Copyright
©2012 Baishideng.
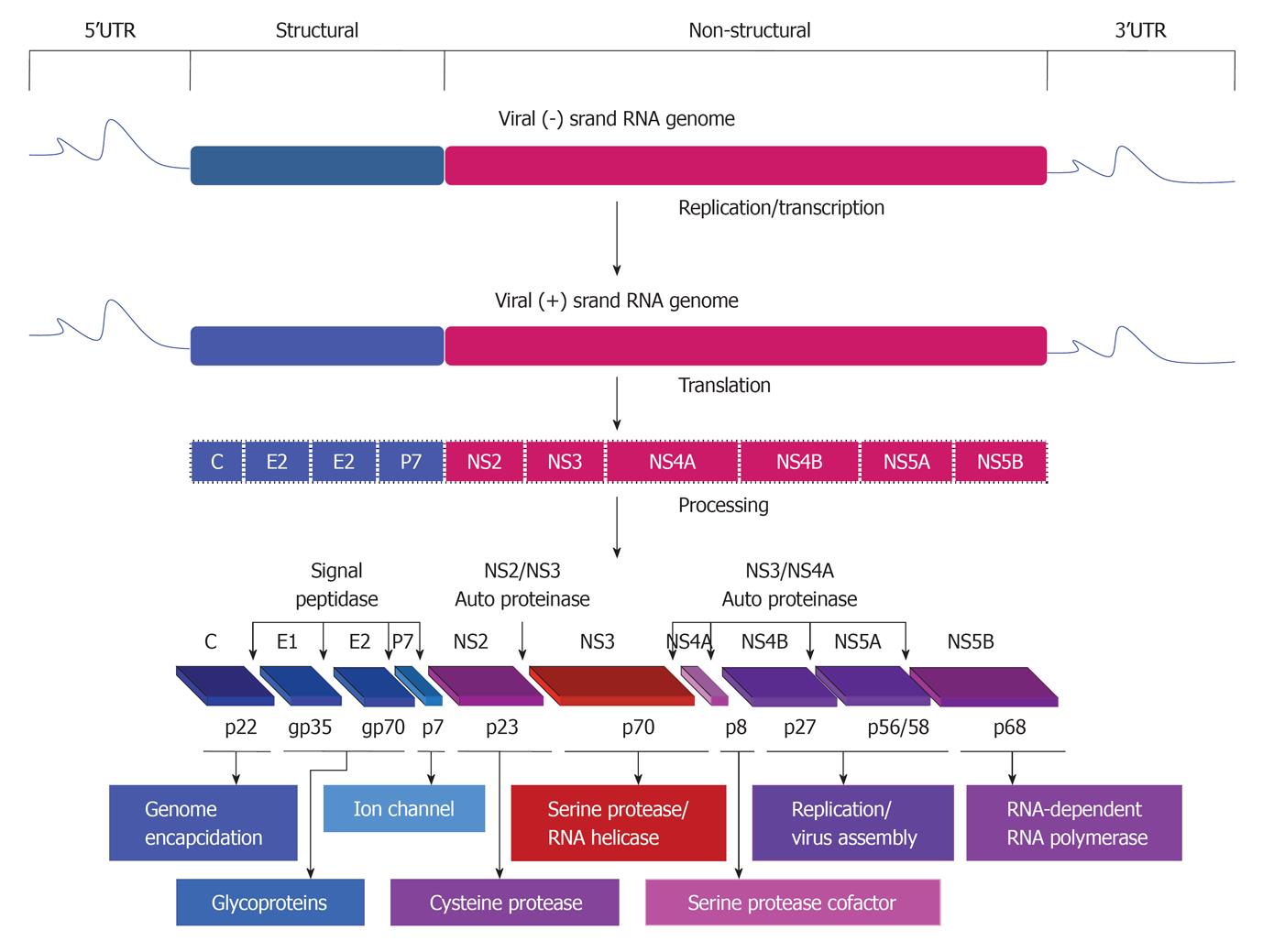
Figure 1 Hepatitis C virus genomic organization,replication,translation and generation of functional proteins.
Proteins encoded by the hepatitis C virus (HCV) genome. HCV is formed by an enveloped particle harboring a plus-strand RNA of about 9.6 kb. The genome carries a long open-reading frame (ORF) encoding a polyprotein precursor of 3010 amino acids. Translation of the HCV ORF is directed via a 5’ nontranslated region (NTR) functioning as an internal ribosome entry site; it permits the direct binding of ribosomes in close proximity to the start codon of the ORF. The HCV polyprotein is cleaved co- and post-translationally by cellular and viral proteases into ten different products, with the structural proteins core (C), envelop 1 (E1) and envelop 2 (E2) located in the N-terminal third, whereas, the nonstructural (NS2, NS3, NS4A, NS4B, NS5A, NS5B) replicative proteins are located in the remainder. Putative functions of the cleavage products are shown.
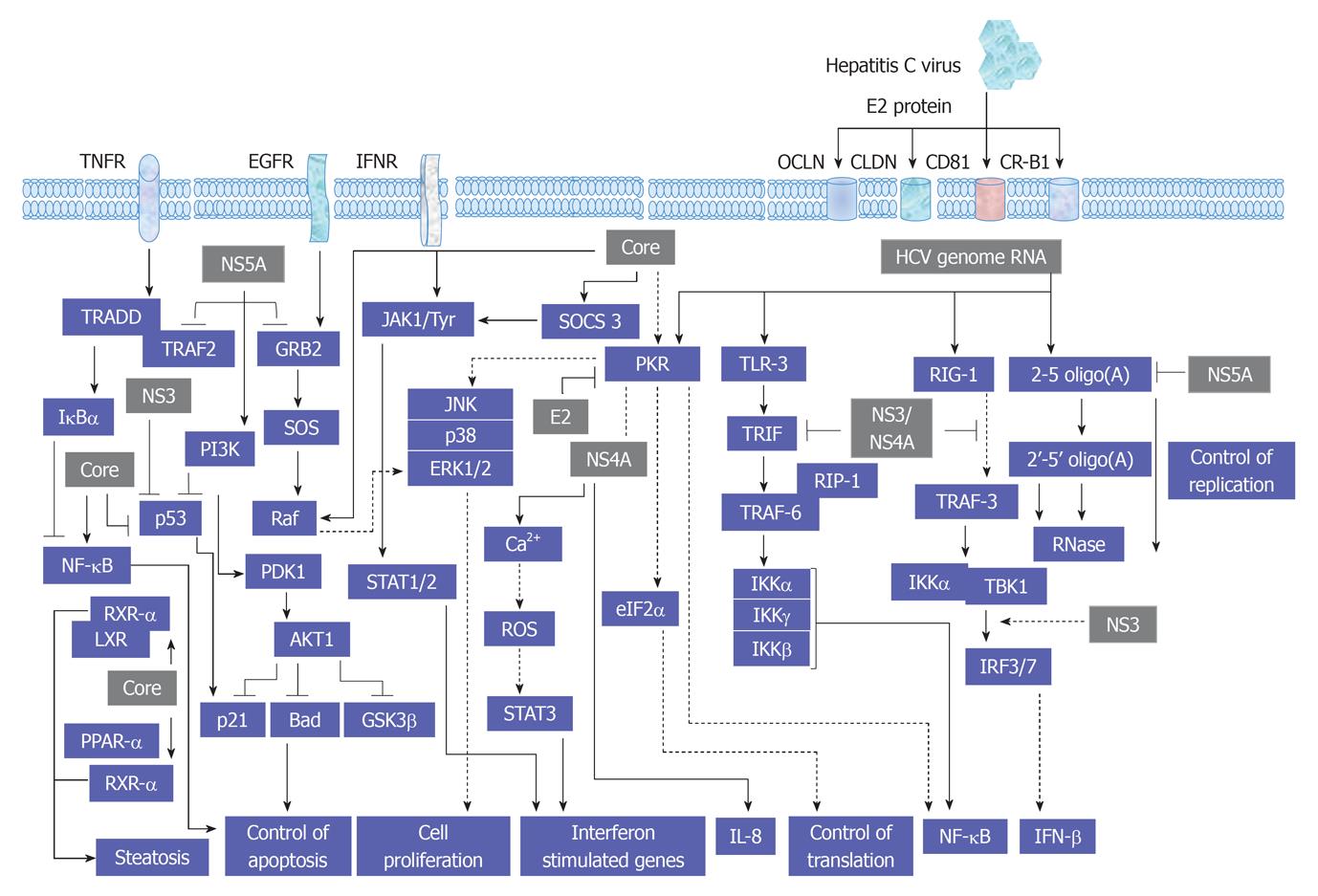
Figure 2 A proposed model for the consequences resulting from the interference of hepatitis C virus with signal transduction processes in host cells.
HCV: Hepatitis C virus; TNFR: Tumor necrosis factor receptor; JAK: Janus kinase; EGFR: Endothelial growth factor receptor; IFN: Interferon; IFNR: IFN receptor; TRAF: Tumor necrosis factor associated factor; TRADD: TNFR-associated protein with death domain; JAK: Janus kinase; JNK: c-Jun N-terminal kinase; SOCS: Suppressor of cytokine signaling; PI3K: Phosphatidylinositol-3-kinase; ERK: Extracellular regulated protein kinase; RIP: Receptor-interacting protein; ROS: Reactive oxygen species; STAT: Signal transducers and activators of transcription; NF-κB: Nuclear factor κB; IL: Interleukin; PPAR: Peroxisome proliferator-activated receptor.
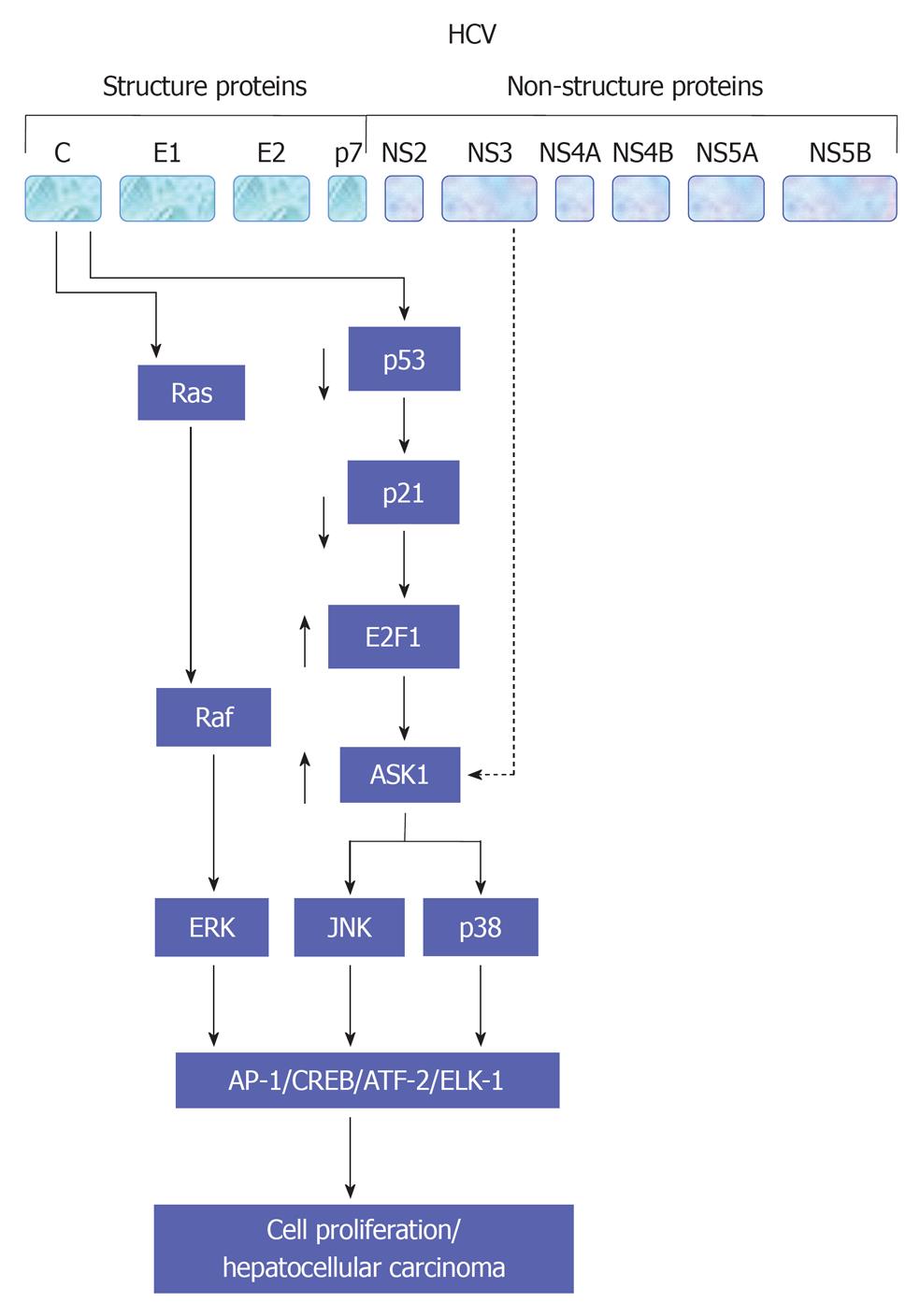
Figure 3 A schematic view for the molecular mechanisms,which are involved in the regulation of hepatitis C virus-induced associated cell proliferation.
A central role for mitogen-activated protein kinase signaling pathways in the modulation of hepatitis C virus-associated hepatocellular carcinoma is also demonstrated. HCV: Hepatitis C virus; AP-1: Activator protein-1; JNK: c-Jun N-terminal kinase; CREB: cAMP response element-binding protein; ERK: Extracellular regulated protein kinase.
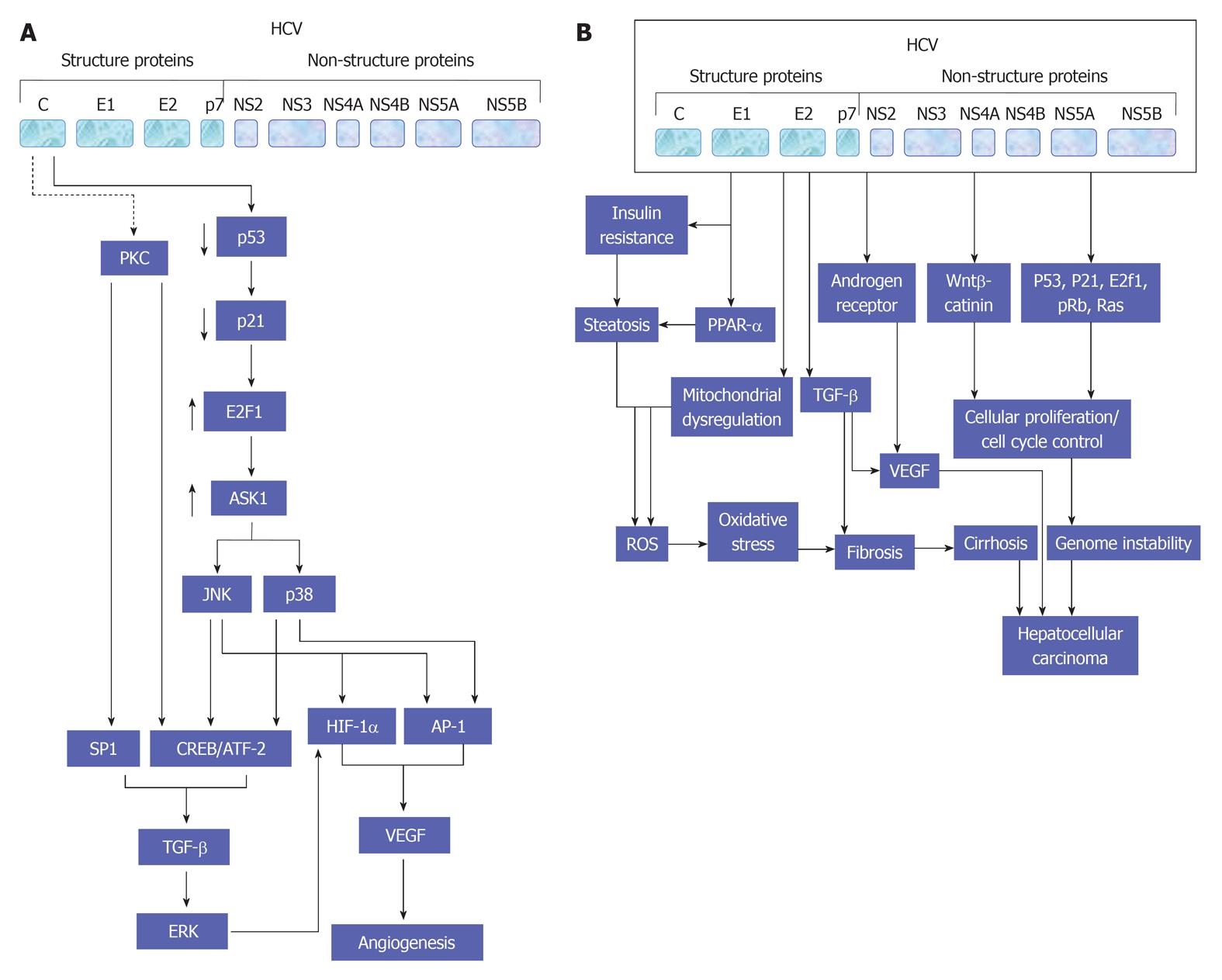
Figure 4 A proposed model for hepatitis C virus - mediated effects in liver cells and the modulatory role of transforming growth factor β in the regulation of both angiogenesis (A) and hepatocellular carcinoma (B).
PKC: Protein kinase C; HCV: Hepatitis C virus; AP-1: Activator protein-1; CREB: cAMP response element-binding protein; JNK: c-Jun N-terminal kinase; ERK: Extracellular regulated protein kinase; TGF: Transforming growth factor; VEGF: Vascular endothelial growth factor; ROS: Reactive oxygen species; PPAR: Peroxisome proliferator-activated receptor.
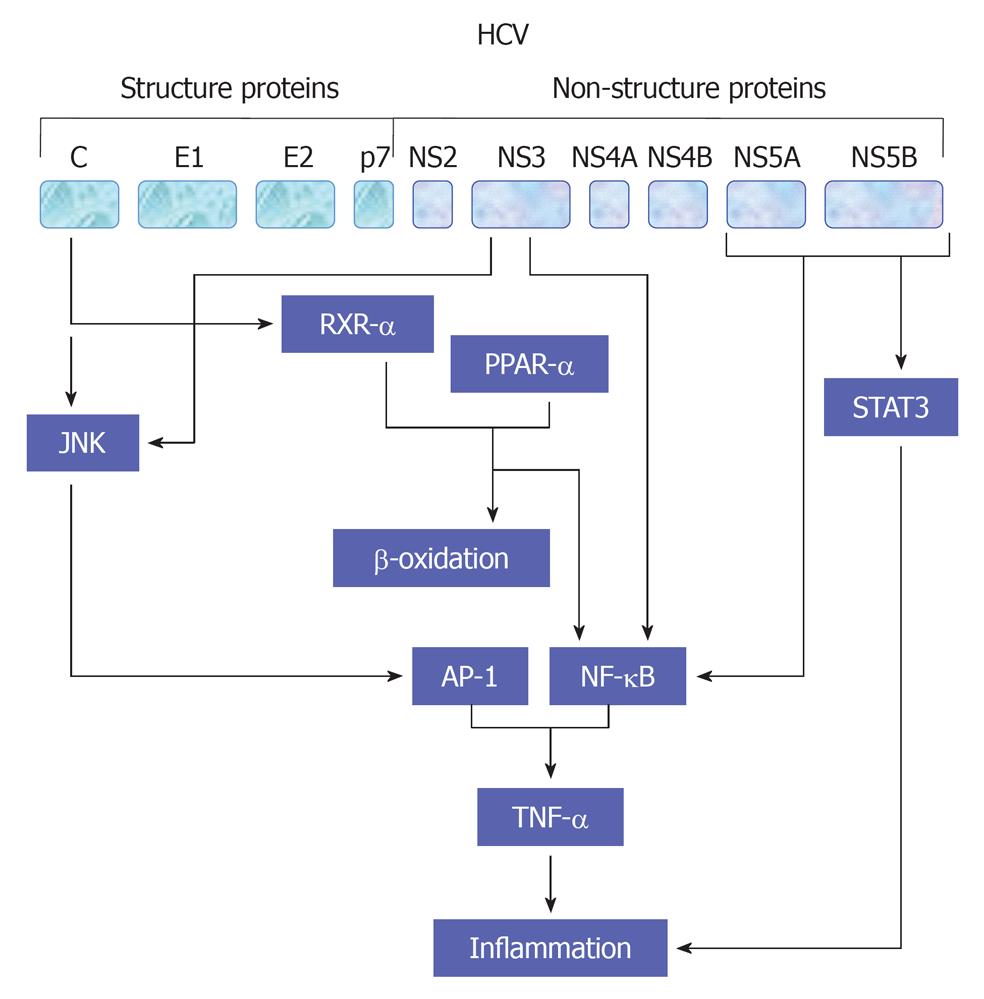
Figure 5 A suggested model for hepatitis C virus -associated inflammation and the role of tumor necrosis factor α in the regulation of this mechanism.
HCV: Hepatitis C virus; JNK: c-Jun N-terminal kinase; AP-1: Activator protein-1; NF-κB: Nuclear factor κB; PPAR: Peroxisome proliferator-activated receptor; TNF: Tumor necrosis factor; STAT: Signal transducers and activators of transcription.
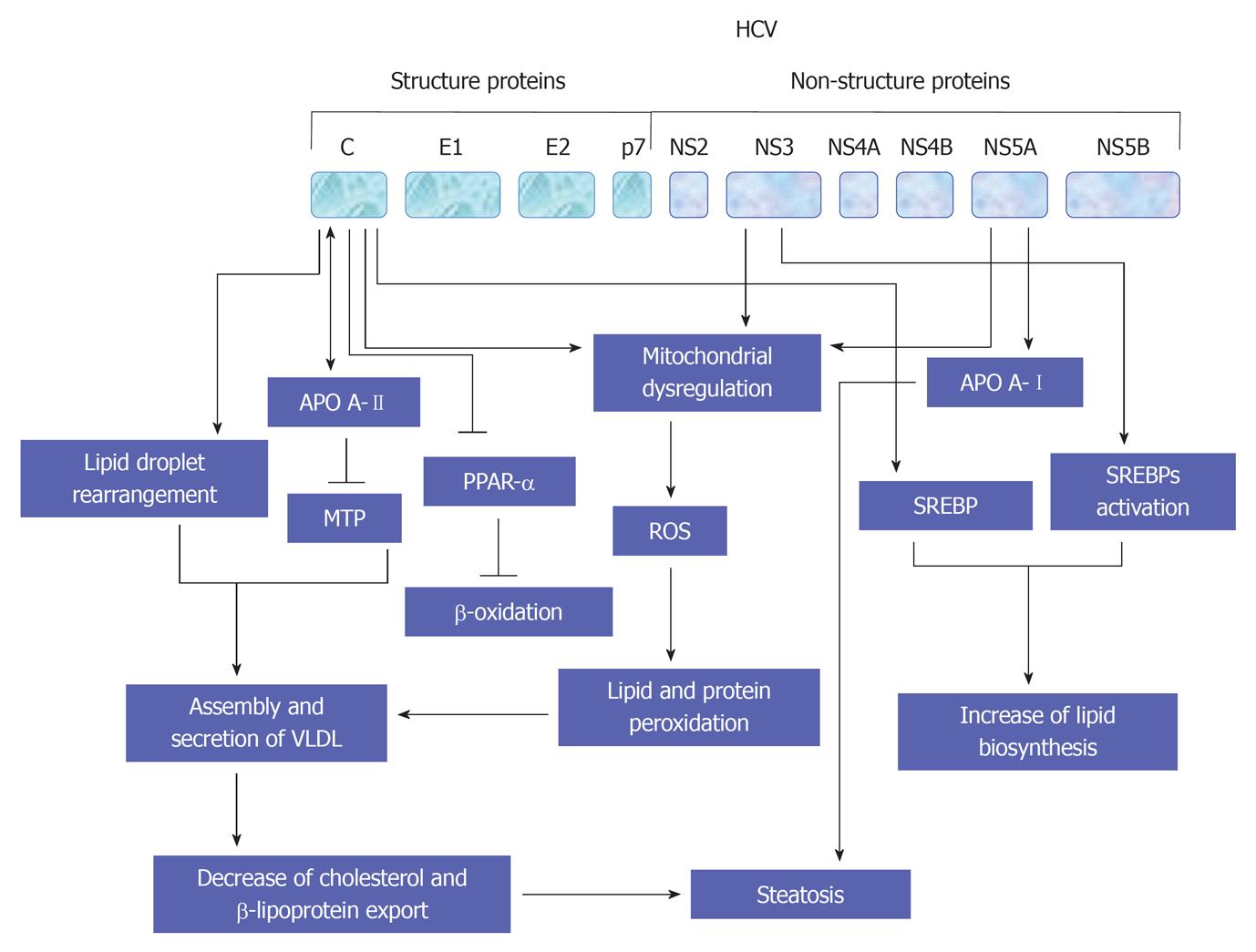
Figure 6 Proposed model for the molecular mechanisms, which are involved in the regulation of hepatitis C virus-associated steatosis.
HCV: Hepatitis C virus; ROS: Reactive oxygen species; PPAR: Peroxisome proliferator-activated receptor.
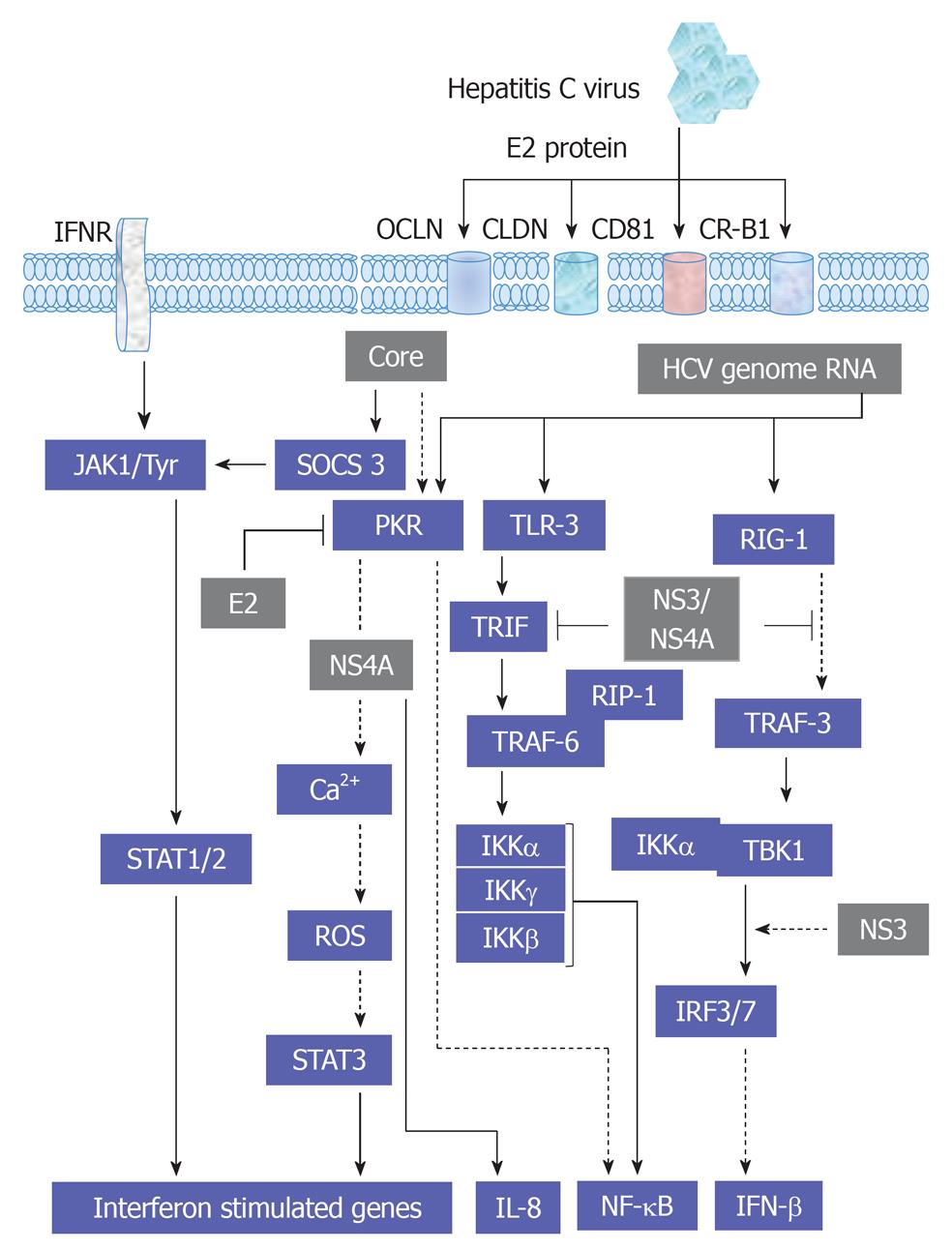
Figure 7 A schematic overview for the interference of hepatitis C virus with cytokines-associated Janus kinase/signal transducers and activators of transcription and cytokines-associated pathways.
HCV: Hepatitis C virus; JAK: Janus kinase; EGFR: Endothelial growth factor receptor; IFN: Interferon; IFNR: IFN receptor; TRAF: Tumor necrosis factor associated factor; JAK: Janus kinase; SOCS: Suppressor of cytokine signaling; RIP: Receptor-interacting protein; ROS: Reactive oxygen species; STAT: Signal transducers and activators of transcription; NF-κB: Nuclear factor κB; IL: Interleukin; PPAR: Peroxisome proliferator-activated receptor.
Figure 8 A proposed view for molecular mechanisms, which are involved in the modulation of hepatitis C virus-mediated insulin resistance during the course of hepatitis C virus infection.
TNF: Tumor necrosis factor; HCV: Hepatitis C virus; SOCS: Suppressor of cytokine signaling; ROS: Reactive oxygen species; STAT: Signal transducers and activators of transcription; IRS: Insulin receptor substrates.
- Citation: Hassan M, Selimovic D, El-Khattouti A, Ghozlan H, Haikel Y, Abdelkader O. Hepatitis C virus-host interactions: Etiopathogenesis and therapeutic strategies. World J Exp Med 2012; 2(2): 7-25
- URL: https://www.wjgnet.com/2220-315X/full/v2/i2/7.htm
- DOI: https://dx.doi.org/10.5493/wjem.v2.i2.7