Published online Nov 27, 2015. doi: 10.5411/wji.v5.i3.99
Peer-review started: June 27, 2015
First decision: July 28, 2015
Revised: October 23, 2015
Accepted: November 13, 2015
Article in press: November 17, 2015
Published online: November 27, 2015
Processing time: 153 Days and 20 Hours
There is a major transformation in gene expression between mature B cells (including follicular, marginal zone, and germinal center cells) and antibody secreting cells (ASCs), i.e., ASCs, (including plasma blasts, splenic plasma cells, and long-lived bone marrow plasma cells). This significant change-over occurs to accommodate the massive amount of secretory-specific immunoglobulin that ASCs make and the export processes itself. It is well known that there is an up-regulation of a small number of ASC-specific transcription factors Prdm1 (B-lymphocyte-induced maturation protein 1), interferon regulatory factor 4, and Xbp1, and the reciprocal down-regulation of Pax5, Bcl6 and Bach2, which maintain the B cell program. Less well appreciated are the major alterations in transcription elongation and RNA processing occurring between B cells and ASCs. The three ELL family members ELL1, 2 and 3 have different protein sequences and potentially distinct cellular roles in transcription elongation. ELL1 is involved in DNA repair and small RNAs while ELL3 was previously described as either testis or stem-cell specific. After B cell stimulation to ASCs, ELL3 levels fall precipitously while ELL1 falls off slightly. ELL2 is induced at least 10-fold in ASCs relative to B cells. All of these changes cause the RNA Polymerase II in ASCs to acquire different properties, leading to differences in RNA processing and histone modifications.
Core tip: B cell differentiation to antibody secreting cells is a highly regulated, complex process facilitated by factors such as interferon regulatory factor 4, Blimp-1, OCA-B, Xbp1, and mammalian target of rapamycin. This results in a switch in immunoglobulin mRNA processing from the membrane-bound to the secretory-specific form, occurring when ELL2 releases RNAP-II pausing during transcription elongation and causes exon skipping and proximal poly(A) site choice.
- Citation: Smith SM, Carew NT, Milcarek C. RNA polymerases in plasma cells trav-ELL2 the beat of a different drum. World J Immunol 2015; 5(3): 99-112
- URL: https://www.wjgnet.com/2219-2824/full/v5/i3/99.htm
- DOI: https://dx.doi.org/10.5411/wji.v5.i3.99
If a man does not keep pace with his companions, perhaps it is because he hears a different drummer. Let him step to the music which he hears….Henry David Thoreau.
B cells mature in the bone marrow, having undergone a series of DNA rearrangements to produce the uniquely rearranged immunoglobulin (Ig) molecules (H2L2) on their surfaces, the B cell receptor (BCR). Also on the B cell surfaces are: CD79 alpha and beta Ig co-activators for the BCR; CD19, which acts as a co-receptor with BCR; CD21, the complement receptor 2 for C3d; pattern recognition receptors like Toll-like receptors 2 and 4; and MHC-Class II molecules. The mature B cells travel to the lymph nodes or the spleen and reside in niches awaiting stimulation.
Upon stimulation the B cell radically alters its program of gene expression and “hears a different drummer”, turning into an antibody producing factory. If the B cells reside in and are stimulated in the marginal zone by T-independent antigen engagement of the BCRs, through the toll-like receptors, or Ig plus C3d, they will differentiate into antibody secreting cells (ASCs) with a high probability of differentiating into short-lived plasma-blasts. Activated marginal zone ASCs persist for only a few days after activation. They die rapidly either through an inability to deal with internal reactive oxygen species formed because of the large amount of secretory-specific antibody molecules they produce and/or because they fail to upregulate receptors for survival signals.
B cells that initially travel to the follicles require a more complex set of reactions in order to be stimulated by antigen. Engagement of the B cell surface CD40 occurs via contact with T cells carrying surface CD40 Ligand (CD154). Secretion of cytokines including interleukin (IL)-2, -4, and -5 by T cells further activates the B cells. CD40 is a member of the tumor necrosis factor superfamily of receptors and engagement results in B cell activation, isotype switching, and somatic hyper-mutation upon passing through a germinal center. Those B cells then differentiate into ASCs or memory cells. The CXCR4+ ASCs from B cells stimulated in follicles can home to specific CXCL12+ niches in the bone marrow and become long-lived ASCs. Long life for ASCs depends on soluble factors like BAFF and APRIL made by the bone marrow stroma and a touch of autophagy to repair damage in the endoplasmic reticulum (ER)[1].
How individual activated B cells choose between division, death, ASC development and class switching is unknown, and the molecular basis of this heterogeneity is still a mystery[2]. The relationship between the short-lived cycling plasma-blasts and the long-lived ASCs also remains unclear. The long-lived ASCs have the highest B-lymphocyte-induced maturation protein 1 (Blimp-1) expression, which might then explain the decreased levels of c-myc and proliferation in them[3]. We addressed these issues more fully in a recent review[4].
Regardless of the source of the B cells (MZ or FO) the activation pathways to ASCs share a number of transcription factors that alter the expression pathways and pave the way to secretion of antibody. Several genes have been implicated in both the activated B cell transcriptional network and the ASC network, such as interferon regulatory factor 4 (Irf4) and Pou2af1 (OCA-B); meanwhile, ELL2, c-Fos, Prdm1 (Blimp-1) and Xbp1 are implicated only in the ASC network[5]. Of these, only ELL2, Irf4 and Pou2af1 (OCA-B) have been shown to act directly on the Ig genes.
Irf4, also known as Pip, is unique amongst many others within its class of Irfs. Irfs have important roles within the immune responses. Irf1 and Irf2 were the first Irfs to be recognized for their novel immunomodulation and hematopoietic effects. Studies of Irfs prompted later discovery of other members in this class. The class now includes Irf3, Irf4, Irf7 and Irf8[6,7]. Irf4 was discovered via analysis of the specific Ets-transcriptions factors it interacts with; Irf4 binds to PU.1, an Ets-transcription factor, and together they form a functional ternary activating complex[8].
B cells experience class-switching recombination along with particular changes to cellular Ig specific transcription factors due to Irf4 regulation. The formation and changes to the germinal center are most highly observed when centrocyte levels are decreased and Ig specific transcription factors become abundant. The progression from germinal center maintenance to germinal center-specific transcription is the final step in the B cell cascade before differentiation can occur. Irf4 meditated differentiation directs B lymphocytes to become memory B cells or ASCs[9]. ELL2, a transcription elongation factor discussed below, is highly expressed in ASCs vs B cells, and Irf4 binds to the ELL2 promoter to induce high levels of ELL2 mRNA[10]. When Irf4 is conditionally knocked out, germinal center formation is profoundly compromised[11]. The proximate cause of the differentiation from B lymphocyte to ASC is Blimp-1. When Blimp-1 is upregulated, the cell is directed to differentiate. With further observation, the ultimate cause is, in fact, Irf4. Irf4 upregulation causes the downstream increase of Blimp-1 via PU.1, Irf4 ternary complex activation. This suggests Irf4 is the major orchestrator of B cell to ASC differentiation.
Originally, Irfs were thought to have all bound to a shared constitutive DNA consensus sequence, but later Irf4 and Irf8 were shown to have much lower affinities to these standard DNA sequence motifs. Due to the lower DNA binding affinity of Irf4 and Irf8, Ets-transcription factors are required to facilitate their DNA binding. Irf4 and Irf8 share similar Ets-transcription factor protein binding domains, and therefore the same Ets-transcription factors are used by both of them. Ets-transcription factors PU.1 and Spi-B have been shown to bind to specific DNA-binding motifs that then recruit Irf4 and Irf8[12]. PU.1 and Spi-B both promote binding to the 3’ enhancers of κIg and λIg light chains. Since both Irf4 and Irf8 are recruited by these factors, there is competition between the two similar Irfs. The outcomes of the Irf competition are starkly different, since B cell to ASC transition will not occur with an abundance of Irf8[13,14].
Irf4 and Irf8 not only compete for the Ets-transcription factors, but also promote expression for repressors of the other’s factors. In doing so, high levels of Irf8 would prompt decreased levels of Irf4, causing greater expression of Irf8-dependent genes. Irf8 dependent genes include Bcl6 and Pax5, which are high in B lymphocytes. Irf8-dependent genes repress Aicda and Blimp-1 expression, which are products of Irf4-dependent transcription[15]. With increased Irf4, the exact opposite occurs, where Irf4-dependent genes such as Aicda and Blimp-1 are expressed. This in turn represses Irf8-dependent gene transcription. Irf8 dependent gene Pax5 is repressed by Blimp-1, which is a negative transcription regulator in B lymphocytes. Presence of Blimp-1 represses Pax5 and c-myc. Repression of c-myc ceases cellular proliferation and causes an overall reduction of surface IgM[16]. The repression of Pax5 results in Xbp1 activation, which causes an increased production of unfolded protein response (UPR) components[17].
Irf4 has also been shown to drive Zbtb20 expression in B cells. Zbtb20, also known as Zfp288, DPZF and HOF, is a complex Bcl6 homologue that is a tramtrack, bric-à-brac, and zinc finger protein[18,19]. Ectopic expression of Zbtb20 induced terminal B cell differentiation to ACS. Along with promoting differentiation, Zbtb20 expression in plasma cells induces cell survival and blocks cell cycle progression. Zbtb20 is directly downstream and regulated by Irf4, and acts independently of Blimp-1[20].
Blimp-1 is encoded by the Prdm1 gene and plays a crucial role in the differentiation of B cells to ASCs, and thus the switch from expression of membrane bound antibody molecules to secreted antibody molecules[21]. The human homolog, PRD1-BF1, was discovered by Keller and Maniatis[22] by isolating a clone from a cDNA library encoding a protein that binds to the PRD1 site of the β-IFN promoter. Its ORF presents krüppel-like zinc finger DNA-binding motifs as well as proline and acidic regions resembling those of other known transcription factors, which indicates that Blimp-1 is a transcriptional regulator[23]. Blimp-1 mRNA expression is low in B cells and only present in late stages of differentiation[23]. Through Northern blots, it has been seen that Blimp-1 accumulation increases 5-fold in cells stimulated with IL-2 and IL-5. B cells transfected with Blimp-1 mature, although not all the way, to a point of exhibiting qualities of early ASCs[23]. It was further shown that upon knocking out Blimp-1, secretion of Ig was severely reduced or failed[24]. Regardless of the levels of Blimp-1, however, B cells will not differentiate in the absence of Xbp1[25], and B cells lacking Blimp-1 are unable to normally induce Xbp1 mRNA as well as unable to normally process the Xbp1 protein[26]. This shows that Blimp-1 acts upstream of Xbp1 in the development of ASCs[25]. B cells deficient in Blimp-1, transfected with Blimp-1 on a retrovirus, were able to secrete IgM, but Blimp-1-/- cells transfected with Xbp1 were not, proving that Xbp1 is not able on its own to drive differentiation if Blimp-1 is absent, thus indicating that Blimp-1 plays additional roles in plasmacytic differentiation[26].
Blimp-1 blocks transcription of a large set of genes[27]. C-myc is known to block terminal B cell differentiation[28]. Ectopically expressing Blimp-1 in pre-B cell lines represses c-myc promoter activity[29] and causes deacetylation of histone H3 associated with the c-myc promoter[30]. By analyzing DNA microarrays after inducible expression of Blimp-1, it was shown that Blimp-1 represses genes associated with progression of the cell cycle as well as synthesis and repair of DNA[27]. Blimp-1 was also shown to repress the gene expression program involved in B cell identity[27]. Blimp-1 represses Pax5[27], which is known to repress Xbp1[31]. This indicates that Blimp-1 induces expression of Xbp1 by repressing its repressor, Pax5[32]. It has also been shown that Blimp-1 represses the transcription elongation factor ELL3. When the ELL3 promoter was cloned, co-transfection with Prdm1 significantly repressed activity in B lymphocytes[33]. Blimp-1 was also seen to shut down Ig class switching by repressing expression of genes required in this process as well as inhibiting signals that serve to activate switch region Ig transcription[27].
Shaffer et al[27] showed that Blimp-1 participates in a negative feedback loop with BCR and Bcl6. BCR signaling restrains terminal differentiation of B cells by suppressing Blimp-1, while expression of Blimp-1 was reciprocally shown to block BCR signaling via the downregulation of its components[27]. Bcl6 is required for the differentiation of Germinal Center B cells[34]. Blimp-1 also represses Bcl6, while overexpression of Bcl6 represses Blimp-1 and thus ASC differentiation[35]. This feedback loop provides very tight control over the decision of a B cell to become an ASC, for while Bcl6 is expressed in a GC B cell, expression of Blimp-1 and thus plasmacytic differentiation is blocked, but as soon as Blimp-1 is activated, Bcl6 is repressed and differentiation begins[27]. It was also shown that Irf4 deficient cells were unable to differentiate and lacked expression of Blimp-1, indicating that Irf-4 directly activates the expression of Blimp-1[36]. In addition to Irf4, c-Fos also influences Blimp-1 expression. A proto-oncogene, c-Fos is a transcriptional regulator that operates on DNA in an indirect fashion via its interaction with other transcriptional activators, such as c-Jun[37]. Ectopic expression of c-Fos with c-Jun induces Blimp-1 expression[38]. H2-c-Fos B cells, once stimulated with LPS, were found to proliferate at a higher level than normal B cells after LPS stimulation and induced enough Blimp-1 for terminal differentiation[39].
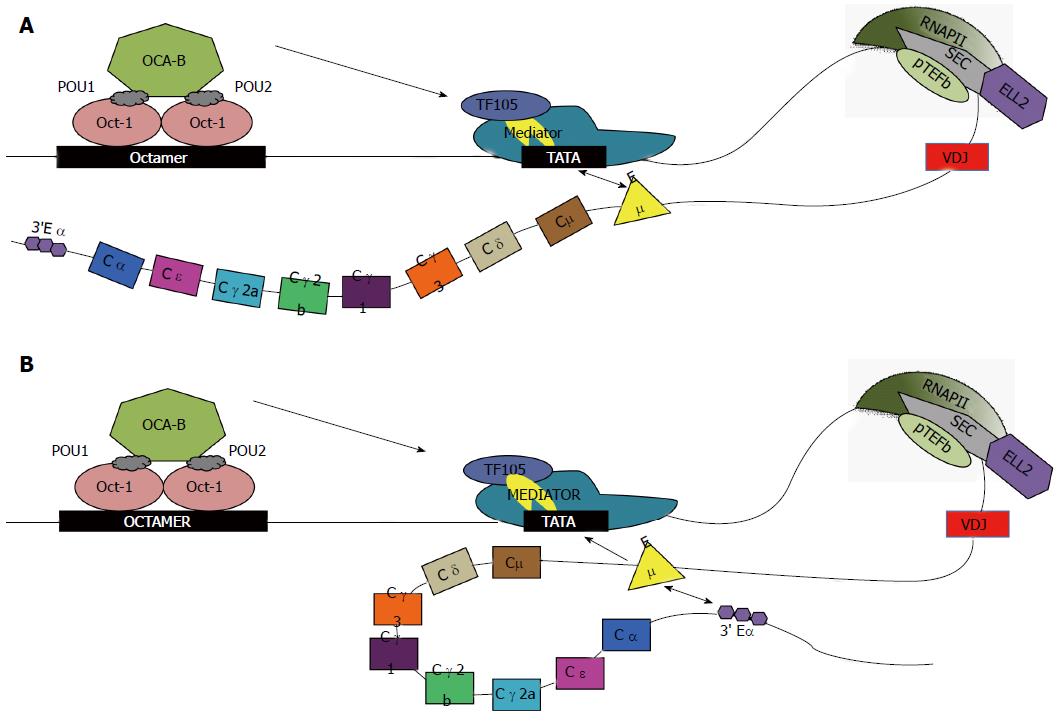
OCA-B, aka Pou2af1, BOB.1, Bob-1, OBF-1, or OBF.1, a-coactivator from B cells that increases Ig promoter transcription, was discovered by Luo et al[40] using the fractionation by ion-exchange chromatography of an oligonucleotide matrix-bound fraction. They isolated a novel B cell coactivator of Oct-1 and Oct-2. Binding of OCA-B to the octamer sequence of IgH is indirect and facilitated by Oct-1 and Oct-2 DNA binding[41] and as depicted in Figure 1. Oct-1 and Oct-2 contain POU domains, POU-1 and POU-2, respectively[42]. These POU domains are sufficient to mediate the interaction between Oct-1 and Oct-2 with OCA-B[41]. OCA-B has been shown to have no effect on the initial transcription of Ig genes, or play a role in the development of early B cells; however, mice deficient in OCA-B nonetheless exhibited impaired immune response[43].
OCA-B increases the effectiveness of Oct-1 and Oct-2 activity on Ig promoters[40]. Ectopically expressing OCA-B stimulates transcription of an IgH promoter in a HeLa nuclear extract[40]. Oct-2 mutants with deletions in one of the two activation domains were generated and were shown to have a reduction of ability to stimulate an artificial octamer-dependent promoter[41].
Several observations surrounding the phenotypes of OCA-B-/- mice made by Kim et al[44] help reveal more information involving its function. Knockouts are able to produce the same levels of IgM mRNA and protein as the WT mice, as well as produce normal numbers of mature surface IgM+/IgD+ splenic B cells[44]. The cells had slightly reduced levels of proliferation following LPS stimulation, but the proliferative response to stimulation by anti-IgM crosslinking was greatly reduced - a response that was almost completely rescued to WT level when IL-4 was added[44]. These results suggest that B cell differentiation and expression of IgM is not affected by knocking out OCA-B, and LPS and IL-4 pathways are for the most part intact. But these mice produce reduced serum levels of secondary Ig isotypes; the numbers of surface Ig-expressing cells and IgG2b, IgG3, and IgG1 secreting cells are not different between the knockout and WT. The rates of secretion per cell, however, are much lower in the OCA-B-/- mice, suggesting that the knockouts are able to undergo the isotype-switching processes, but are incapable of efficiently expressing these switched Ig genes. Interestingly, knockout mice lack splenic germinal centers as well as germinal centers in lymph nodes[45]. There was also seen to be an increase in OCA-B expression in normal germinal center B cells[45].
Mice deficient in OCA-B also displayed a 2-4 fold decrease in splenic B cells, which suggests that it is required for splenic B cell maturation[43]. While there was a reduction in levels of mature B cells, cells of early differentiation stages remained unaffected[43]. OCA-B has been demonstrated to repress the development of the transitional Syndecan-1int cell by decreasing the division-based rate of differentiation[46]. In later B cell development, OCA-B is required to promote differentiation into cells that exhibit high rates of Ig secretion[46]. This role of OCA-B in plasmacytic differentiation is in part due to its interaction with Blimp-1. OCA-B-/- cells do not express Blimp-1 in vitro in response to CD40L and IL-4, and the genes that Blimp-1 is known to repress, such as Pax5 and Bcl6, are consequently expressed at high levels in these differentiating knockout cells[46]. In addition to its interaction with Blimp-1, OCA-B has also been shown to regulate immunosuppressive miRNA expression by the conserved octamer motif in the promoter of miR-146a[47]. In the absence of OCA-B, expression of miR-146a and miR-210 is greatly reduced, an interesting finding considering both have been found to suppress NF-κB signaling[47].
A comparison of the > 100 sequences of promoter regions for Igh V regions in the mouse genome shows only the simple consensus of an octamer binding sequence (ATGCAAT) and an INR or initiation region[48]. Some Igh genes contain, while some lack, a TATA box, which would be bound by TBP and basal transcription factors. OCA-B interacts with TAF105, a lymphocyte variant of TFIID[49], and is upregulated in ASCs[50] (see Figure 1). The deletion of either TAF105 or OCA-B alone in vivo does not block Ig secretion[51,52]. Meanwhile, a set of enhancers [3’ Igh (alpha) enhancers, HS1-4] are found far downstream of the whole Igh gene cluster with long range effects for heavy chain class switching and V-D-J recombination[53,54]. Our studies and those of others with transfected Igh genes showed full regulation of the secretory vs membrane alternative RNA processing choice with constructs that lacked the HS1-4 enhancers but retained the Emu/EH enhancer, see for example[55-58]. The Ig heavy chain EH/Emu enhancer stimulates transcription from functional promoters in B lymphocytes[53] but not other cell types.
The EH/Emu enhancer region most likely loops back to the promoter and communicates with it as shown in Figure 1; it contains binding sites for the indicated transcription factors. It appears that for squelching of lymphocyte-specific transcription in non-lymphoid cells, the binding of the repressive nuclear factor-μNR to the Igh enhancer prevents nuclear matrix attachment by interfering with the positively acting matrix attachment region proteins such as MAR-BP1, which drive transcription in B cells[59]. Igh 3’ enhancer-bound OCA-B and promoter-bound TFII-I mediate promoter-enhancer interactions, in both cis and trans, that are important for Igh transcription. This suggests an important function for OCA-B in Igh 3’ enhancer function in vivo that may be important for high levels of secretory-specific mRNA production[60].
Antibody molecules are first expressed on the surface of maturing B cells as membrane spanning receptors for immunogens and are known as the B cell receptor or BCR. Engagement of the BCR by cognate antigen on a mature B cell leads to activation of transcription and cell growth as described above. After the activation process, the pre-mRNA transcribed from the rearranged Ig heavy chain gene is alternatively processed to produce the secretory-specific form of Igh mRNA, reviewed in[4]. Not only is there a shift to use of the proximal poly(A) site but also a large increase in the overall amount of mRNA, with a less than 2-fold increase in RNAP-II loading on the gene[61]. This indicates that RNA processing increases the quality and the quantity of the mRNA made from the Igh locus. The transition in Igh mRNA processing serves as a hallmark for the differentiation of the B cell to a plasma blast or an ASC and is a harbinger of major changes in cellular architecture and transcription allowing the Ig protein to be secreted[62]. The RNA polymerase in the ASC is traveling to the beat of drum different from that in a B cell. Transcription elongation sets the tempo and acts as the drum major.
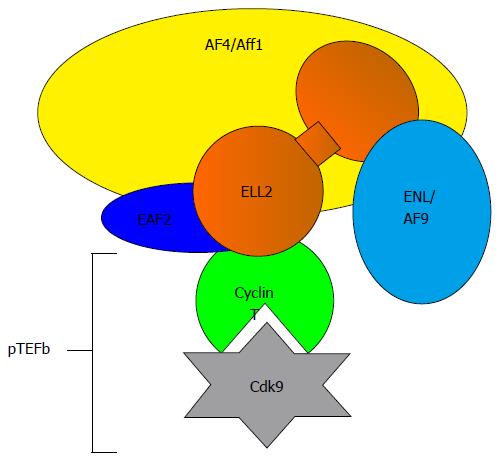
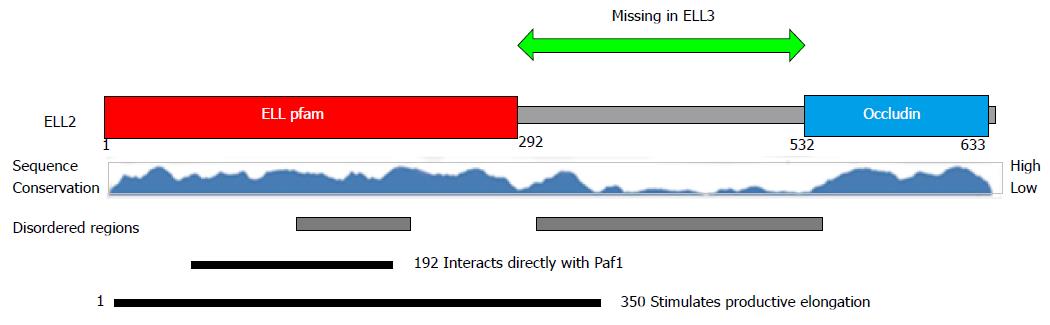
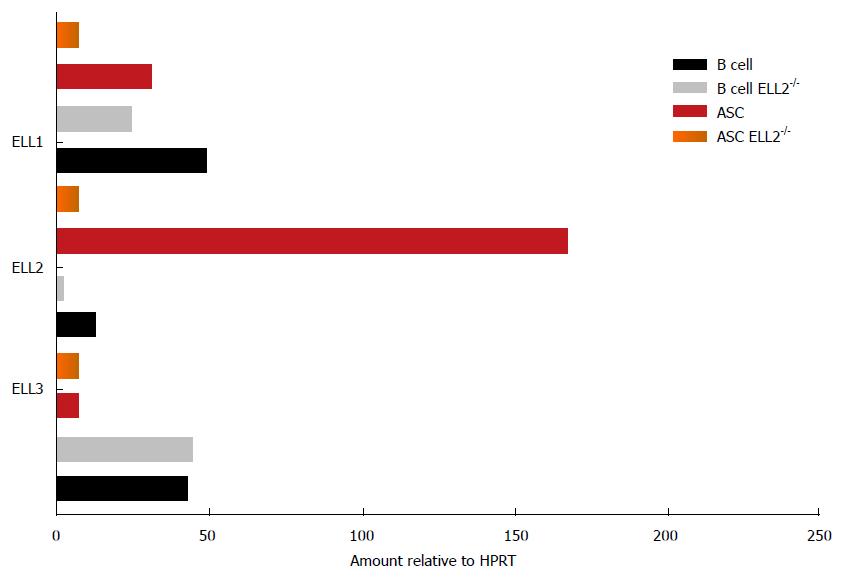
All three members of the ELL gene family, 1, 2, 3, are involved in transcription elongation in the super elongation complex, SEC, see Figure 2. ELL1 was cloned from multiple lineage leukemia cells when its COOH terminal half was found to be a fusion partner with MLL, a histone H3 K4 methylase[63]. Based on the available literature, ELL1 may play its biggest role in DNA repair and small RNA synthesis[64,65]. Other family members were cloned because of their homology to ELL1. ELL3, at 397 amino acids long, differs in sequence from ELL1 and ELL2 (602 and 633 amino acids respectively) and lacks the central disordered region depicted in Figure 3, but retains the majority of the NH2-terminal productive elongation domain and the occludin homology/p53 interacting domain. Both ELL1 and ELL3 have been shown to sequester p53 and abrogate its activities[66,67]. ELL2 was not tested for this activity. ELL3 was first described as testis specific[68], but subsequently it was shown to play a role in the epithelial-mesenchymal transition[69] and to mark enhancers in ES cells, priming for future gene activation[70]. ELL2 replaces ELL3, which predominates in embryonic cells[69] and B cells; ELL3 levels are diminished after stimulation to ASC differentiation even in ELL2 conditional knockouts[62] see Figure 4. Its role in B cells is as yet undefined.
ELL1 was unable to substitute for ELL2 in driving proximal poly(A) site choice in the Igh locus[71]. ELL1 and 2 differ primarily in the sequence of the disordered region starting at amino acid 292 in Figure 3. Thus each ELL can be expected to have unique interactions and functions in transcription elongation based on its unique sequences. For example, Mediator subunit 26 drives the association of ELL1 with snRNA gene promoters[72]. Using yeast two-hybrid assays, we have shown that conserved portions of the central disordered, proline-rich regions of ELL1 and 2 bind specific proteins[73]; the absence of this region in ELL3 dictates that it will have different associations.
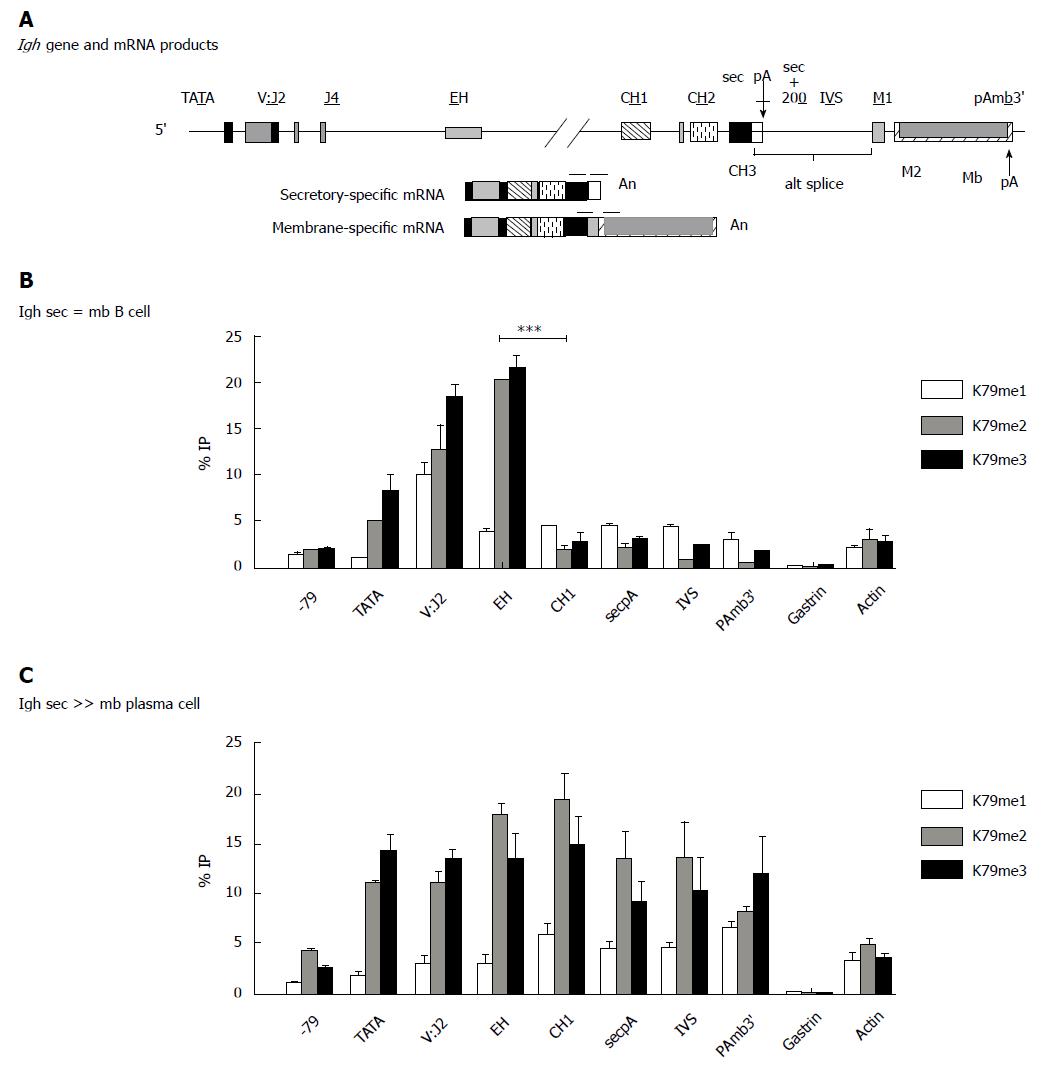
We have shown that ELL2 modifies the RNA polymerases in ASCs[10,61,71]; this causes RNAP-II to traverse the genes in a manner that is unlike that in a B cell and hence the RNAP-IIs in ASCs “travel to the beat of a different drum”. ELL2 has important and now well established roles in releasing paused RNAP-II in Human Immunodeficiency Virus (HIV) infection and in multiple myeloma[4,74]. There is a > 6-fold rise in the level of ELL2 ASCs (see Figure 4)[10,62,71,73,75], mediated by the Irf4 transcription factor[76-78]. There is also a increase in ELL2 mediated by Blimp-1 expression[76]. We showed that ELL2 drives alternative RNA processing [exon skipping and first poly(A) site choice] to influence the expression of the secretory-specific form of Igh mRNA at the expense of the membrane form[10] diagrammed in Figure 5A. This occurs because more mature mRNA results from every pass of the RNAP-II; processivity is increased by ELL2. Studies of the ELL2 promoter (-1142 to + 154) show Irf4 and NF-κB p65 responsive sites[62], cyclic AMP response elements, and binding sites for the viral onco-protein Tax made in HTLV infection[79]. In the SEC, ELL2 associates with the positive transcription factor P-TEFb, AFF4, and other proteins found in fusions with MLL in cancer that facilitate H3K4 methylation[80], see Figure 2.
In the case of model ASCs vs B cells, more ELL2 and P-TEFb are recruited to the RNAP-II on the identical Igh gene; there is a correspondingly higher level of ser-2 phosphorylation of the carboxyl-terminal domain (CTD) of RNAP-II nearer the promoter[61,71]. The scope of modifications of the histones on the Igh gene is different from that seen in B cells and ASCs. We saw more H3K79 di- and tri-methylation as well as H3K4 methylation 3’ of the internal heavy chain enhancer in ASCs, which is indicative of a more open chromatin configuration[71] (see Figure 5). H3K79 methylation has also been linked to alterations in splicing[81]. All of these changes in chromatin would favor use of promoter proximal poly(A) sites, like that of the secretory Igh poly(A) site and skipping of the splice sites that would be necessary for the production of the Igh membrane-encoding form of Igh. Ironically, an elongation factor causes the production of a shorter Igh mRNA.
Our B-cell specific ELL2 conditional knockout mice (ell2loxp/loxp CD19cre/+aka ELL2cKO)[62] exhibit normal numbers of splenic B cells but curtailed primary and secondary humoral responses both in NP-ficoll and NP-KLH immunized animals. In ELL2 cKO mice relative to ELL2+/+ animals: CD138+/ B220 lo ASCs in spleen were reduced; there were fewer IgG1+ antibody producing cells in the bone marrow (i.e., long-lived plasma cells); splenic B cells stimulated by LPS ex vivo were ¼ as likely to produce B220loCD138+ cells (ASCs) than from control splenic B-cells. The “pseudo ASCs” that arise in the ELL2 cKO have a paucity of secreted Igh, and distended, abnormal appearing ER by electron microscopy. The amounts of Ig kappa, activating transcription factor 6 (Atf6), BCMA (Tnfrsf1), BiP, Cyclin B2, OCA-B, and Xbp1 mRNAs, unspliced and spliced, are severely reduced in the ELL2 cKOs[62]. Thus we showed that ELL2 is essential for antibody synthesis and export.
The complex expression pattern of the three ELL family members in B cells and ASCs both in ELL2+/+ and the ELL2-/- conditional knockouts is shown in Figure 4. The knockout of ELL2 influences its own and ELL1 mRNAs but not that of ELL3, which declines following LPS stimulation to ASCs regardless of the presence of ELL2[62].
A combination of genome-wide high-throughput sequencing methods and drug treatments that inhibit P-TEFb have suggested that P-TEFb-driven release of paused RNAP-II from promoter-proximal regions to begin productive elongation is a widespread and necessary step in transcription. Studies have shown that inhibition of P-TEFb, and by extension SEC, that prevents RNAP-II release, blocks almost all transcription[9,29,30]. Thus, all active genes experience a potentially rate-limiting pausing step in the transcription cycle and require SEC activity for gene body transcription. However, this pause step causes a significant accumulation of promoter-proximally paused RNAP-II only at a subset of active genes in untreated cells (40%-70%, depending on the method and cell type)[3,9,29,31-33]. Presumably SEC activity is simply not limiting on the remainder of genes that do not show accumulation of paused RNAP-II. This finding indicates that a pausing- and SEC-dependent release step could become a rate-limiting and potentially regulatory step at all active genes[82].
Transcription and RNA processing are controlled in cells by the cooperating processes of modifications to the CTD of RNAP-II, addition of elongation and RNA processing factors to the RNAP-II complex, and by chromatin modifications[83]. Negative elongation factor (NELF) and DRB sensitive factor (DSIF) are recruited to an RNAP-II when it pauses just after initiation of transcription. DSIF is composed of the highly conserved Spt4 and Spt5 subunits, which have been shown to have unique parts to play at different phases of Ig class switch recombination[84] and in germinal center B cells[85]. NELF is found only in paused metazoan, not yeast transcription complexes[86]. Recruitment to the paused RNAP-II of P-TEFb, composed of cyclin T and cdk9, and its associated factors like ELL into a super elongation complex[87], results in phosphorylation of DSIF and the ser-2 of the carboxyl-terminal end of RNAP-II. NELF is also phosphorylated by P-TEFb, releasing it from the now elongation-competent RNAP-II complex. It is clear from studies with HIV tat and tar that recruitment of P-TEFb with ELL2 also facilitates interactions of the five-subunit polymerase associated factor (paf) with RNAP-II; paf then recruits the polyadenylation factors[88]. This would favor promoter proximal polyadenylation. Studies using RNAP-II mutants or drugs to slow elongation show that reduced transcription rates are coupled with alternative exon inclusion while speeding up the polymerase causes exon skipping[89]. In addition, transcription factors have been shown to control the use of alternative exons and control splicing patterns, presumably by differentially setting up the RNAP-II complex or its elongation rate[90].
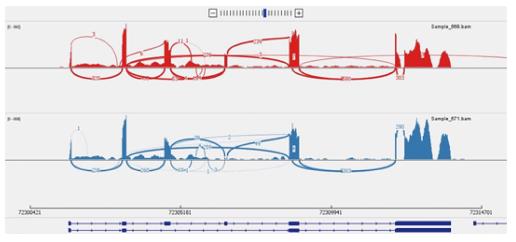
Using deep mRNA sequencing, the knockdown of ELL2 by siRNA in a plasma cell line was shown to influence several other genes besides Igh secretory specific mRNA processing, namely several splicing factors, cyclin B2 (Ccnb2), and the B cell maturation antigen (Tnfrsf17) aka BCMA. Long term survival of plasma cells is impaired by the lack of BCMA in a knockout mouse[91]. But loss of BCMA alone in -/- mice does not alter humoral responses (T-independent or T-dependent) nor the formation of short-lived plasma cells, yet loss of ELL2 in mice does[62]. Benson et al[92] saw changes in splicing in a number of genes involved in mRNA processing; this would have had other far reaching secondary effects beyond that of ELL2 on transcription. Dissociating the direct vs indirect effects of ELL2 is key to understanding its role in changing the RNAP-II and RNA processing patterns on a given gene in ASCs. In the ELL2 conditional knockout mice we saw not only reduced Igh processing to the secretory-specific form, but also deficiency in light chain mRNA synthesis and decreased expression of UPR genes, especially in Xbp1. We also saw changes in the splicing of some ELL2 target genes as illustrated in the Sashimi plot shown in Figure 6 for Xaf1, a gene involved in apoptosis of cells[93].
Elongation factors like ELL2 not only change RNA processing patterns but they also increase the processivity of RNAP-II[94]. As a consequence, they can cause higher production of mature mRNA from a precursor and boost mRNA yields without increasing RNAP-II loading. For example, using a cyclin B2 promoter, addition of ELL2 cDNA had a greater than 7-fold enhancement in the luciferase reporter systems relative to a Blimp-1 promoter[62]. It is also worth noting that many of the genes in primary B cells affected by the loss of ELL2 after stimulation to ASCs are genes expressed at high mRNA abundance (Atf6, BiP, Igh, IgL, Pou2af1, Xbp1), genes for which efficient pre-mRNA to mature mRNA processing would be important. Interestingly, in the ELL2 knockout, there was a 5-fold decrease in the expression of Pou2af1 (OBF-1/ BOB-1/OCA-B) mRNA, a putative downstream target of Xbp1[95]. We also saw that ELL2 could enhance luciferase yields from a promoter carrying the UPR elements, so the effect on Pou2af1 could be direct as well as indirect through decreased Xbp1.
The increase of ELL2 in ASCs drives alternative RNA processing and leads to an increase in secretory Igh mRNA[10]. The substantial amount of Ig chains being produced must first be processed efficiently into antibodies by the ER. Differentiating B cells adapt to the added stress of processing the increased amount of antibody by inducing the UPR, a signaling cascade prompted by ER stress that upregulates ER chaperone and folding enzymes expression (reviewed in[96]). Even after initiation of terminal B cell differentiation and efficient elongation take place, regulators of the UPR and alleviators of ER stress are needed to ensure that successful ASC development occurs.
In mature ASCs, the ER response is unique from that seen in other cells[97]. The UPR in many cells typically has three arms, the Ire1/Xbp1 pathway, an Atf6 pathway, and the PERK pathway[98]. But PERK knockout mice secrete normal amounts of Ig, while PERK protein expression is not changed significantly between B cells and ASCs[99,100]. In addition, Atf6 is not necessary for the development of ASCs; thus when B cells are stimulated to secrete antibody, the primary pathway for ER remodeling appears to reside in the Ire1 to Xbp1 pathway[101].
Aggregation and then auto phosphorylation of Ire1 causes it to acquire the ability to specifically cleave and then splice Xbp1 mRNA; the newly spliced Xbp1 RNA species encodes a novel Xbp1 protein with transcriptional activity on its own promoter and other UPR promoters containing the UPR element UPRE[99]. In an Xbp1 conditional deletion, the mice show defects in ASC development[25] and low levels of secretory Ig[102]. But it has been argued that the consequences of Xbp1 deletion alone are relatively mild[103]. ASCs are present in normal frequencies in resting and immunized animals, and Ig secretion is reduced but not eliminated in conditional Xbp1 knockouts. Thus the gene regulatory program controlling ASC differentiation may proceed relatively normally in the absence of Xbp1[103].
On further analysis, the low levels of Igh mRNA in Xbp1-/- mice result from the 8-fold increased levels of Ire1-P over control; the highly abundant Ire1-P cleaves the Igh mu secretory mRNA[104]. This is a process similar to the previously described pathway[105] in which Ire1-P can act to cleave its own mRNA, as well as other RNAs in a process called regulated IRE1-dependent decay[106]. Only Xbp1 mRNA is spliced, not cleaved, by Ire1-P to form a new functional RNA[107]. A double deletion of Xbp1 and Ire1 restores IgM secretion by inhibiting Ig mRNA degradation[104]. Mutations in the Ire1 nuclease function cause only a 2-fold reduction in Ig secretion[108]. Taken together, this leads to a conclusion that some Ig secretion can occur without the unusual cleavage and splicing of Xbp1 and there may be other proteins that allow for the upregulation of the UPR besides the spliced mRNA encoded Xbp1. As we discussed above, ELL2 has a role in enhancing the transcription of other UPR proteins through the UPR element[62] thereby linking production of the Igh secretory mRNA and the build-up of the UPR. Activation of the mammalian target of rapamycin (mTOR) pathway can also bypass Xbp1 for Ig secretion[109].
mTOR is a vital serine/threonine kinase with two known subunit complexes, mTORC1 and mTORC2[110]. Much of the known function of mTOR, primarily in complex one, shows major roles in cellular proliferation[111] and Ig secretion[112]. The main function of mTORC1 is to recognize nutrient levels and mitogenic signals, and with these trigger cellular growth and proliferation. mTORC2 differs as it is nutrient independent and is activated by growth factors[113]. Within its pathway is the tuber sclerosis complex (TSC), which is an inhibitory complex of mTOR. TSC presents itself in two forms, TSC1 and TSC2. The two forms come together as a heterodimeric complex[114,115]. Akt, a protein kinase, is responsible for phosphorylation of TSC2 and is activated after LPS-stimulation[115]. The release of TSC complex inhibition induces mTOR via BCR stimulation. The reversion of TSC1 inhibition of mTORC1 is responsible for protein synthesis in LPS-activated B cells, which is coupled with substantial ER stress[116]. ER stress can activate the UPR, restoring ER stability, or possibly lead to autophagy or apoptosis[117]. During B cell differentiation to ASC, ER remodeling is substantial and exclusively facilitated by the Xbp1 UPR pathway. Skipping of the Xbp1 pathway has been shown to allow B cells to viably differentiate into long-lived ASCs that only secrete small amounts of Igs[103,104]. It was shown that the ER morphology was highly compromised in the Xbp1 knockout after ASC transition. The successful transition, even with compromised ER morphology, from B cell to ASC was shown to be due to the positive regulation of mTOR[109].
Looking directly at mTOR, inhibition of mTORC1 in mice induces macro-autophagy[118]. When activated, mTORC1 promotes cell growth and protein synthesis. Along with mTORC1, TSC1 ablation ex vivo resulted in cell death of developing ASCs[116]. The production of a TSC1 KO, Xbp1 KO and TSC1/Xbp1 DKO has allowed for complete analysis of mTORC1 outcomes independent of parallel pathways. TSC1 KO promoted ASC differentiation with increased mTOR activity along with an unexpected increase in Ire1. Pertaining to the antibody secretion of the ASCs, the expected reduction of IgM and IgG1 levels in the KOs and DKO was observed. A marked increase of IgA titers in the serum of the DKO was also observed. This correlates with the novel finding that mTOR activation can bypass Xbp1 for antibody secretion. To assess the effect of TSC1 KO on the ER, the DKO and the Xbp1 KO were compared after LPS stimulation. It was clear that the double knockout had a less compromised ER, suggesting that mTOR activation and its directed UPR play a crucial role in ER maintenance and remodeling[109].
The modification of RNAP-II elongation by ELL2 in ASCs is dramatic with far-reaching consequences. It is important to further study the direct effects that this modification is having on transcription of Igh and on expression of other ASC genes. What else occurs as a result of RNA polymerases traveling to the beat of a different drum? The significance of understanding these systems lies in the foundation of the correct production and processing of antibodies, a vital part of immune response. Further study will allow for an expanded breadth of understanding concerning this complex system as well as great advances in diagnosis and therapy for autoimmunity and immune-deficiency diseases.
P- Reviewer: Jin B, Weissert R S- Editor: Gong XM L- Editor: A E- Editor: Jiao XK
| 1. | Pyo JO, Yoo SM, Ahn HH, Nah J, Hong SH, Kam TI, Jung S, Jung YK. Overexpression of Atg5 in mice activates autophagy and extends lifespan. Nat Commun. 2013;4:2300. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 428] [Cited by in RCA: 544] [Article Influence: 49.5] [Reference Citation Analysis (0)] |
| 2. | Nutt SL, Hodgkin PD, Tarlinton DM, Corcoran LM. The generation of antibody-secreting plasma cells. Nat Rev Immunol. 2015;15:160-171. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 773] [Cited by in RCA: 982] [Article Influence: 98.2] [Reference Citation Analysis (0)] |
| 3. | Fernández D, Ortiz M, Rodríguez L, García A, Martinez D, Moreno de Alborán I. The proto-oncogene c-myc regulates antibody secretion and Ig class switch recombination. J Immunol. 2013;190:6135-6144. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 4. | Bayles I, Milcarek C. Plasma cell formation, secretion, and persistence: the short and the long of it. Crit Rev Immunol. 2014;34:481-499. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 5. | Shi W, Liao Y, Willis SN, Taubenheim N, Inouye M, Tarlinton DM, Smyth GK, Hodgkin PD, Nutt SL, Corcoran LM. Transcriptional profiling of mouse B cell terminal differentiation defines a signature for antibody-secreting plasma cells. Nat Immunol. 2015;16:663-673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 275] [Cited by in RCA: 302] [Article Influence: 30.2] [Reference Citation Analysis (0)] |
| 6. | Mamane Y, Heylbroeck C, Génin P, Algarté M, Servant MJ, LePage C, DeLuca C, Kwon H, Lin R, Hiscott J. Interferon regulatory factors: the next generation. Gene. 1999;237:1-14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 416] [Cited by in RCA: 434] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 7. | Lu D, Nakagawa R, Lazzaro S, Staudacher P, Abreu-Goodger C, Henley T, Boiani S, Leyland R, Galloway A, Andrews S. The miR-155-PU.1 axis acts on Pax5 to enable efficient terminal B cell differentiation. J Exp Med. 2014;211:2183-2198. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 74] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 8. | Eisenbeis CF, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377-1387. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 372] [Cited by in RCA: 382] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 9. | Klein U, Casola S, Cattoretti G, Shen Q, Lia M, Mo T, Ludwig T, Rajewsky K, Dalla-Favera R. Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol. 2006;7:773-782. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 535] [Cited by in RCA: 592] [Article Influence: 31.2] [Reference Citation Analysis (0)] |
| 10. | Martincic K, Alkan SA, Cheatle A, Borghesi L, Milcarek C. Transcription elongation factor ELL2 directs immunoglobulin secretion in plasma cells by stimulating altered RNA processing. Nat Immunol. 2009;10:1102-1109. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 89] [Cited by in RCA: 105] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 11. | Willis SN, Good-Jacobson KL, Curtis J, Light A, Tellier J, Shi W, Smyth GK, Tarlinton DM, Belz GT, Corcoran LM. Transcription factor IRF4 regulates germinal center cell formation through a B cell-intrinsic mechanism. J Immunol. 2014;192:3200-3206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 83] [Cited by in RCA: 91] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 12. | Brass AL, Zhu AQ, Singh H. Assembly requirements of PU.1-Pip (IRF-4) activator complexes: inhibiting function in vivo using fused dimers. EMBO J. 1999;18:977-991. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 154] [Cited by in RCA: 163] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 13. | Pongubala JM, Nagulapalli S, Klemsz MJ, McKercher SR, Maki RA, Atchison ML. PU.1 recruits a second nuclear factor to a site important for immunoglobulin kappa 3’ enhancer activity. Mol Cell Biol. 1992;12:368-378. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 193] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 14. | Shukla V, Lu R. IRF4 and IRF8: Governing the virtues of B Lymphocytes. Front Biol (Beijing). 2014;9:269-282. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 40] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 15. | Carotta S, Willis SN, Hasbold J, Inouye M, Pang SH, Emslie D, Light A, Chopin M, Shi W, Wang H. The transcription factors IRF8 and PU.1 negatively regulate plasma cell differentiation. J Exp Med. 2014;211:2169-2181. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 105] [Cited by in RCA: 116] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 16. | Lee SC, Bottaro A, Insel RA. Activation of terminal B cell differentiation by inhibition of histone deacetylation. Mol Immunol. 2003;39:923-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 49] [Cited by in RCA: 50] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 17. | Kallies A, Hasbold J, Fairfax K, Pridans C, Emslie D, McKenzie BS, Lew AM, Corcoran LM, Hodgkin PD, Tarlinton DM. Initiation of plasma-cell differentiation is independent of the transcription factor Blimp-1. Immunity. 2007;26:555-566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 204] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 18. | Zhang W, Mi J, Li N, Sui L, Wan T, Zhang J, Chen T, Cao X. Identification and characterization of DPZF, a novel human BTB/POZ zinc finger protein sharing homology to BCL-6. Biochem Biophys Res Commun. 2001;282:1067-1073. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 58] [Cited by in RCA: 66] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 19. | Mitchelmore C, Kjaerulff KM, Pedersen HC, Nielsen JV, Rasmussen TE, Fisker MF, Finsen B, Pedersen KM, Jensen NA. Characterization of two novel nuclear BTB/POZ domain zinc finger isoforms. Association with differentiation of hippocampal neurons, cerebellar granule cells, and macroglia. J Biol Chem. 2002;277:7598-7609. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 66] [Cited by in RCA: 72] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 20. | Chevrier S, Emslie D, Shi W, Kratina T, Wellard C, Karnowski A, Erikci E, Smyth GK, Chowdhury K, Tarlinton D. The BTB-ZF transcription factor Zbtb20 is driven by Irf4 to promote plasma cell differentiation and longevity. J Exp Med. 2014;211:827-840. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 75] [Cited by in RCA: 85] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 21. | Buckland J. BLIMP1, BCL6 and B-cell fate. Nat Rev Immunol. 2002;2:629-629. [RCA] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 22. | Keller AD, Maniatis T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991;5:868-879. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 263] [Cited by in RCA: 293] [Article Influence: 8.6] [Reference Citation Analysis (0)] |
| 23. | Turner CA, Mack DH, Davis MM. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell. 1994;77:297-306. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 599] [Cited by in RCA: 614] [Article Influence: 19.8] [Reference Citation Analysis (0)] |
| 24. | Martins G, Calame K. Regulation and functions of Blimp-1 in T and B lymphocytes. Annu Rev Immunol. 2008;26:133-169. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 292] [Cited by in RCA: 300] [Article Influence: 17.6] [Reference Citation Analysis (0)] |
| 25. | Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300-307. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 985] [Cited by in RCA: 999] [Article Influence: 41.6] [Reference Citation Analysis (0)] |
| 26. | Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607-620. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 619] [Cited by in RCA: 660] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 27. | Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51-62. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 800] [Cited by in RCA: 792] [Article Influence: 34.4] [Reference Citation Analysis (0)] |
| 28. | Prochownik EV, Kukowska J. Deregulated expression of c-myc by murine erythroleukaemia cells prevents differentiation. Nature. 1986;322:848-850. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 209] [Cited by in RCA: 246] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 29. | Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596-599. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 338] [Cited by in RCA: 343] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 30. | Yu J, Angelin-Duclos C, Greenwood J, Liao J, Calame K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol Cell Biol. 2000;20:2592-2603. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 257] [Cited by in RCA: 271] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 31. | Reimold AM, Ponath PD, Li YS, Hardy RR, David CS, Strominger JL, Glimcher LH. Transcription factor B cell lineage-specific activator protein regulates the gene for human X-box binding protein 1. J Exp Med. 1996;183:393-401. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 140] [Article Influence: 4.8] [Reference Citation Analysis (0)] |
| 32. | Matthias P, Rolink AG. Transcriptional networks in developing and mature B cells. Nat Rev Immunol. 2005;5:497-508. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 174] [Cited by in RCA: 174] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 33. | Alexander LEMM, Watters JM, Maurin M, Wright KL. Transcriptional repressor PRDM1/Blimp-1 directly regulates transcriptional elongation factor ELL3 during terminal B-cell differentiation. Cancer Res. 2014;74:2331. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 34. | Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439-448. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 302] [Cited by in RCA: 300] [Article Influence: 10.7] [Reference Citation Analysis (0)] |
| 35. | Shaffer AL, Yu X, He Y, Boldrick J, Chan EP, Staudt LM. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 2000;13:199-212. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 665] [Cited by in RCA: 659] [Article Influence: 26.4] [Reference Citation Analysis (0)] |
| 36. | Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat Immunol. 2011;12:304-311. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 430] [Cited by in RCA: 506] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 37. | Chiu R, Boyle WJ, Meek J, Smeal T, Hunter T, Karin M. The c-Fos protein interacts with c-Jun/AP-1 to stimulate transcription of AP-1 responsive genes. Cell. 1988;54:541-552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 965] [Cited by in RCA: 1067] [Article Influence: 28.8] [Reference Citation Analysis (1)] |
| 38. | Yu Z, Sato S, Trackman PC, Kirsch KH, Sonenshein GE. Blimp1 activation by AP-1 in human lung cancer cells promotes a migratory phenotype and is inhibited by the lysyl oxidase propeptide. PLoS One. 2012;7:e33287. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 23] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 39. | Ohkubo Y, Arima M, Arguni E, Okada S, Yamashita K, Asari S, Obata S, Sakamoto A, Hatano M, O-Wang J. A role for c-fos/activator protein 1 in B lymphocyte terminal differentiation. J Immunol. 2005;174:7703-7710. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 40. | Luo Y, Fujii H, Gerster T, Roeder RG. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231-241. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 194] [Cited by in RCA: 240] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 41. | Luo Y, Roeder RG. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115-4124. [PubMed] |
| 42. | Murphy S, Yoon JB, Gerster T, Roeder RG. Oct-1 and Oct-2 potentiate functional interactions of a transcription factor with the proximal sequence element of small nuclear RNA genes. Mol Cell Biol. 1992;12:3247-3261. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 101] [Cited by in RCA: 82] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 43. | Schubart DB, Rolink A, Kosco-Vilbois MH, Botteri F, Matthias P. B-cell-specific coactivator OBF-1/OCA-B/Bob1 required for immune response and germinal centre formation. Nature. 1996;383:538-542. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 219] [Cited by in RCA: 233] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 44. | Kim U, Qin XF, Gong S, Stevens S, Luo Y, Nussenzweig M, Roeder RG. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for normal production of immunoglobulin isotypes. Nature. 1996;383:542-547. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 188] [Cited by in RCA: 203] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 45. | Greiner A, Müller KB, Hess J, Pfeffer K, Müller-Hermelink HK, Wirth T. Up-regulation of BOB.1/OBF.1 expression in normal germinal center B cells and germinal center-derived lymphomas. Am J Pathol. 2000;156:501-507. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 46. | Corcoran LM, Hasbold J, Dietrich W, Hawkins E, Kallies A, Nutt SL, Tarlinton DM, Matthias P, Hodgkin PD. Differential requirement for OBF-1 during antibody-secreting cell differentiation. J Exp Med. 2005;201:1385-1396. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 47] [Cited by in RCA: 53] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 47. | Lindner JM, Kayo H, Hedlund S, Fukuda Y, Fukao T, Nielsen PJ. Cutting edge: The transcription factor Bob1 counteracts B cell activation and regulates miR-146a in B cells. J Immunol. 2014;192:4483-4486. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 9] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 48. | Johnston CM, Wood AL, Bolland DJ, Corcoran AE. Complete sequence assembly and characterization of the C57BL/6 mouse Ig heavy chain V region. J Immunol. 2006;176:4221-4234. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 131] [Cited by in RCA: 127] [Article Influence: 6.7] [Reference Citation Analysis (0)] |
| 49. | Wolstein O, Silkov A, Revach M, Dikstein R. Specific interaction of TAFII105 with OCA-B is involved in activation of octamer-dependent transcription. J Biol Chem. 2000;275:16459-16465. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 19] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 50. | Qin XF, Reichlin A, Luo Y, Roeder RG, Nussenzweig MC. OCA-B integrates B cell antigen receptor-, CD40L- and IL 4-mediated signals for the germinal center pathway of B cell development. EMBO J. 1998;17:5066-5075. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 51. | Freiman RN, Albright SR, Chu LE, Zheng S, Liang HE, Sha WC, Tjian R. Redundant role of tissue-selective TAF(II)105 in B lymphocytes. Mol Cell Biol. 2002;22:6564-6572. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 10] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 52. | Teitell MA. OCA-B regulation of B-cell development and function. Trends Immunol. 2003;24:546-553. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 49] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 53. | Sen R, Grosschedl R. Memories of lost enhancers. Genes Dev. 2010;24:973-979. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 18] [Cited by in RCA: 19] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 54. | Volpi SA, Verma-Gaur J, Hassan R, Ju Z, Roa S, Chatterjee S, Werling U, Hou H, Will B, Steidl U. Germline deletion of Igh 3’ regulatory region elements hs 5, 6, 7 (hs5-7) affects B cell-specific regulation, rearrangement, and insulation of the Igh locus. J Immunol. 2012;188:2556-2566. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 37] [Cited by in RCA: 42] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 55. | Peterson ML, Bingham GL, Cowan C. Multiple features contribute to the use of the immunoglobulin M secretion-specific poly(A) signal but are not required for developmental regulation. Mol Cell Biol. 2006;26:6762-6771. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 13] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 56. | Peterson ML, Perry RP. Regulated production of mu m and mu s mRNA requires linkage of the poly(A) addition sites and is dependent on the length of the mu s-mu m intron. Proc Natl Acad Sci USA. 1986;83:8883-8887. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 63] [Cited by in RCA: 90] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 57. | Peterson ML, Perry RP. The regulated production of mu m and mu s mRNA is dependent on the relative efficiencies of mu s poly(A) site usage and the c mu 4-to-M1 splice. Mol Cell Biol. 1989;9:726-738. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 87] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 58. | Peterson ML, Gimmi ER, Perry RP. The developmentally regulated shift from membrane to secreted mu mRNA production is accompanied by an increase in cleavage-polyadenylation efficiency but no measurable change in splicing efficiency. Mol Cell Biol. 1991;11:2324-2327. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 23] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 59. | Zong RT, Scheuermann RH. Mutually exclusive interaction of a novel matrix attachment region binding protein and the NF-muNR enhancer repressor. Implications for regulation of immunoglobulin heavy chain expression. J Biol Chem. 1995;270:24010-24018. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 25] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 60. | Ren X, Siegel R, Kim U, Roeder RG. Direct interactions of OCA-B and TFII-I regulate immunoglobulin heavy-chain gene transcription by facilitating enhancer-promoter communication. Mol Cell. 2011;42:342-355. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 55] [Cited by in RCA: 52] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 61. | Shell SA, Martincic K, Tran J, Milcarek C. Increased phosphorylation of the carboxyl-terminal domain of RNA polymerase II and loading of polyadenylation and cotranscriptional factors contribute to regulation of the ig heavy chain mRNA in plasma cells. J Immunol. 2007;179:7663-7673. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 19] [Cited by in RCA: 16] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 62. | Park KS, Bayles I, Szlachta-McGinn A, Paul J, Boiko J, Santos P, Liu J, Wang Z, Borghesi L, Milcarek C. Transcription elongation factor ELL2 drives Ig secretory-specific mRNA production and the unfolded protein response. J Immunol. 2014;193:4663-4674. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 28] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 63. | Shilatifard A, Lane WS, Jackson KW, Conaway RC, Conaway JW. An RNA polymerase II elongation factor encoded by the human ELL gene. Science. 1996;271:1873-1876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 269] [Cited by in RCA: 276] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 64. | Mourgues S, Gautier V, Lagarou A, Bordier C, Mourcet A, Slingerland J, Kaddoum L, Coin F, Vermeulen W, Gonzales de Peredo A. ELL, a novel TFIIH partner, is involved in transcription restart after DNA repair. Proc Natl Acad Sci USA. 2013;110:17927-17932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 36] [Cited by in RCA: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 65. | Smith ER, Lin C, Garrett AS, Thornton J, Mohaghegh N, Hu D, Jackson J, Saraf A, Swanson SK, Seidel C. The little elongation complex regulates small nuclear RNA transcription. Mol Cell. 2011;44:954-965. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 69] [Cited by in RCA: 74] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 66. | Wiederschain D, Kawai H, Gu J, Shilatifard A, Yuan ZM. Molecular basis of p53 functional inactivation by the leukemic protein MLL-ELL. Mol Cell Biol. 2003;23:4230-4246. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 35] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 67. | Wiederschain D, Kawai H, Shilatifard A, Yuan ZM. Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage. J Biol Chem. 2005;280:24315-24321. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 68. | Miller T, Williams K, Johnstone RW, Shilatifard A. Identification, cloning, expression, and biochemical characterization of the testis-specific RNA polymerase II elongation factor ELL3. J Biol Chem. 2000;275:32052-32056. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 59] [Cited by in RCA: 61] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 69. | Ahn HJ, Cha Y, Moon SH, Jung JE, Park KS. Ell3 enhances differentiation of mouse embryonic stem cells by regulating epithelial-mesenchymal transition and apoptosis. PLoS One. 2012;7:e40293. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 16] [Cited by in RCA: 16] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 70. | Lin C, Garruss AS, Luo Z, Guo F, Shilatifard A. The RNA Pol II elongation factor Ell3 marks enhancers in ES cells and primes future gene activation. Cell. 2013;152:144-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 81] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 71. | Milcarek C, Albring M, Langer C, Park KS. The eleven-nineteen lysine-rich leukemia gene (ELL2) influences the histone H3 protein modifications accompanying the shift to secretory immunoglobulin heavy chain mRNA production. J Biol Chem. 2011;286:33795-33803. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 18] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 72. | Takahashi H, Takigawa I, Watanabe M, Anwar D, Shibata M, Tomomori-Sato C, Sato S, Ranjan A, Seidel CW, Tsukiyama T. MED26 regulates the transcription of snRNA genes through the recruitment of little elongation complex. Nat Commun. 2015;6:5941. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 38] [Cited by in RCA: 38] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 73. | Arumemi F, Bayles I, Paul J, Milcarek C. Shared and discrete interacting partners of ELL1 and ELL2 by yeast two-hybrid assay. Adv Biosci Biotech. 2013;4. [RCA] [DOI] [Full Text] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 74. | Zhou Q, Li T, Price DH. RNA polymerase II elongation control. Annu Rev Biochem. 2012;81:119-143. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 426] [Cited by in RCA: 469] [Article Influence: 36.1] [Reference Citation Analysis (0)] |
| 75. | Milcarek C. Hide and go seek: activation of the secretory-specific poly (A) site of Igh by transcription elongation factors. In: Grabowski P. RNA Processing. InTech 2011; . |
| 76. | Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee AH, Qian SB, Zhao H, Yu X, Yang L, Tan BK, Rosenwald A. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity. 2004;21:81-93. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 1] [Reference Citation Analysis (0)] |
| 77. | Shaffer AL, Emre NC, Lamy L, Ngo VN, Wright G, Xiao W, Powell J, Dave S, Yu X, Zhao H. IRF4 addiction in multiple myeloma. Nature. 2008;454:226-231. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 600] [Cited by in RCA: 566] [Article Influence: 33.3] [Reference Citation Analysis (0)] |
| 78. | Sciammas R, Li Y, Warmflash A, Song Y, Dinner AR, Singh H. An incoherent regulatory network architecture that orchestrates B cell diversification in response to antigen signaling. Mol Syst Biol. 2011;7:495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 91] [Cited by in RCA: 103] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 79. | Mann MC, Strobel S, Fleckenstein B, Kress AK. The transcription elongation factor ELL2 is specifically upregulated in HTLV-1-infected T-cells and is dependent on the viral oncoprotein Tax. Virology. 2014;464-465:98-110. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 10] [Cited by in RCA: 10] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 80. | Lin C, Smith ER, Takahashi H, Lai KC, Martin-Brown S, Florens L, Washburn MP, Conaway JW, Conaway RC, Shilatifard A. AFF4, a component of the ELL/P-TEFb elongation complex and a shared subunit of MLL chimeras, can link transcription elongation to leukemia. Mol Cell. 2010;37:429-437. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 508] [Cited by in RCA: 466] [Article Influence: 31.1] [Reference Citation Analysis (0)] |
| 81. | Suzuki MG, Ito H, Aoki F. Effects of RNAi-mediated knockdown of histone methyltransferases on the sex-specific mRNA expression of Imp in the silkworm Bombyx mori. Int J Mol Sci. 2014;15:6772-6796. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 6] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 82. | Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol. 2015;16:167-177. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 670] [Cited by in RCA: 625] [Article Influence: 62.5] [Reference Citation Analysis (0)] |
| 83. | Allemand E, Batsché E, Muchardt C. Splicing, transcription, and chromatin: a ménage à trois. Curr Opin Genet Dev. 2008;18:145-151. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 80] [Cited by in RCA: 86] [Article Influence: 5.1] [Reference Citation Analysis (0)] |
| 84. | Stanlie A, Begum NA, Akiyama H, Honjo T. The DSIF subunits Spt4 and Spt5 have distinct roles at various phases of immunoglobulin class switch recombination. PLoS Genet. 2012;8:e1002675. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 35] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 85. | Shlomchik MJ, Weisel F. Germinal center selection and the development of memory B and plasma cells. Immunol Rev. 2012;247:52-63. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 299] [Cited by in RCA: 322] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 86. | Sansó M, Fisher RP. Pause, play, repeat: CDKs push RNAP II’s buttons. Transcription. 2013;4:146-152. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 87. | Smith E, Lin C, Shilatifard A. The super elongation complex (SEC) and MLL in development and disease. Genes Dev. 2011;25:661-672. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 251] [Cited by in RCA: 275] [Article Influence: 19.6] [Reference Citation Analysis (0)] |
| 88. | Chou S, Upton H, Bao K, Schulze-Gahmen U, Samelson AJ, He N, Nowak A, Lu H, Krogan NJ, Zhou Q. HIV-1 Tat recruits transcription elongation factors dispersed along a flexible AFF4 scaffold. Proc Natl Acad Sci USA. 2013;110:E123-E131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 75] [Article Influence: 5.8] [Reference Citation Analysis (0)] |
| 89. | Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. RNA. 2004;10:1489-1498. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 377] [Cited by in RCA: 367] [Article Influence: 17.5] [Reference Citation Analysis (0)] |
| 90. | Nogues G, Kadener S, Cramer P, Bentley D, Kornblihtt AR. Transcriptional activators differ in their abilities to control alternative splicing. J Biol Chem. 2002;277:43110-43114. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 138] [Cited by in RCA: 140] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 91. | O’Connor BP, Raman VS, Erickson LD, Cook WJ, Weaver LK, Ahonen C, Lin LL, Mantchev GT, Bram RJ, Noelle RJ. BCMA is essential for the survival of long-lived bone marrow plasma cells. J Exp Med. 2004;199:91-98. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 735] [Cited by in RCA: 849] [Article Influence: 40.4] [Reference Citation Analysis (0)] |
| 92. | Benson MJ, Aijö T, Chang X, Gagnon J, Pape UJ, Anantharaman V, Aravind L, Pursiheimo JP, Oberdoerffer S, Liu XS. Heterogeneous nuclear ribonucleoprotein L-like (hnRNPLL) and elongation factor, RNA polymerase II, 2 (ELL2) are regulators of mRNA processing in plasma cells. Proc Natl Acad Sci USA. 2012;109:16252-16257. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 33] [Cited by in RCA: 34] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 93. | Qiao L, Gu Q, Dai Y, Shen Z, Liu X, Qi R, Ma J, Zou B, Li Z, Lan HY. XIAP-associated factor 1 (XAF1) suppresses angiogenesis in mouse endothelial cells. Tumour Biol. 2008;29:122-129. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 11] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 94. | Shilatifard A. Factors regulating the transcriptional elongation activity of RNA polymerase II. FASEB J. 1998;12:1437-1446. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 86] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 95. | Shen Y, Hendershot LM. Identification of ERdj3 and OBF-1/BOB-1/OCA-B as direct targets of XBP-1 during plasma cell differentiation. J Immunol. 2007;179:2969-2978. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 27] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 96. | Gass JN, Gunn KE, Sriburi R, Brewer JW. Stressed-out B cells? Plasma-cell differentiation and the unfolded protein response. Trends Immunol. 2004;25:17-24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 92] [Cited by in RCA: 92] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 97. | Ma Y, Shimizu Y, Mann MJ, Jin Y, Hendershot LM. Plasma cell differentiation initiates a limited ER stress response by specifically suppressing the PERK-dependent branch of the unfolded protein response. Cell Stress Chaperones. 2010;15:281-293. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 112] [Cited by in RCA: 111] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 98. | Takayanagi S, Fukuda R, Takeuchi Y, Tsukada S, Yoshida K. Gene regulatory network of unfolded protein response genes in endoplasmic reticulum stress. Cell Stress Chaperones. 2013;18:11-23. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 68] [Cited by in RCA: 68] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 99. | Gass JN, Gifford NM, Brewer JW. Activation of an unfolded protein response during differentiation of antibody-secreting B cells. J Biol Chem. 2002;277:49047-49054. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 233] [Cited by in RCA: 239] [Article Influence: 10.4] [Reference Citation Analysis (0)] |
| 100. | Gass JN, Jiang HY, Wek RC, Brewer JW. The unfolded protein response of B-lymphocytes: PERK-independent development of antibody-secreting cells. Mol Immunol. 2008;45:1035-1043. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 106] [Article Influence: 6.2] [Reference Citation Analysis (0)] |
| 101. | Aragon IV, Barrington RA, Jackowski S, Mori K, Brewer JW. The specialized unfolded protein response of B lymphocytes: ATF6α-independent development of antibody-secreting B cells. Mol Immunol. 2012;51:347-355. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 47] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 102. | Tirosh B, Iwakoshi NN, Glimcher LH, Ploegh HL. XBP-1 specifically promotes IgM synthesis and secretion, but is dispensable for degradation of glycoproteins in primary B cells. J Exp Med. 2005;202:505-516. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 62] [Cited by in RCA: 66] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 103. | Taubenheim N, Tarlinton DM, Crawford S, Corcoran LM, Hodgkin PD, Nutt SL. High rate of antibody secretion is not integral to plasma cell differentiation as revealed by XBP-1 deficiency. J Immunol. 2012;189:3328-3338. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 96] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 104. | Benhamron S, Hadar R, Iwawaky T, So JS, Lee AH, Tirosh B. Regulated IRE1-dependent decay participates in curtailing immunoglobulin secretion from plasma cells. Eur J Immunol. 2014;44:867-876. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 77] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 105. | Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323-331. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 839] [Cited by in RCA: 779] [Article Influence: 48.7] [Reference Citation Analysis (0)] |
| 106. | Tirasophon W, Lee K, Callaghan B, Welihinda A, Kaufman RJ. The endoribonuclease activity of mammalian IRE1 autoregulates its mRNA and is required for the unfolded protein response. Genes Dev. 2000;14:2725-2736. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 192] [Cited by in RCA: 213] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 107. | Lee K, Tirasophon W, Shen X, Michalak M, Prywes R, Okada T, Yoshida H, Mori K, Kaufman RJ. IRE1-mediated unconventional mRNA splicing and S2P-mediated ATF6 cleavage merge to regulate XBP1 in signaling the unfolded protein response. Genes Dev. 2002;16:452-466. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 790] [Cited by in RCA: 836] [Article Influence: 36.3] [Reference Citation Analysis (0)] |
| 108. | Iwawaki T, Akai R, Kohno K. IRE1α disruption causes histological abnormality of exocrine tissues, increase of blood glucose level, and decrease of serum immunoglobulin level. PLoS One. 2010;5:e13052. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 84] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 109. | Benhamron S, Pattanayak SP, Berger M, Tirosh B. mTOR activation promotes plasma cell differentiation and bypasses XBP-1 for immunoglobulin secretion. Mol Cell Biol. 2015;35:153-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 69] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 110. | Sarbassov DD, Ali SM, Sengupta S, Sheen JH, Hsu PP, Bagley AF, Markhard AL, Sabatini DM. Prolonged rapamycin treatment inhibits mTORC2 assembly and Akt/PKB. Mol Cell. 2006;22:159-168. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1994] [Cited by in RCA: 2104] [Article Influence: 110.7] [Reference Citation Analysis (0)] |
| 111. | Menon S, Manning BD. Common corruption of the mTOR signaling network in human tumors. Oncogene. 2008;27 Suppl 2:S43-S51. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 234] [Cited by in RCA: 266] [Article Influence: 17.7] [Reference Citation Analysis (0)] |
| 112. | Powell JD, Delgoffe GM. The mammalian target of rapamycin: linking T cell differentiation, function, and metabolism. Immunity. 2010;33:301-311. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 407] [Cited by in RCA: 389] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 113. | Fayard E, Xue G, Parcellier A, Bozulic L, Hemmings BA. Protein kinase B (PKB/Akt), a key mediator of the PI3K signaling pathway. Curr Top Microbiol Immunol. 2010;346:31-56. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 65] [Cited by in RCA: 139] [Article Influence: 9.3] [Reference Citation Analysis (0)] |
| 114. | Donahue AC, Fruman DA. Distinct signaling mechanisms activate the target of rapamycin in response to different B-cell stimuli. Eur J Immunol. 2007;37:2923-2936. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 63] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 115. | Weichhart T, Costantino G, Poglitsch M, Rosner M, Zeyda M, Stuhlmeier KM, Kolbe T, Stulnig TM, Hörl WH, Hengstschläger M. The TSC-mTOR signaling pathway regulates the innate inflammatory response. Immunity. 2008;29:565-577. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 581] [Cited by in RCA: 639] [Article Influence: 37.6] [Reference Citation Analysis (0)] |
| 116. | Goldfinger M, Shmuel M, Benhamron S, Tirosh B. Protein synthesis in plasma cells is regulated by crosstalk between endoplasmic reticulum stress and mTOR signaling. Eur J Immunol. 2011;41:491-502. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 117. | Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519-529. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4586] [Cited by in RCA: 5044] [Article Influence: 280.2] [Reference Citation Analysis (0)] |
| 118. | Kamada Y, Funakoshi T, Shintani T, Nagano K, Ohsumi M, Ohsumi Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J Cell Biol. 2000;150:1507-1513. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 832] [Cited by in RCA: 908] [Article Influence: 36.3] [Reference Citation Analysis (0)] |