Published online Aug 4, 2017. doi: 10.5492/wjccm.v6.i3.172
Peer-review started: February 17, 2017
First decision: April 14, 2017
Revised: May 2, 2017
Accepted: May 12, 2017
Article in press: May 15, 2017
Published online: August 4, 2017
Processing time: 166 Days and 14.1 Hours
To detect blood withdrawal for patients with arterial blood pressure monitoring to increase patient safety and provide better sample dating.
Blood pressure information obtained from a patient monitor was fed as a real-time data stream to an experimental medical framework. This framework was connected to an analytical application which observes changes in systolic, diastolic and mean pressure to determine anomalies in the continuous data stream. Detection was based on an increased mean blood pressure caused by the closing of the withdrawal three-way tap and an absence of systolic and diastolic measurements during this manipulation. For evaluation of the proposed algorithm, measured data from animal studies in healthy pigs were used.
Using this novel approach for processing real-time measurement data of arterial pressure monitoring, the exact time of blood withdrawal could be successfully detected retrospectively and in real-time. The algorithm was able to detect 422 of 434 (97%) blood withdrawals for blood gas analysis in the retrospective analysis of 7 study trials. Additionally, 64 sampling events for other procedures like laboratory and activated clotting time analyses were detected. The proposed algorithm achieved a sensitivity of 0.97, a precision of 0.96 and an F1 score of 0.97.
Arterial blood pressure monitoring data can be used to perform an accurate identification of individual blood samplings in order to reduce sample mix-ups and thereby increase patient safety.
Core tip: Blood samplings for point-of-care analysis are essential procedures performed in large quantities in hospital wards every day. Whereas many guidelines and good practices exist, human error may still occur and additional safeguards are needed to avoid mix-ups. Using data from arterial blood pressure monitoring, which regularly is present in critical patients for whom errors would be most severe, different features, even the absence of information, may be used for analysis. We developed a novel approach accounting for lack of data in arterial blood pressure monitoring to determine the exact time of blood withdrawal for better sample dating and patient identification.
- Citation: Peter J, Klingert W, Klingert K, Thiel K, Wulff D, Königsrainer A, Rosenstiel W, Schenk M. Algorithm-based arterial blood sampling recognition increasing safety in point-of-care diagnostics. World J Crit Care Med 2017; 6(3): 172-178
- URL: https://www.wjgnet.com/2220-3141/full/v6/i3/172.htm
- DOI: https://dx.doi.org/10.5492/wjccm.v6.i3.172
Pre-analytical procedures in point-of-care diagnostics are prone to different types of safety relevant errors. Most of them are caused by human failure the “often interrupted” and stressful surrounding of intensive care units. The most common error in the process of handling and analyzing venous blood samples is patient misidentification[1]. Critical patient identification errors are observed in up to 1 per 1000 procedures or specimens[2-4]. The correct dating and matching of blood samples is significant for an adequate patient care and helps to avoid life threatening situations caused by mix-ups[1,5,6].
Precise sample labeling (e.g., barcode labels) is the most obvious way of prevention. Establishing guidelines for sample handling might be another. However, noncompliance with these guidelines, introducing additional effort of documentation and cross-checking, remains a major problem[1,7,8].
Technical safeguards against human failure would be a forward-looking strategy in this setting. The estimation of additional parameters for patient identification in the analytical phase may help to identify sampling mix-ups[9]. But those all face the limitations of post-analytical data analysis. Approaches to prevent mixed-up samples therefore have to be technical solutions becoming effective in the pre-analytical phase[10].
Arterial blood sampling from the arterial line leads to an unavoidable and characteristic pattern (e.g., artificial blood pressure, no pulsation) in blood pressure monitoring[11,12]. With respect to vital sign monitoring, this pattern is useless and is often overlooked. But the missing of blood pressure data contains useful information. Monitoring data may be missing not at random (MNAR) or be missing at random (MAR)[13]. Data MAR like the missing of a single measurement in a defined sequence may reduce sample size or degree of freedom for analysis but probably will not bias the result. Data MNAR, like missing data in specific conditions, could add a strong bias to analyses[14]. Dealing with missing data can be performed in different ways: Missing data can just be excluded, the last value carried forward, related information can be used to estimate the missing value, imputation can be performed on logical rules or indicator variables can be used to represent the state of missing information. However, mean imputation is still the most common method to process missing data[15]. In the case of blood withdrawal, missing ABP information is NMAR caused by the specific procedure. Therefore, analyzing this pattern may help to identify the patient- and time-dependent blood sampling procedure and thus provide a valuable marker for pre-analytical safety approaches.
The aim of this study was to use available and missing information from arterial pressure monitoring to detect the exact time of blood withdrawal. Those calculated points in time can be used as a reference for dating blood samples and improve patient safety by validating a blood sample with a performed blood withdrawal from the selected patient.
A proof-of-concept approach is implemented to demonstrate the capabilities of the developed algorithm in real-time.
Test and validation data were obtained from studies performed at the Department of General, Visceral and Transplant Surgery at the University Hospital Tübingen, Germany, in an experimental ICU setting for animal studies.
The arterial blood pressure was measured with a PiCCO-catheter system (MAQUET Holding B.V. and Co. KG, Germany) placed in the femoral artery using Seldinger technique and connected to a medical three-way tap allowing for blockage of the blood flow and attaching a syringe or vacutainer for blood withdrawal. For ABP measurements, a pressurized saline infusion bag was used to provide a standing fluid pillar for the sensor.
The mean (ABPm), diastolic (ABPd) and systolic (ABPs) arterial pressure were displayed with an IntelliVue MP50-Monitor (Koninklijke Philips Electronics N.V., Netherlands), exported via a serial connection and processed with the TICoMS monitoring and control framework and stored in a PostgreSQL database at 1 Hz[16].
The detection of blood withdrawals using pressure monitoring was tailored to a usual blood withdrawal process from an arterial access. During blood withdrawal, the three-way tap was rotated and pressure measurement via the standing fluid pillar was blocked off. Then the sample was collected. Afterwards the three-way tap was returned to its measurement position and the catheter was flushed. Blood withdrawals were performed by physicians, scientists, medical students and lab technicians. The blood sample was measured in a blood gas analysis device (ABL800 FLEX, Radiometer Medical ApS, Denmark).
By rotating the three-way tap to obtain a blood sample, the pressure measurement was decoupled from the physiological state and set to an artificial level. Therefore, a static mean pressure ABPm with no physiological pulsation was observed on the patient monitor, leading to an absence of measurements for ABPs and ABPd.
Using this observation, the detection algorithm was designed and implemented with Matlab 2016a (The MathWorks, Inc., United States). The direct connection to the database and integration into the processing pipeline allowed real-time application.
In a first step, raw data from the patient monitor were processed and analyzed to detect deviations from the current state and to calculate indicator tags for each parameter at 1 Hz. If multiple observations were detected within this timeframe, the most recent measurement was used for further processing.
The detection was based on a scoring function that was normalized between 0 and 1. To calculate the scoring function all used parameters (ABPm, ABPs, ABPd) can be weighted individually, thus adapting their influence to the total score. To obtain the normalized score the sum of all parameters weights used (w↓i) (must be 1. For detection of blood withdrawals, the weight of all parameters was chosen to be the same and the scoring was therefore based on the normalized sum of the individual scores for each parameter:
S = (SABPm + SABPs + SABPd)/3.
The scoring function therefore represents a score between 0 and 1, whereas 1 means that all indicators for a blood withdrawal are present and 0 that no indication for such an event was given. This allows an easy adaption of the algorithm and including additional parameters.
When the scoring function was calculated, a simple threshold was used to determine if a blood withdrawal event was present. The threshold for the scoring function was set to STh = 0.7, and if S > STh a manipulation was assumed.
To obtain a more robust result and avoid false detections for cases where only a single measurement exceeds the threshold, a series of 10 successive points in time must reach the threshold STh to be accounted for as a blood withdrawal event.
For ABPm, the score SABPm was based on a deviation from the mean observation of the last 10 min, ignoring missing data. If the deviation was more than 10 mmHg from the mean the individual score was 1 otherwise 0.
For ABPs and ABPd the availability of the values was used for the scores SABPs and SABPd, respectively. Therefore, the tags from the first algorithmic step can be used and accounted for in the score as 1 if the measurement currently was missing or 0 if the measurement was present.
Due to multiple points in time where the threshold may be reached, a consensus time was calculated. The first point in a successive order of 10 scores exceeding the threshold was used.
First, existing data sets from previous study trials were used to calculate the exact times of blood withdrawals retrospectively. The arterial pressure measurements from the trials were read from the database and processed in successive order for each stored point in time. Each detected event was automatically plotted with a Matlab script as a graphic chart of a 10-min window to perform a visual inspection and validation of the variables and the scoring function.
Because the used blood gas analyzer was connected to the TICoMS infrastructure as the used medical framework as well, the analysis results and their dates were also processed and stored during the performed trials and thus available for retrospective analysis. The exact times of blood gas analyses were extracted and used as a reference for the blood withdrawal times. For each known analysis point the event was processed in the same way as the events detected by the algorithm: Plotting the corresponding pressure measurements and calculating a detection score. The resulting points in time and plots where then used to determine correct hits by the algorithm and match the detections correctly to the blood gas analysis and other events.
Second, a real-time version of the algorithm was tested alongside two study trials performed with the experimental setup described above. This gave the opportunity to observe the blood withdrawal events and the analytic algorithm concurrently without delay. The integration and real-time processing capabilities of the proposed method within the used medical software framework was evaluated.
For evaluation of the algorithm’s performance, all known blood gas measurements are matched to the detected events and counted as hits, missed blood gas analysis events are counted as misses. Additional detections were visually inspected and compared to other performed procedures to determine if an actual manipulation is present. Such events are additional blood withdrawals for laboratory and activated clotting time analysis. These events are accounted for as additional hits by the algorithm and denoted as other, whereas all other disturbances erroneously detected by the proposed algorithm are classified as false positives (FP) and denoted as errors. The performance of the detection is evaluated in terms of sensitivity or true positive rate, which represents the fraction of detected events of the total number of events:
TPR = (hits + other)/(hits + other + misses).
Precision or positive predictive value:
PPV = (hits + other)/(hits + other + errors).
And the F1 score as the harmonic mean of sensitivity and precision:
F1 = [2 × (hits + other)]/[2 × (hits + other + misses + errors)]
The retrospective analysis was performed by calculating the scoring function and thus the exact times of blood withdrawal events from the numerical values stored in the database. Independently, the timestamps in the database for the results of the blood gas analysis were exported.
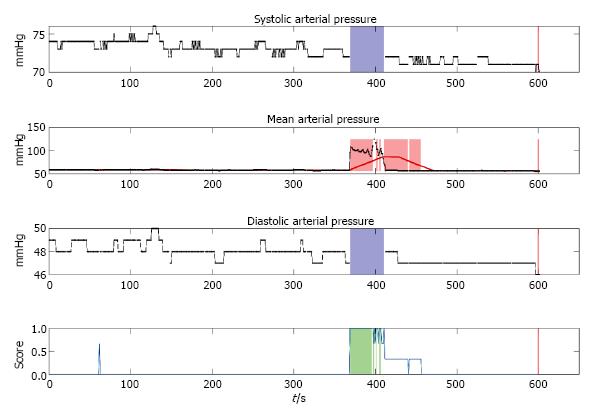
Each event detected by the algorithm was processed to store the exact times in a text file, containing all dates and times of the events and as an image where the ABPm, ABPs, ABPd measurements and the calculating scoring function are plotted below each other. An example for such a detection event plot is shown in Figure 1. In this figure 10 min windows of the three measured parameters ABPs, ABPm, ABPd and the calculated scoring function are displayed. For ABPs and ABPd time frames with missing data are highlighted with a blue box in their respective graphs. For ABPm the calculated moving average pressure is plotted with a red line. Exceeding the defined deviation from the mean pressure is highlighted with a red box in the graph. Scoring function threshold exceedance for a consecutive order of 10 measurements is highlighted with green boxes.
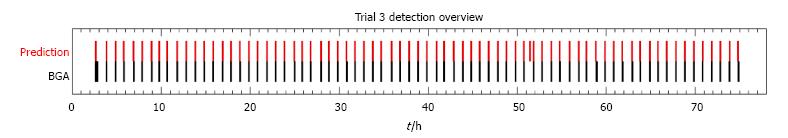
Additionally, after processing all data from a single trial, a summarized plot for all events was generated to provide an overview of the performed blood gas analyses in comparison to the detected events. Such an overview is shown in Figure 2 for trial number 3. The plot displays the detected events and the performed blood gas analyses along the time axis of the study trial. In the upper row, red bars are used to show the points of blood withdrawal detections. The lower row represents the time of known blood gas analysis measurements. The blood gas analyses were performed regularly and except for one analysis at the beginning, all events were successfully detected by the algorithm. An additional detection is shown around hour 51 of the trial. Furthermore, the delay between blood withdrawal and the storing of the analysis results can be observed, as the red bars precede the black bars slightly.
The results of the retrospective analyses for all trials and the algorithmic performance for each individual trial and in total are shown in Table 1. Each column represents a single trial with the number of performed BGAs, the number of manipulations detected by the algorithm, the hits and misses for the BGAs, other events and false positive detections. The statistical measures for each trial, sensitivity, precision and F1 score are shown below. The rightmost column shows the combined result of all analyzed trials. A total number of 434 BGAs was present in the observed data. The algorithm detected 514 events, of which 422 were BGAs. Thus, BGAs could be successfully detected in 422 of 434 cases (97.23%). Sixty-four other observed events like blood withdrawal for laboratory and activated clotting time analysis were detected. In 18 cases, the algorithm performed a wrongful detection of an event, whereas only fluctuations in the pressure curve were present. Overall, a sensitivity of 0.97, a precision of 0.96 and an F1 score of 0.97 were achieved. The algorithm proved to successfully detect blood withdrawals by a broad variety of caregivers at different levels of professional experience.
| Trial | 1 | 2 | 3 | 4 | 5 | 6 | 7 | Total |
| Number of BGAs | 72 | 34 | 75 | 73 | 66 | 40 | 74 | 434 |
| Detected Manipulations | 77 | 36 | 74 | 73 | 78 | 57 | 109 | 514 |
| Detected BGAs (hits) | 69 | 33 | 73 | 70 | 66 | 38 | 73 | 422 |
| Missed BGAs (misses) | 3 | 1 | 2 | 3 | 0 | 2 | 1 | 12 |
| Other events (other) | 5 | 2 | 1 | 1 | 8 | 15 | 32 | 64 |
| False detections (errors) | 3 | 1 | 0 | 2 | 4 | 4 | 4 | 18 |
| Sensitivity | 0.96 | 0.97 | 0.97 | 0.96 | 1.00 | 0.95 | 0.99 | 0.97 |
| Precision | 0.96 | 0.97 | 1.00 | 0.97 | 0.94 | 0.91 | 0.95 | 0.96 |
| F1 Score | 0.96 | 0.97 | 0.99 | 0.97 | 0.97 | 0.93 | 0.97 | 0.97 |
The real-time version of the algorithm was successfully integrated in the medical software framework for two additional trials and could successfully detect blood withdrawals and manipulations from the arterial catheter in real-time, thus showing the fundamental feasibility of the proposed method for clinical application as a precaution measure against mix-ups.
A highly accurate algorithm was developed to detect blood withdrawals and other manipulation events in patients with established arterial blood pressure monitoring. This algorithm identified more than 97 percent of the performed samplings for blood gas analyses.
Using a 10-min sliding window for mean calculation of the measured ABPm allowed for dynamic adaption to the current patient state and changing blood pressures. Therefore, the detection capabilities were maintained stable for long observation times (96 h in this study). In all trials a total number of only 18 false positive detections occurred. Even if the algorithm detected such additional events, there was a strong bias between false positive and false negative rates for the task of withdrawal detections. Successful detection of all real events was the uppermost important goal, as it was required for a correct sample dating a matching for the patient. False negative rate was therefore the critical statistical size as an undetected blood sample cannot be dated and processed. On the other hand, false positives, hence detections without indication did occur but caused no significant harm due to their low frequency of occurrence. However, an increased frequency of false positive detections yields the risk of selecting wrong events. But this error was limited to a definable timeframe. Only the last 10 min were evaluated for detection of blood withdrawal events, so only events within this time frame are relevant.
Characteristic pattern of blood withdrawals occurring on arterial blood pressure were used for algorithmic detection of the events. Neither data imputation for missing measurements nor ignoring missing values was performed. Instead, by using tags indicating when information was missing this knowledge of systolic and diastolic pressure was preserved. In the context of arterial blood withdrawal this data was NMAR but absent due to the artificial pressure level provided by the pressurized infusion bag. The combination of present information with tags derived from the missing systolic and diastolic measurements yields this useful detection algorithm.
Due to general anesthetics, patient movement was not present in study conditions but should be evaluated for a general clinical application. However, explicitly using the information obtained from the absence of the systolic and diastolic pressures, random events like movements should not lead to such a loss of data, thus not exceeding the threshold for the scoring function.
With a known exact time of blood withdrawal, the blood sample can automatically be dated back to the moment of withdrawal if the measurement was processed by a hospital information system. As shown in the analysis, for the chosen 10 min window the event can be dated back from the exact measurement times to obtain the withdrawal point. If longer times for handling the blood samples are needed, the observed timeframe should be enlarged or even shifted, for example ignoring the most recent minutes for the analysis at all, as processing and transporting time of the blood sample provides a lower boundary.
Additional parameters can easily be included in the detection algorithm with an individual scoring weight to adapt for specific procedures or additional detection performance. If for example an arterial temperature was measured as well, the temperature change resulting from flushing the catheter with saline solution, can be detected as a characteristic event as well and be included in the scoring function.
For improved versions of the detection algorithm and additional studies there are several directions further research might be headed to. Instead of using the observed numerical values for ABPm, ABPs and ABPd, the pressure curve itself may be analyzed. Using such high resolution raw data, calculation of the numerical values for systolic, diastolic and mean pressure may be specifically tailored to process the state of missing data, therefore refining the detection by improving the decision when a systolic and diastolic measurement can be calculated. This might be further extended by integrating models for heart beat detection and to establish a better estimate for deviations from the expected heart rate and variations in to decide if a physiological measurement was present.
The application of the provided solution to a hospital ward with multiple patients could be performed by integration in point-of-care devices like a blood gas analyzer. As suggested by Huijsmans et al[9], usage of additional parameters in the analytical phase should be considered for better sample identification and reducing mix-ups. Using the novel information of automatically calculated times for blood withdrawals, a list of admitted patients can be limited to those, where such an event was detected. This would reduce the error potential of wrong data entries or mix-ups in patient identification from the entire patient database. A full list of all patients should still be available in a second menu, providing an override if detection was not possible. However, this additional step would force attention of the caregiver, being aware of currently performing an override and focusing on the correct patient selection in this no longer standard procedure. This can significantly improve patient identification as the most commonly observed problem in different studies by Wallin, Wagar and Vallenstein et al[1-3].
Additionally, after successful patient identification, a selection of the detected points of blood withdrawal could be performed and directly be stored with the analysis result, allowing for a more accurate timing of the blood sample. The time between blood withdrawal and analysis is known when automated detection is performed. A warning could then be shown if a predefined time between sampling and measurement is exceeded.
In this paper, we provide a sample application and a first step on the way to automated systems in clinical care settings interacting with point-of-care devices. Detection of blood withdrawal is just an example for the broad range of possibilities that may lead to solutions which will help caregivers and reduce their workload or provide additional safeguards in stressful situations addressing the major problem of noncompliance with guidelines observed by Iboje et al[7].
Major challenges are still to be faced in terms of data access, interconnection of medical devices and dealing with consequently overwhelming big data of patient information. These tasks need to be solved in cooperation with established knowledge from computer science, tailored to the specific needs of the medical sector. By unleashing such great potential, many repetitive or standardized tasks can be automated or computer-assisted checks and protection systems against mistakes could be implemented to increase patient safety and reduce the risk of potential errors for caregivers in stressful situations caused by high workload.
Monitoring of arterial blood pressure with arterial catheters is commonly performed in critical patients. Regular arterial blood withdrawals are performed to assess the patient state.
Interconnection of medical devices and automated systems in the medical sector are still experimental. Combining computer science and medicine is still a challenge. Whereas automation and guidance and advanced safeguards are common in other fields of application.
A novel approach for the detection of blood withdrawal in patients with an arterial catheter for arterial blood pressure monitoring is described. By interconnecting a patient monitor to a point-of-care diagnostic device, the selection of patient data can be narrowed to patients with plausible sampling events.
Detection of blood withdrawal may be a useful feature integrated in medical monitors and blood gas analysis devices to increase patient safety by allowing a better, automated sample dating and the reduction of the risk for sample mix-ups in hospital wards. However, for a practical implementation additional validation steps are required and medical devices like the blood gas analyzer must be adapted by the manufacturer to allow a pre-selection for patients with recently detected blood withdrawals.
Arterial blood pressure (ABP), Mean Arterial blood pressure (MAP or ABPm), systolic ABPs, diastolic ABPd, blood gas analysis BGA, arterial blood gas (ABG).
The authors prepared and evaluated the algorithm of automatic detection of blood withdrawals by using data from continuous direct blood pressure monitoring. The algorithm may be useful to recognize and eliminate errors in recording blood withdrawal events from the specific patients. The algorithm provides reasonable precision and prediction rates. The methods used were adequate. The manuscript is well-written and will be interesting for intensive care specialists.
Manuscript source: Invited manuscript
Specialty type: Critical care medicine
Country of origin: Germany
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P- Reviewer: Beltowski J S- Editor: Song XX L- Editor: A E- Editor: Lu YJ
| 1. | Wallin O, Söderberg J, Van Guelpen B, Stenlund H, Grankvist K, Brulin C. Blood sample collection and patient identification demand improvement: a questionnaire study of preanalytical practices in hospital wards and laboratories. Scand J Caring Sci. 2010;24:581-591. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 31] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 2. | Wagar EA, Tamashiro L, Yasin B, Hilborne L, Bruckner DA. Patient safety in the clinical laboratory: a longitudinal analysis of specimen identification errors. Arch Pathol Lab Med. 2006;130:1662-1668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 3. | College of American Pathologists, Valenstein PN, Raab SS, Walsh MK. Identification errors involving clinical laboratories: a College of American Pathologists Q-Probes study of patient and specimen identification errors at 120 institutions. Arch Pathol Lab Med. 2006;130:1106-1113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 4. | Nutting PA, Main DS, Fischer PM, Stull TM, Pontious Ma. Problems in laboratory testing in primary care. J of the American Medical Association. 1996;275:635-639. [DOI] [Full Text] |
| 5. | Plebani M, Carraro P. Mistakes in a stat laboratory: types and frequency. Clin Chem. 1997;43:1348-1351. [PubMed] |
| 6. | Aslan B, Stemp J, Yip P, Gun-Munro J. Method precision and frequent causes of errors observed in point-of-care glucose testing: a proficiency testing program perspective. Am J Clin Pathol. 2014;142:857-863. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Ibojie J, Urbaniak SJ. Comparing near misses with actual mistransfusion events: a more accurate reflection of transfusion errors. Br J Haematol. 2000;108:458-460. [PubMed] |
| 8. | van Dongen-Lases EC, Cornes MP, Grankvist K, Ibarz M, Kristensen GB, Lippi G, Nybo M, Simundic AM; Working Group for Preanalytical Phase (WG-PRE), European Federation of Clinical Chemistry and Laboratory Medicine (EFLM). Patient identification and tube labelling - a call for harmonisation. Clin Chem Lab Med. 2016;54:1141-1145. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 25] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 9. | Huijsmans CJ, Heilmann FG, van der Zanden AG, Schneeberger PM, Hermans MH. Single nucleotide polymorphism profiling assay to exclude serum sample mix-up. Vox Sang. 2007;92:148-153. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 10. | Aron R, Dutta S, Janakiraman R, Pathak PA. The Impact of Automation of Systems on Medical Errors: Evidence from Field Research. ISR. 2011;22:429-446. [RCA] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 84] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 11. | Aleks N, Russell SJ, Madden MG, Morabito D, Staudenmayer K, Cohen M, Manley GT. Probabilistic detection of short events, with application to critical care monitoring. Proceedings of the Advances in Neural Information Processing Systems. 2009;49-56. |
| 12. | Bauer M, Gather U, Imhoff M. The identification of multiple outliers in online monitoring data. Dortmund: University of Dortmund, Technical Report SFB 475 1999; . |
| 13. | Gelman A, Hill J. Missing-data imputation. Data Analysis Using Regression and Multilevel/Hierachical Models. Cambridge: Cambridge University Press 2006; 529-544. |
| 14. | Osborne JW. Dealing with Missing or Incomplete Data: Debunking the Myth of Emptiness. Best Practices in Data United States: Cleaning SAGE Publications, Inc 2013; 105-138. [DOI] [Full Text] |
| 15. | Johnson AE, Ghassemi MM, Nemati S, Niehaus KE, Clifton DA, Clifford GD. Machine Learning and Decision Support in Critical Care. Proc IEEE Inst Electr Electron Eng. 2016;104:444-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 236] [Cited by in RCA: 191] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 16. | Peter J, Klingert W, Königsrainer A, Rosenstiel W, Bogdan M, Schenk M. TICoMS - A modular and message-based framework for monitoring and control of medical devices. Comp Med Sy. 2014;473-474. [DOI] [Full Text] |