Copyright
©The Author(s) 2016.
World J Exp Med. May 20, 2016; 6(2): 37-54
Published online May 20, 2016. doi: 10.5493/wjem.v6.i2.37
Published online May 20, 2016. doi: 10.5493/wjem.v6.i2.37
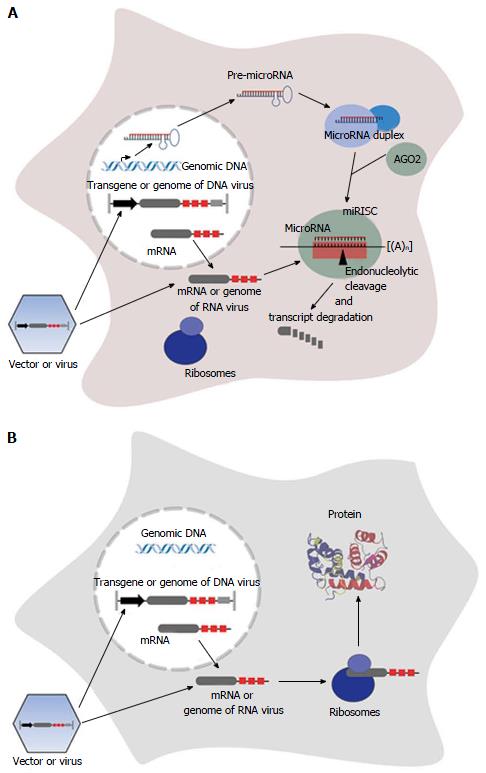
Figure 1 Principle of microRNA-mediated suppression of transgene expression and viral replication.
After transduction/infection of a cell, vector DNA/viral DNA is transcribed in the nucleus (for lentiviral vectors integrated into host DNA). The transcript/viral RNA containing artificial miR-TS (red boxes) is transported into the cytoplasm. If corresponding microRNA is expressed (A), it binds to miR-TS and the target RNA is endonucleolytically cleaved and degraded. If certain microRNAs are not expressed (B), the RNA is translated into protein. The protein shown in the three-dimensional structure is dimeric S100A1[180]. AGO: Argonaute; miRISC: MicroRNA-induced silencing complex.
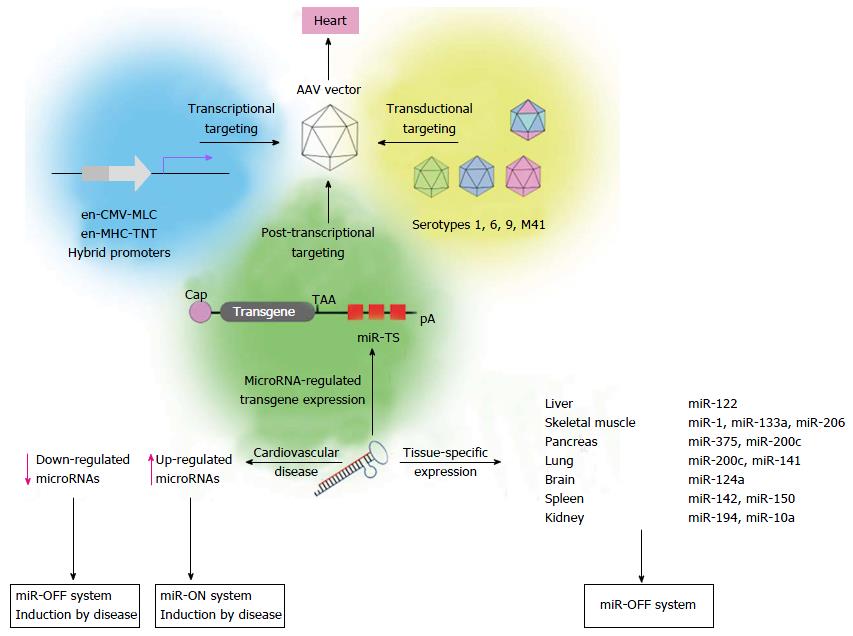
Figure 2 Adeno-associated virus vector-based microRNA targeting strategies to improve cardiac - specific gene therapy.
Cardiac-specific gene transfer of AAV vectors can be obtained by different targeting strategies, including transcriptional targeting using cardiac hybrid promoters[68,122], transductional targeting[109-113,187,188] or microRNA-regulated post-transcriptional transgene suppression. miR-OFF system (right site): Expression of miR-TS-bearing transgenes can be selectively suppressed in non-cardiac tissue by microRNAs, which are expressed in these tissues, but not in the heart. For the miR-OFF system (left site): If microRNAs are selectively down-regulated in cardiovascular diseases, transgene expression may be restricted to these cells by use of miR-TS corresponding to down-regulated microRNAs. If microRNAs are up-regulated in cardiovascular diseases, corresponding miR-TS can be used in the miR-ON system[178] for regulation of a repressor protein to restrict transgene expression to respective cells. Shown microRNAs are specifically expressed in mouse tissues (unpublished). AAV: Adeno-associated virus; pA: Polyadenylation site; CMV: Cytomegalovirus; MLC: Myosin light chain; MHC: Myosin heavy chain; TNT: Troponin T.
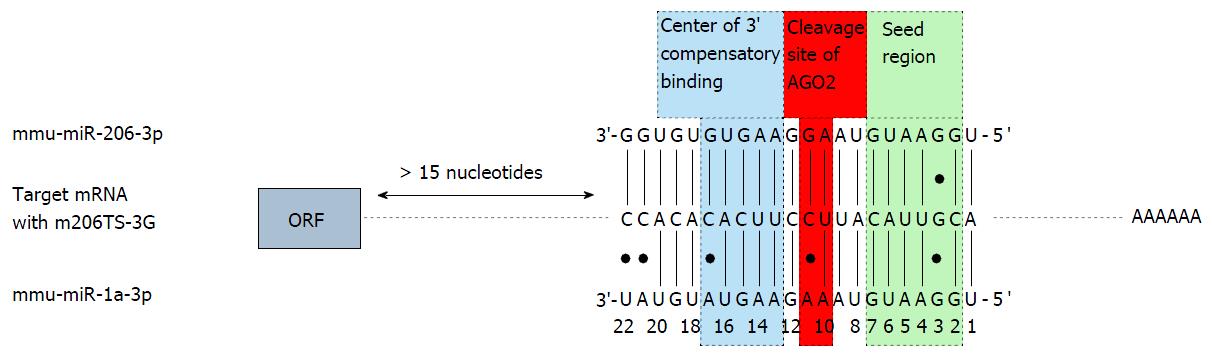
Figure 3 Sequence alignment of murine miR-206, miR-1 and mutated miR-206TS, referred to as m206TS-3G.
The last of these sequences contains a C to G transition compared to miR-206TS at microRNA nucleotide position 3. Vertical dashes, Watson-Crick pairing; dots, mismatches; green boxed capital letters, seed region. Potential cleavage site of AGO2 between nucleotide 10 and 11 is boxed in red. The center of the potential 3’ compensatory microRNA binding is boxed in blue. Nucleotide positions are numbered according to microRNA sequence. ORF: Open reading frame; AGO: Argonaute.
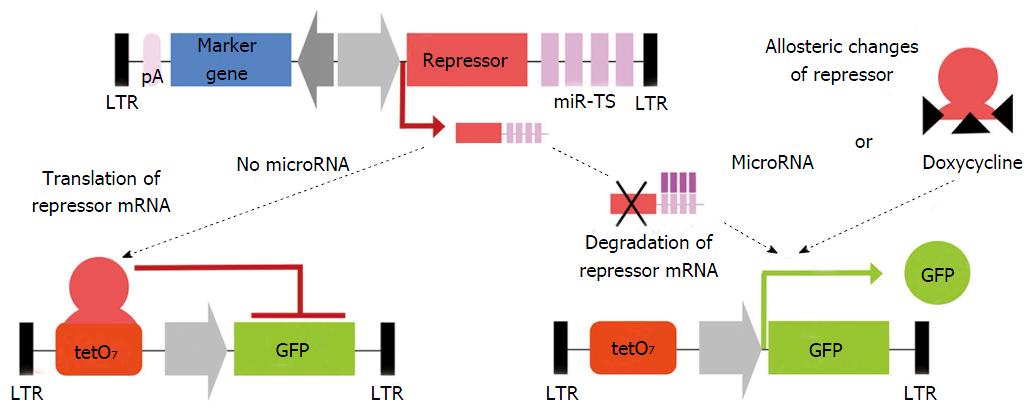
Figure 4 Schematic of the miR-ON system using lentiviral vectors in the indicated conditions.
Two independent lentiviral vectors containing a repressor protein and a GFP reporter with operator sequences for the repressor protein, respectively. The repressor protein, either tetracycline repressor (tTR) or tTR-Kruppel-associated box, constitutively binds to the tetO sequence, thereby suppressing reporter gene expression. If the repressor mRNA containing miR-TS is not translated into native protein due to microRNA-mediated degradation of the repressor mRNA or is unable to bind tetO sequence because of doxycycline-induced allosteric changes of the repressor, GFP expression is switched on. GFP expression is inhibited if the repressor is translated due to lack of an inhibiting microRNA, thereby allowing the repressor to bind to the operator sequence. Arrows indicate orientation of transcription. Modified according to[178]. LTR: Long terminal repeats; miR-TS: Artificial microRNA target sites; pA: Polyadenylation site; tetO7: Seven tandemly arranged tetracycline operator sequences; GFP: Green fluorescent protein.
- Citation: Geisler A, Fechner H. MicroRNA-regulated viral vectors for gene therapy. World J Exp Med 2016; 6(2): 37-54
- URL: https://www.wjgnet.com/2220-315X/full/v6/i2/37.htm
- DOI: https://dx.doi.org/10.5493/wjem.v6.i2.37