus: effective control of virus escape and reversion to neurovirulent phenotype. J Virol. 2012;86:5647-5659. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 51] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 174. | Pham AM, Langlois RA, TenOever BR. Replication in cells of hematopoietic origin is necessary for Dengue virus dissemination. PLoS Pathog. 2012;8:e1002465. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 77] [Cited by in RCA: 84] [Article Influence: 6.5] [Reference Citation Analysis (0)] |
| 175. | Tsetsarkin KA, Liu G, Kenney H, Bustos-Arriaga J, Hanson CT, Whitehead SS, Pletnev AG. Dual miRNA targeting restricts host range and attenuates neurovirulence of flaviviruses. PLoS Pathog. 2015;11:e1004852. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 25] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 176. | Kamrud KI, Coffield VM, Owens G, Goodman C, Alterson K, Custer M, Murphy MA, Lewis W, Timberlake S, Wansley EK. In vitro and in vivo characterization of microRNA-targeted alphavirus replicon and helper RNAs. J Virol. 2010;84:7713-7725. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 22] [Cited by in RCA: 23] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 177. | Annalisa Lattanzi, Bernhard Gentner, Daniela Corno, Tiziano Di Tomaso, Pieter Mestdagh, Frank Speleman, Luigi Naldini, Angela Gritti. Dynamic Activity of miR-125b and miR-93 during Murine Neural Stem Cell Differentiation In Vitro and in the Subventricular Zone Neurogenic Niche. PLoS One. 2013;8:e67411. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 27] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 178. | Amendola M, Giustacchini A, Gentner B, Naldini L. A double-switch vector system positively regulates transgene expression by endogenous microRNA expression (miR-ON vector). Mol Ther. 2013;21:934-946. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 25] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 179. | Lau S, Rylander Ottosson D, Jakobsson J, Parmar M. Direct neural conversion from human fibroblasts using self-regulating and nonintegrating viral vectors. Cell Rep. 2014;9:1673-1680. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 32] [Cited by in RCA: 33] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 180. | Wright NT, Varney KM, Ellis KC, Markowitz J, Gitti RK, Zimmer DB, Weber DJ. The three-dimensional solution structure of Ca(2+)-bound S100A1 as determined by NMR spectroscopy. J Mol Biol. 2005;353:410-426. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 71] [Cited by in RCA: 81] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 181. | Card PB, Hogg RT, Gil Del Alcazar CR, Gerard RD. MicroRNA silencing improves the tumor specificity of adenoviral transgene expression. Cancer Gene Ther. 2012;19:451-459. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 11] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 182. | Papapetrou EP, Kovalovsky D, Beloeil L, Sant’angelo D, Sadelain M. Harnessing endogenous miR-181a to segregate transgenic antigen receptor expression in developing versus post-thymic T cells in murine hematopoietic chimeras. J Clin Invest. 2009;119:157-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 31] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 183. | Matsui H, Hegadorn C, Ozelo M, Burnett E, Tuttle A, Labelle A, McCray PB, Naldini L, Brown B, Hough C. A microRNA-regulated and GP64-pseudotyped lentiviral vector mediates stable expression of FVIII in a murine model of Hemophilia A. Mol Ther. 2011;19:723-730. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 59] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 184. | Åkerblom M, Sachdeva R, Barde I, Verp S, Gentner B, Trono D, Jakobsson J. MicroRNA-124 is a subventricular zone neuronal fate determinant. J Neurosci. 2012;32:8879-8889. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 152] [Cited by in RCA: 161] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 185. | O’Neill SM, Hinkle C, Chen SJ, Sandhu A, Hovhannisyan R, Stephan S, Lagor WR, Ahima RS, Johnston JC, Reilly MP. Targeting adipose tissue via systemic gene therapy. Gene Ther. 2014;21:653-661. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 30] [Cited by in RCA: 45] [Article Influence: 4.1] [Reference Citation Analysis (0)] |
| 186. | Greig JA, Peng H, Ohlstein J, Medina-Jaszek CA, Ahonkhai O, Mentzinger A, Grant RL, Roy S, Chen SJ, Bell P. Intramuscular injection of AAV8 in mice and macaques is associated with substantial hepatic targeting and transgene expression. PLoS One. 2014;9:e112268. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 48] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 187. | Jessup M, Greenberg B, Mancini D, Cappola T, Pauly DF, Jaski B, Yaroshinsky A, Zsebo KM, Dittrich H, Hajjar RJ. Calcium Upregulation by Percutaneous Administration of Gene Therapy in Cardiac Disease (CUPID): a phase 2 trial of intracoronary gene therapy of sarcoplasmic reticulum Ca2+-ATPase in patients with advanced heart failure. Circulation. 2011;124:304-313. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 568] [Cited by in RCA: 579] [Article Influence: 41.4] [Reference Citation Analysis (0)] |
| 188. | Yang L, Jiang J, Drouin LM, Agbandje-McKenna M, Chen C, Qiao C, Pu D, Hu X, Wang DZ, Li J. A myocardium tropic adeno-associated virus (AAV) evolved by DNA shuffling and in vivo selection. Proc Natl Acad Sci USA. 2009;106:3946-3951. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 168] [Cited by in RCA: 165] [Article Influence: 10.3] [Reference Citation Analysis (0)] |