Published online Dec 9, 2024. doi: 10.5409/wjcp.v13.i4.100493
Revised: September 28, 2024
Accepted: October 16, 2024
Published online: December 9, 2024
Processing time: 73 Days and 4.2 Hours
Thus far, genetic analysis of patients clinically diagnosed with glycogen storage diseases (GSDs) in Thailand has not been reported.
To evaluate the clinical and biochemical profiles, molecular analysis and long-term outcomes of Thai children diagnosed with hepatic GSD.
Children aged < 18 years diagnosed with hepatic GSD and followed up at King Chulalongkorn Memorial Hospital were recruited. Whole-exome sequencing (WES) was performed to identify the causative gene variants. Medical records were assessed.
All eight children with histopathologically confirmed diagnosis were classified by WES into subtypes Ia (n = 1), III
Hepatomegaly, transaminitis, and hypoglycemia are the hallmarks of GSD confirmed by liver histopathology. Molecular analysis can confirm the diagnosis or classify the subtype that might benefit from personalized treatment, prognosis, and long-term care.
Core Tip: Hepatic glycogen storage diseases (GSDs) are rare but treatable conditions. While liver histopathology is helpful for diagnosis, it cannot differentiate between GSD subtypes. Data on long-term outcomes with extensive nutritional management are limited. This study evaluates the clinical and biochemical profiles, molecular analysis, and long-term outcomes of Thai children with hepatic GSDs, identifying two novel causative variants. The findings indicate that extensive nutritional management, including frequent uncooked cornstarch supplementation, a high-protein diet, and a low lactose-fructose diet, yields favorable outcomes across GSD subtypes. However, tailored management, particularly for GSD types III and VI, can further enhance quality of life and minimize complications.
- Citation: Vanduangden J, Ittiwut R, Ittiwut C, Phewplung T, Sanpavat A, Sintusek P, Suphapeetiporn K. Molecular profiles and long-term outcomes of Thai children with hepatic glycogen storage disease in Thailand. World J Clin Pediatr 2024; 13(4): 100493
- URL: https://www.wjgnet.com/2219-2808/full/v13/i4/100493.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v13.i4.100493
Glycogen storage diseases (GSDs) are rare inherited disorders that affect glycogen metabolism, with a global incidence of approximately < 1 in 100000[1]. The pathogenesis of GSDs involves defects in the enzymes responsible for breaking down glycogen, resulting in the storage of glycogen in mainly the liver, muscle, and heart[1]. GSDs have 23 subtypes that can be broadly classified into hepatic GSDs or muscle GSDs depending on the primary system affected. Hepatic GSDs include types 0a, I, III, IV, VI, and IX and Fanconi–Bickel syndrome that are inherited in an autosomal recessive manner, except for type IXa, which is an X-linked recessive disorder. Genetic analysis is ultimately responsible for the final identification of GSD subgroups given the diversity of clinical and laboratory findings[1]. Because GSDs are treatable conditions, prompt and accurate diagnosis are essential for the commencement of targeted therapy tailored to the subtypes, preventing permanent injury and improving the quality of life of the patients.
Worldwide, molecular genetic studies of GSD subtypes have increased, particularly in several Asian nations including China[2-4], India[5,6], and Pakistan[7]. Thus far, no studies have conducted genetic analysis of patients clinically diagnosed with GSDs in Thailand. Therefore, this study sought to delineate the clinical and genetic attributes of hepatic GSDs in Thai children. By conducting comprehensive assessments of clinical profiles, laboratory studies—including liver histology—and long-term outcomes following treatment, this research not only enhances our understanding of GSDs in the Thai population but also contributes valuable insights into the phenotypic variability, potential genetic mutations, and therapeutic responses associated with these disorders. Ultimately, the findings could inform improved diagnostic and treatment strategies for GSDs in Thailand and similar contexts, paving the way for future research and better healthcare outcomes for affected individuals.
The study population included children aged < 18 years who were diagnosed with hepatic GSDs at the time of their initial diagnosis and regularly followed up at King Chulalongkorn Memorial Hospital, Thailand, between January 2010 and September 2023.
Written informed assent and/or informed consent for this study were obtained from the patients and/or their parents. In addition, genetic counseling and informed consent were provided to all participants and their guardians before collecting blood samples for genetic analysis. This study was approved by the Institute Research Board at Chulalongkorn University, Bangkok, Thailand (No. 0678/65).
Data were extracted from the electronic medical records, which included the clinical profiles at the time of presentation, biochemical profiles (fasting blood glucose, uric acid, complete blood count, liver function test, lipids, creatinine, lactate dehydrogenase, lactate, creatine kinase, and coagulogram), radiological parameters, liver histopathology, nutritional and medical therapy, and outcomes at the end of the follow-up period.
In every outpatient visit, the growth parameters of all patients were monitored, and the weight-for-length/height in children aged < 5 years and the Z-scores for height and body mass index (BMI) were employed for children aged 5–19 years, according to the growth chart that was established by the World Health Organization.
Growth and nutrition status: Overweight was defined as the weight-for-length/height > 2 SD in children aged < 5 years and BMI-for-age > 1 SD in children aged 5–19 years. Obesity was defined as weight-for-length/height > 3 SD in children aged < 5 years and BMI-for-age > 2 SD in children aged 5–19 years. Wasting was defined as weight-for-length/height < −2 SD in children aged < 5 years and BMI-for-age < −2 SD in children aged 5–19 years. Stunting referred to length/height-for-age < −2 SD.
Biochemical results: Hypoglycemia is defined if blood sugar less than 60 mg/dL. Hyperuricemia is defined if serum uric acid is 5.9 mg/dL for < 9 years (both genders), 6.1 mg/dL for 9–11 years (both genders), 7.0 mg/dL (males), and 6.2 mg/dL (females) for 12–14 years. Dyslipidemia is defined if cholesterol ≥ 200 mg/dL or low density lipoprotein ≥ 130mg/dL, high density lipoprotein < 40 mg/dL, triglyceride ≥ 100 mg/dL in children aged less than 9 years and triglyceride ≥ 130 mg/dL in children aged between 9-19 years. Transaminase is defined if aspartate aminotransferase or alanine aminotransferase > 40 U/L. Hyperbilirubinemia is defined if total bilirubin > 2mg/dL. Anemia is defined if hemoglobin < 11 g/dL for age 6 months-5 years, < 11.5 g/dL for age > 5-12 years and female adolescent, < 12 g/dL for male adolescent.
After obtaining informed consent, 3 mL of peripheral blood was collected from patients and their available parents. Genomic DNA was extracted from peripheral blood leukocytes using the Puregene blood kit (Qiagen, Hilden, Germany). Whole-exome sequencing (WES) was performed. Briefly, DNA was used to create a library for Illumina sequencing, specifically focusing on the exome (coding regions of genes). The library was enriched for exonic regions using a specific kit. Finally, the captured libraries were sequenced using a NextSeq2000 instrument. The sequencing data for single-nucleotide variants and insertions/deletions (indels) were analyzed. The variants were filtered based on the following criteria: (1) Location within exons or nearby introns of GSD-related genes[4]; (2) Nonsynonymous changes (altering the protein sequence); (3) Rarity in the Genome Aggregation Database (GnomAD); (4) Low frequency in Thai exome controls; (5) (for missense variants) Predicted to be damaging by SIFT and Polyphen protein prediction tools, and (6) Relevance to the symptoms of hepatic GSDs.
As mentioned, the bioinformatic pipeline employed for WES data analysis involved a multi-step filtering process to identify potentially pathogenic variants. All single nucleotide variants and indels were initially filtered based on their location within exons or flanking introns of genes relevant to GSD. Only non-synonymous variants were considered, and those with a minor allele frequency less than 1% in the 1000 Genomes Project and 0.1% in the GnomAD were retained. Additionally, variants were excluded if they were identified in more than 10 alleles in a control cohort of 5432 Thai exomes. For missense variants, predictions from SIFT and Polyphen were used to assess their potential impact on protein function. Finally, variants were prioritized if they were associated with the patients’ phenotype or known to be involved in the disease under investigation. This rigorous filtering approach ensured the identification of the most likely pathogenic variants for further validation.
Continuous and categorical data were presented as median values, interquartile ranges, and proportions or percentages. Fisher’s exact test was used to compare discrete data, and the Mann–Whitney U test was used to analyze continuous data. A P-value of < 0.05 indicated significance. STATA version 18 was used for all statistical analyses. The figures were made using GraphPad Prism version 9.2.0.
A total of eight patients who were diagnosed with GSDs were recruited. Molecular analysis confirmed the diagnosis of GSDs and subtype identification. The median age at presentation was 2.18 (1.57–3.05) years, and 37.5% were male. However, the median time of diagnosis and molecular analysis were 1.56 (0.9–2.4) and 3.41 (2.78–6.49) years from the presentation, respectively. Two patients (25%) were from families of consanguineous couples. Based on the results of the molecular analysis, one patient each was classified into subtypes Ia (12.5%) and IX (12.5%), and three patients were classified into subtypes III (37.5%) and VI (37.5%) (Table 1). Ten variants were identified in G6PC (n = 1), AGL (n = 4), PYGL (n = 5), and PHKA2 (n = 1). The compound heterozygous c.1611+1G>C and c.1735G>A variants in AGL identified in patient 4 were novel (Table 1). A comprehensive comparison of two novel AGL variants, including their allele frequencies in control databases, potential functional effects, and American College of Medical Genetics and Genomics (ACMG) classifications are demonstrated in Table 2[8]. Remarkably, the known c.2467C>T (p.Gln823Ter) mutation in PYGL[9] appears to be a potential hotspot, present in all three patients with PYGL mutations.
| Patient number | Gene | WES | Variant | Amino acid | Exon/ | Zygosity | Inheriting parent of allele1 | Transcript | hg19 coordinate | Ref. |
| GSD Ia | ||||||||||
| 5 | G6PC | Duo | c.648G>T | p.Leu216Leu | Exon 5 | Homozygous or Hemizygous2 | Mother | NM_000151.4 | chr17:41063017 G>T | [21] |
| GSD III | ||||||||||
| 3 | AGL | Trio | c.2578delG | p.Val860LeufsTer8 | Exon 20 | Homozygous | Father and mother | NM_000642.3 | chr1:100350156 delG | Clinvar |
| 4 | AGL | Singleton | c.1611+1G>C | N/A | Intron 12 | Heterozygous | Unknown | NM_000642.3 | chr1:100343385 G>C | Novel4 |
| c.1735G>A | p.Glu579Lys | Exon 13 | Heterozygous | Unknown | NM_000642.3 | chr1:100345602 G>A | Novel4 | |||
| 6 | AGL | Trio | c.1735+1G>T | N/A | Intron 13 | Homozygous | Father and mother | NM_000642.3 | chr1:100345603 G>T | [22] |
| GSD VI | ||||||||||
| 2 | PYGL | Trio | c.514C>T | p.Arg172Ter | Exon 4 | Heterozygous | Father | NM_002863.5 | chr14:51398405 G>A | [9] |
| c.2467C>T | p.Gln823Ter | Exon 20 | Heterozygous | Mother | NM_002863.5 | chr14:51372187 G>A | [9] | |||
| 7 | PYGL | Singleton | c.2467C>T | p.Gln823Ter | Exon 20 | Heterozygous | Unknown | NM_002863.5 | chr14:51372187 G>A | [9] |
| c.932G>A | p.Arg311His | Exon 8 | Heterozygous | Unknown | NM_002863.5 | chr14:51383747 C>T | [23] | |||
| 8 | PYGL | Trio | c.1726C>T | p.Arg576Ter | Exon 14 | Heterozygous | Father | NM_002863.5 | chr14:51378916 G>A | [24]5 |
| c.2467C>T | p.Gln823Ter | Exon 20 | Heterozygous | Mother | NM_002863.5 | chr14:51372187 G>A | [9] | |||
| GSD IX | ||||||||||
| 1 | PHKA2 | Trio | c.2746C>T | p.Arg916Trp | Exon 25 | hemizygous | Mother | NM_000292.3 | chrX:18924673 G>A | [25] |
| Variant | GRCh37/hg19 gnomAD | GRCh38/hg38 gnomAD | 5432 Thai exomes | Potential functional impact | Reported in other GSD cases | ACMG classification |
| c.1611+1G>C (p.?) | Not identified | Not identified | Not identified | Potentially disruptive to splicing due to its location at the splice donor site. May lead to aberrant mRNA processing and protein expression | Not reported | Likely pathogenic (PVS1, PM2) |
| c.1735G>A (p.Glu579Lys) | Not identified | Identified in 1 out of 1461034 alleles | Not identified | Missense mutation resulting in a Glu579Lys amino acid change within the transferase catalytic domain. May affect enzyme activity or protein stability | Not reported | Variant of uncertain significance (PM2, PP3) |
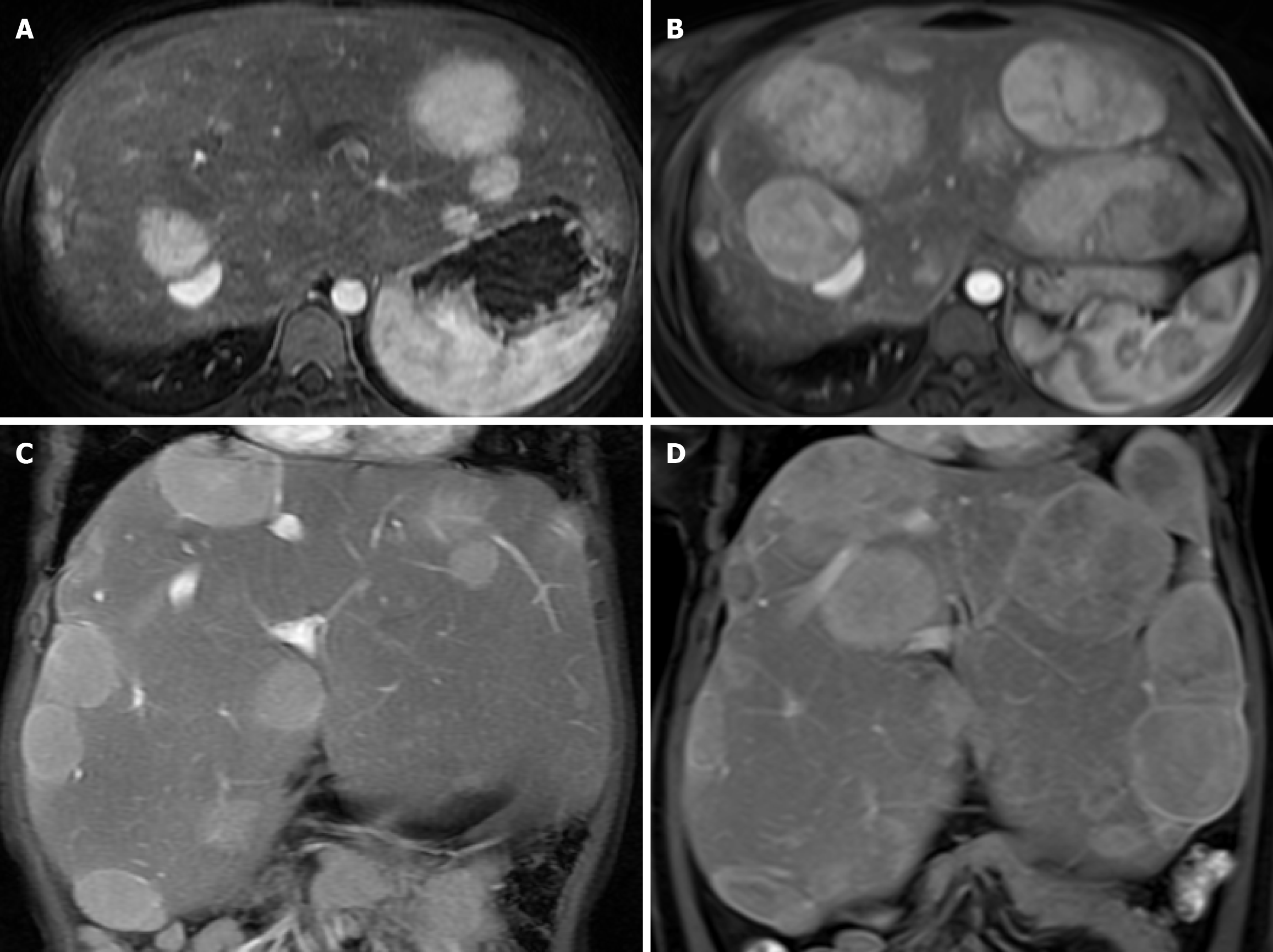
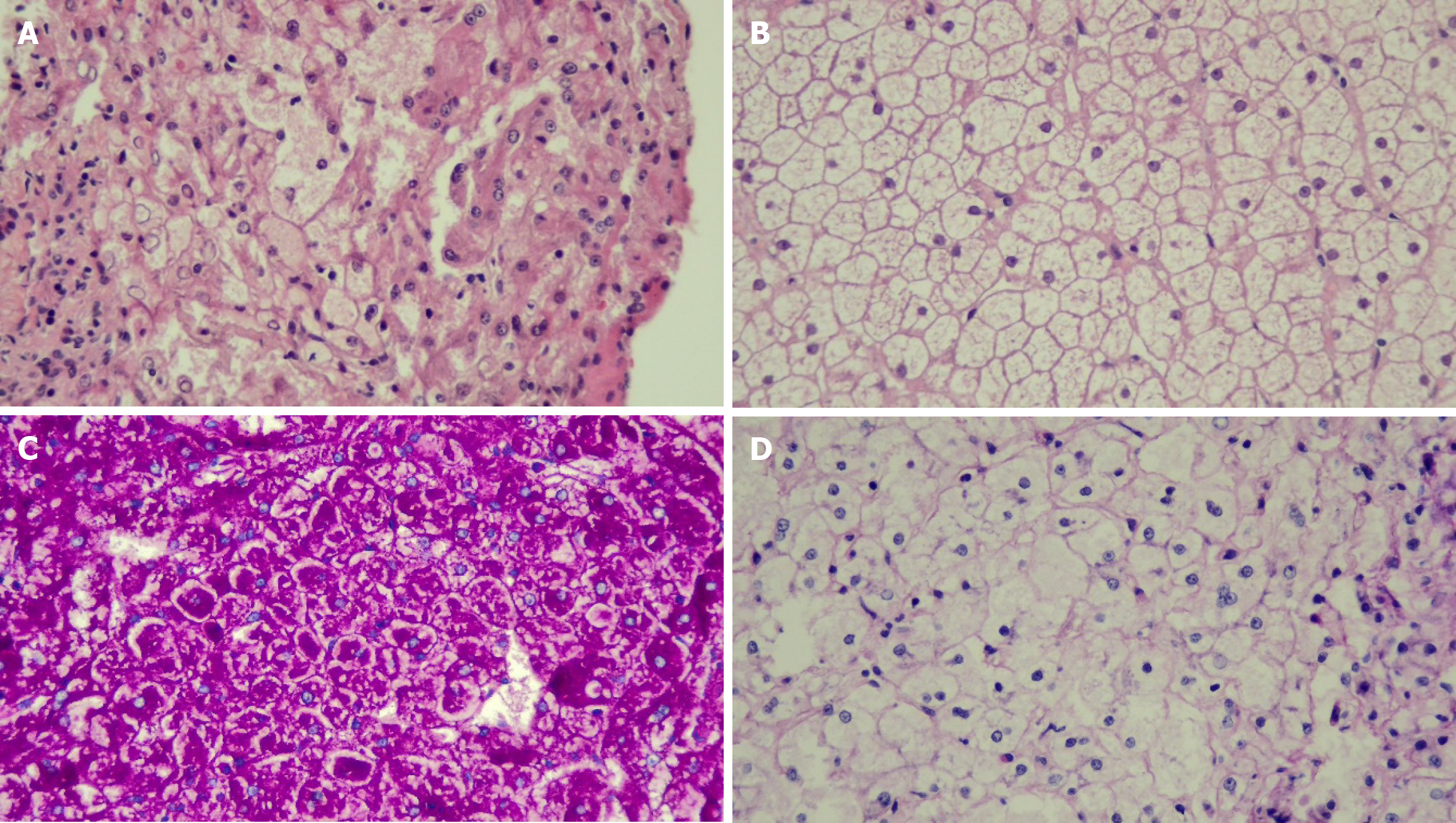
The first manifestations were abdominal distension from hepatomegaly (100%), doll-like facies (37.5%), wasting (25%), overweight (12.5%), stunting (62.5%), gastrointestinal bleeding (12.5%), epistaxis (12.5%), and jaundice (12.5%). Biochemical profiles at presentation were hypoglycemia (100%), transaminitis (100%), hyperuricemia (50%), anemia (25%), hypercholesterolemia (62.5%), hypertriglyceridemia (100%), and direct hyperbilirubinemia (50%) (Table 3). All patients had normal renal function. Liver ultrasonography revealed hepatomegaly with diffuse increased parenchyma echogenicity in all patients. One patient had multiple liver tumors, with magnetic resonance imaging revealing the feature of hepatic adenomatosis (Figure 1). Liver pathology exhibited hepatocyte ballooning with positive periodic acid-Schiff staining and diastase sensitivity in all patients (Figure 2).
| Patient number | Sex | Age at clinical Dx (year) | Time from clinical Dx to molecular Dx (year) | First presentation | Growth and nutritional status | Biochemical result | Follow-up time (year) | Clinical and biological profiles at the last follow-up | |||||||||||
| AST (U/L) | ALT (U/L) | BS (mg/dL) | TC (mg/dL) | TG (mg/dL) | Uric Acid (mg/dL) | Growth and nutritional status | AST (U/L) | ALT(U/L) | BS (mg/dL) | TC (mg/dL) | TG (mg/dL) | Uric Acid (mg/dL) | |||||||
| GSD I | |||||||||||||||||||
| 5 | F | 14.7 | 12.52 | Hepatomegaly, recurrent epistaxis | Wasting/ stunting | 118 | 67 | 53 | 210 | 376 | 8.6 | 13.8 | Stunting | 33 | 28 | 58 | 179 | 396 | 9.7 |
| GSD III | |||||||||||||||||||
| 3 | F | 3.02 | 8.49 | Hepatomegaly | Normal | 4302 | 1341 | 48 | 241 | 622 | 5.1 | 11.5 | Overweight | 510 | 375 | 72 | 205 | 124 | 8 |
| 4 | F | 1.43 | 12.22 | Hepatomegaly | Normal | 629 | 547 | 20 | 138 | 119 | 3.3 | 12.3 | Normal | 33 | 28 | 78 | 137 | 63 | 5.8 |
| 6 | M | 2.62 | 3.06 | Hepatomegaly, doll-like facies | Stunting | 1878 | 854 | 30 | 180 | 231 | 6.1 | 3.1 | Normal | 626 | 408 | 35 | 184 | 203 | 6.8 |
| GSD VI | |||||||||||||||||||
| 2 | F | 1.75 | 9.24 | Hepatomegaly | Wasting | 746 | 736 | 56 | 305 | 335 | 6.9 | 12.5 | Normal | 15 | 14 | 83 | 137 | 89 | 6.1 |
| 7 | F | 3.08 | 2.08 | Hepatomegaly | Stunting | 72 | 64 | 92 | 256 | 189 | 4.3 | 2.1 | Normal | 55 | 40 | 72 | 226 | 104 | 5.9 |
| 8 | M | 1.35 | 1.10 | Hepatomegaly, doll-like facies | Overweight/ stunting | 920 | 665 | 55 | 246 | 303 | 3.9 | 1.8 | Overweight | 72 | 90 | 86 | 170 | 100 | 5.4 |
| GSD IX | |||||||||||||||||||
| 1 | M | 1.7 | 2.75 | Hepatomegaly, doll-like facies | Stunting | 1531 | 439 | 34 | 145 | 159 | 5.3 | 6.48 | Overweight/stunting | 161 | 78 | 84 | 145 | 97 | 5.2 |
Because genetic analysis was unavailable at the time of diagnosis and the diagnosis of GSD was initially based on clinical presentations and liver histopathology, the mainstay of treatment was to normalize blood sugar and other biochemical profiles. Cornstarch supplementation was advocated in all patients, given 1–7.2 g/kg/day twice to five times a day depending on the result of random blood glucose, in conjunction with a high-protein diet (1.5–3 g/kg/day) and low-lactose–fructose ingestion. One of them (patient 5, GSD type I) had hyperuricemia that required allopurinol (Table 3). All patients had regular follow-up appointments every 3–6 months. Nearly all patients demonstrated good compliance with the uncooked cornstarch prescription, except for patients 1 (GSD type I), 2 (GSD type VI), and 5 (GSD type XI). Despite poor compliance, patient 2 (GSD type VI) had normal liver enzyme levels, lipid profiles, and growth at the last follow-up. By contrast, patients 1 (GSD type I) and 5 (GSD type XI) had better biochemical profiles but exhibited stunted growth from diagnosis through the last follow-up. Patients 3, 4, and 6 (GSD type III) experienced frequent hypoglycemia, requiring cornstarch supplementation of up to 7.2 g/kg/day (patient 3) and up to five times a day. Despite good compliance, patients 3 and 6 had persistent transaminitis and dyslipidemia, and patient 3 was also overweight.
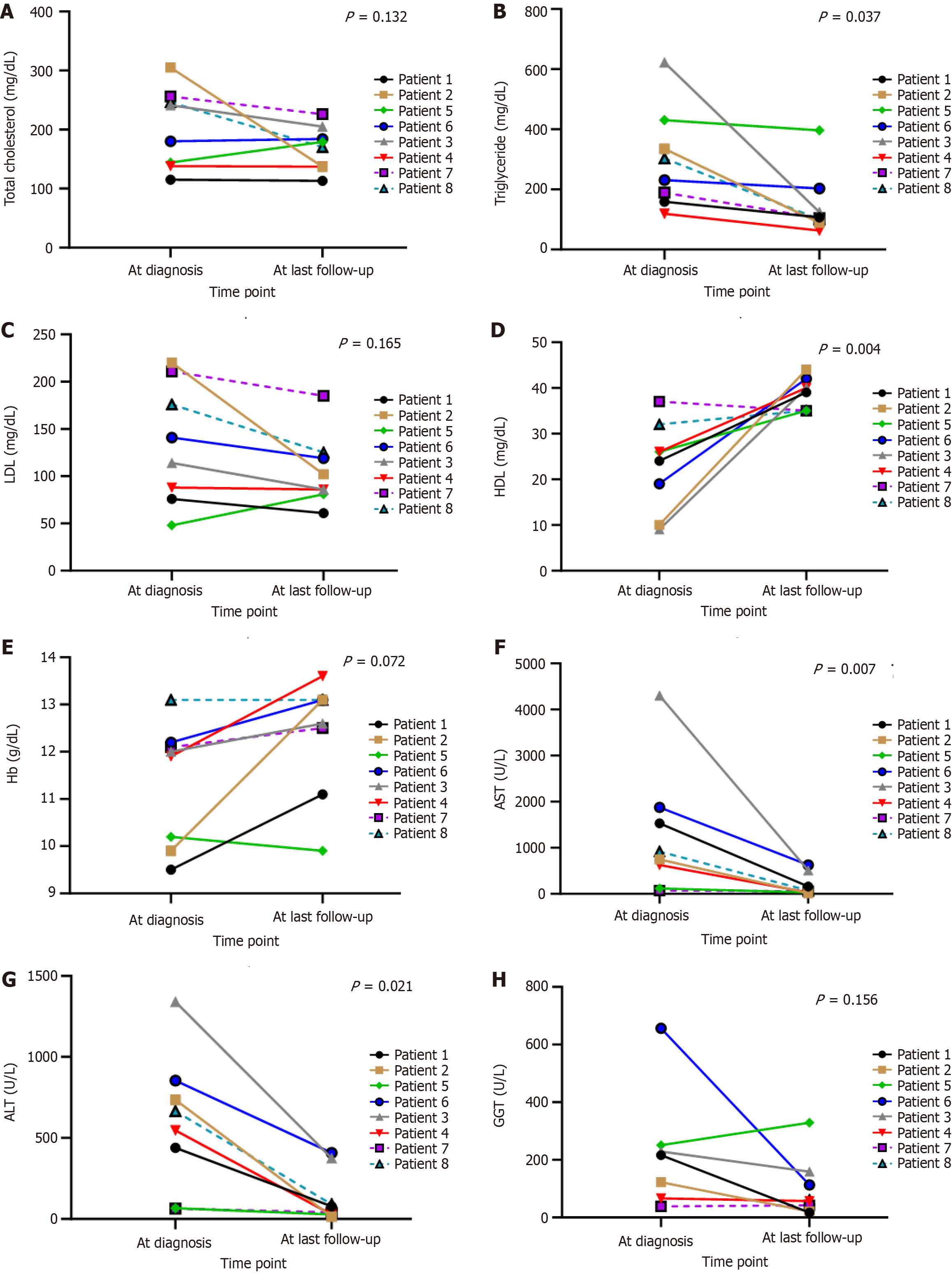
All eight patients remain alive, with a median follow-up period of 9.59 (1.20–10.56) years. For the outcome at the end of follow-up, patients 1 and 5 still had stunting (25%), but none had wasting. However, patients 1, 3, and 8 were overweight, with BMI Z-score 1.2 SD (37.5%). Liver function, blood sugar, and lipid profiles improved in all patients. Transaminitis turned to normal in 3 (37.5%) patients. Blood sugar improved in all patients; however, two of them (25%) still had hypoglycemia with prolonged fasting time (patients 3 and 6; GSD type III). Patients 2 and 4 diagnosed with GSD types III and VI could omit uncooked corn starch before bedtime as determined by their genetic subtype analysis without hypoglycemia from their random blood glucose. Hypercholesterolemia was observed in 2 (25%) patients, and hypertriglyceridemia was noted in 2 (25%) patients. Hyperuricemia was found in 4 (50%) patients (Table 4 and Figure 3). One patient with GSD type I had increasing size of the hepatic adenomatosis but normal liver enzyme, requiring liver transplantation (waiting list) (Figure 1). For other comorbidities, none had hypertension, chronic kidney disease, delayed motor development, seizure due to hypoglycemia, or hypertrophic cardiomyopathy.
| Blood test | At the diagnosis | At the last follow-up | P value
|
| Hb (mg/dL) | 11.9 (10.1-12.2) | 12.9 (11.8-13.1) | 0.072 |
| WBC | 11140 (9975-14020) | 9415 (8600-10690) | 0.046 |
| Platelet | 386000 (325500-457500) | 377500 (327000-448000) | 1 |
| Blood sugar (mg/dL) | 55 (30-87) | 72 (58-83.5) | 0.248 |
| Total cholesterol | 241 (138-256) | 174.5 (137-194.5) | 0.132 |
| Triglyceride (mg/dL) | 231 (159-335) | 105.5 (94.5-163.5) | 0.037 |
| LDL (mg/dL) | 141 (88-211) | 94 (83.5-122) | 0.165 |
| HDL (mg/dL) | 24 (10-32) | 39.5 (35-41) | 0.004 |
| Uric acid (mg/dL) | 5.1 (3.9-6.5) | 6 (5.6-7.55) | 0.148 |
| ALT (U/L) | 665 (67-854) | 59 (28-232.5) | 0.021 |
| AST (U/L) | 833 (374-1704) | 63.5 (33-335.5) | 0.007 |
| Total bilirubin (mg/dL) | 0.39 (0.24-0.99) | 0.53 (0.43-0.70) | 0.462 |
| Direct bilirubin (mg/dL) | 0.22 (0.10-0.41) | 0.22 (0.16-0.28) | 1 |
| Globulin (mg/dL) | 3.1 (2.8-3.4) | 3.4 (2.85-4.05) | 0.354 |
| Albumin (mg/dL) | 4.3 (4.0-4.5) | 4.4 (4.25-4.4) | 0.335 |
| GGT | 169 (66.2-251) | 59 (31.5-136) | 0.156 |
| Blood urea nitrogen | 12 (8-14) | 10.5 (9.5-13.5) | 0.771 |
| Creatinine | 0.25 (0.21-0.4) | 0.41 (0.26-0.55) | 0.072 |
To the best of our knowledge, this study is the first to demonstrate the clinical and molecular characteristics of hepatic GSDs in Thai children. The most common hepatic GSDs in this study are types III and VI. The known c.2467C>T (p.Gln823Ter) mutation in PYGL appears to be a potential hotspot, present in all three individuals with GSD type VI. All patients presented with hepatomegaly, hypoglycemia, hypercholesterolemia and transaminitis. Liver histopathology was used to confirm the diagnosis with high accuracy. The improvement in lipid profiles, growth parameters, and liver enzymes in most patients proposed that nutritional management had yielded favorable long-term outcomes. In addition, molecular analysis successfully classified GSD subtypes and identified two novel AGL variants, expanding the mutational spectrum.
In the present study, the hallmarks of GSDs included the triad of hepatomegaly, transaminitis, and hypoglycemia that were consistent with the studies in India and Pakistan by Kumar et al[6] (n = 57) and Ahmed et al[7] (n = 55), in which all of them had marked abdominal distension probably from hepatomegaly. Liang et al[4] studied in Chinese children (n = 49), and all of them had the triad, and Beyzaei et al[10] (n = 14) studied in Iranian cases of GSDs, and all of them had hepatomegaly and transaminitis. As a result, the triad is useful to narrow the diagnosis that should be confirmed using liver biopsy or molecular analysis. In regions where molecular analysis is inaccessible, liver biopsy might be considered for timely management. However, guidelines for the specific management according to hepatic GSD subtypes have been established for hepatic GSD types I[11], III[12], IV[13], VI[14], and IX[14] that help physicians target or personalize management. In addition, long-term complications appear to differ among types, for example, hepatic adenoma can occur in GSD types I, III, and IV[15]. Thus, monitoring complications of GSD subtypes is cost-benefit and saves time leading to a better quality of life of patients. Many mimic diseases from GSDs should be considered, such as congenital disorder of glycosylation, lysosomal disorders, and mitochondrial disorders[16-18]. Consequently, in centers with limited resources, molecular analysis must be subsequently performed by sending specimens to facilities capable of confirming the diagnosis and classifying subtypes for tailored treatment[19]. In addition, understanding patients’ genetic background is beneficial for counseling and family planning.
In this study, most Thai children with GSDs had types III and VI, in contrast with findings from China[2-4,8], Iran[10], India[5,6], Parkistan[7], Spain[19], and Italy[20] where GSD type VI is relatively rare. GSD type III, common in many countries, is associated with recurrent hypoglycemic seizures, leading to delayed development. However, in this study, Thai children with GSD type III did not experience seizures or demonstrate significant developmental delays. Notably, one child with GSD type III (with a novel causative variant) achieved good clinical and biochemical outcomes, and uncooked cornstarch supplementation was stopped during adolescence. For GSD type IX, our patient exhibited improved biochemical profiles but continued to experience growth retardation at age 9, consistent with reports from the ACMG[14]. This finding contrasts with that of a larger study in China where nearly all patients with GSD type IXa (n = 17) had mild disorders and favorable outcomes, often achieving normal growth by adolescence without cornstarch supplementation[2]. Patients with GSD type VI, although less reported in Asia, generally have better outcomes than those with GSD types I, III, and XI. Clinical symptoms and biochemical profiles often improve into adulthood with specific nutritional therapy, which helps preserve growth, improve glycemic control, and prevent liver complications. In this study, three children with GSD type VI had normal growth, and two had normal liver enzymes and lipid profiles after extensive nutritional management. One child, who had poor compliance with uncooked cornstarch but adhered to a high-protein diet, also achieved good clinical and biochemical outcomes, indicating that tapering treatment for GSD type VI may be reasonable. In addition, the known c.2467C>T (p.Gln823Ter) mutation in PYGL appears to be a potential hotspot, present in all three individuals with GSD type VI, and could be considered the screening mutation for patients suspected of GSDs in Thailand.
Nutritional management is the mainstay of treatment, particularly cornstarch supplementation for GSD type I because of frequent fasting hypoglycemia. However, cornstarch might not be necessary if patients with GSD type III reach adolescence[12], and only bedtime cornstarch might be considered in GSD types VI and IX. In addition to nutritional management, a comprehensive, long-term care approach involving the surveillance of disease sequelae based on GSD subtypes is recommended. Therefore, the precise diagnosis of GSD subtypes can enhance the quality of life, not only by enabling more targeted nutritional management but also by addressing the potential involvement of other organs[12,13].
In this, all patients received similar nutritional management and long-term surveillance with multidisciplinary teams in all aspects of growth, metabolic, cardiac, and liver complications. The treatment outcome was favorable, with nearly normal growth and significant improvement in the biochemical profiles of lipid and liver enzymes. However, after genetic analysis, two of our patients were diagnosed with GSD types III and VI and experienced frequent hypoglycemia during their toddler years. As they grow (aged 11 and 14 years), hypoglycemia subsequently improves, allowing them to discontinue both daytime and nighttime cornstarch supplementation, with only occasional home blood sugar monitoring. These findings highlight the genetic heterogeneity of GSDs, affirming the use of WES in the diagnosis and clinical decision-making. A high-protein diet is also advocated in patients with GSD types VI and XI as a protective effect against overweight/obesity and insulin resistance[20]. This dietary approach was advised for all patients with GSD in this study, with favorable outcomes; however, its generalizability across all GSD types warrants further study.
This study has several limitations, primarily due to the small sample size, which resulted in low statistical power and limitations in the generalizability of the findings to all patients with GSDs. A multicenter study focusing on Thai children with GSDs could provide valuable insights, particularly if it includes investigations into genotype–phenotype correlations, explores novel therapeutic approaches, and evaluates long-term outcomes, particularly given the increased availability of genetic analysis in recent times. In addition, hypoglycemia, a hallmark of the disease, may be present from birth and can go undetected, potentially affecting child development—a factor not deeply explored in this study. Finally, the age at diagnosis and treatment duration among patients may confound the observed improvements in patients’ growth and biochemical profiles.
The findings of this study indicate that key features of liver GSDs encompass hepatomegaly, transaminitis, and hypoglycemia. Stunting or growth retardation is commonly observed. Although a liver biopsy can confirm the diagnosis, it is unable to identify the specific GSD subtype. Therefore, molecular diagnosis using WES plays a crucial role in determining the GSD subtype, facilitating tailored treatment for patient subgroups. After the treatment of our patients, notable enhancements were observed in blood sugar levels, aspartate transaminase, alanine transaminase, and lipid profiles. Moreover, height and BMI improved after treatment. This underscores the significance of effective nutritional management in optimizing patient outcomes.
| 1. | Weinstein DA, Steuerwald U, De Souza CFM, Derks TGJ. Inborn Errors of Metabolism with Hypoglycemia: Glycogen Storage Diseases and Inherited Disorders of Gluconeogenesis. Pediatr Clin North Am. 2018;65:247-265. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 52] [Article Influence: 7.4] [Reference Citation Analysis (0)] |
| 2. | Zhang J, Yuan Y, Ma M, Liu Y, Zhang W, Yao F, Qiu Z. Clinical and genetic characteristics of 17 Chinese patients with glycogen storage disease type IXa. Gene. 2017;627:149-156. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Dong R, Wei X, Zhang K, Song F, Lv Y, Gao M, Wang D, Ma J, Gai Z, Liu Y. Genotypic and phenotypic characteristics of 12 chinese children with glycogen storage diseases. Front Genet. 2022;13:932760. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 4. | Liang Y, Du C, Wei H, Zhang C, Zhang M, Hu M, Fang F, Luo X. Genotypic and clinical analysis of 49 Chinese children with hepatic glycogen storage diseases. Mol Genet Genomic Med. 2020;8:e1444. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 20] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 5. | Poojari V, Shah I, Shetty NS, Mirani S, Tolani D. Clinical profile and outcome of glycogen storage disease in Indian children. Trop Doct. 2021;51:189-192. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 6. | Kumar TV, Bhat M, Narayanachar SG, Narayan V, Srikanth AK, Anikar S, Shetty S. Molecular and clinical profiling in a large cohort of Asian Indians with glycogen storage disorders. PLoS One. 2022;17:e0270373. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 7. | Ahmed S, Akbar F, Ali AJ, Afroze B. Clinical, pathological and molecular spectrum of patients with glycogen storage diseases in Pakistan. J Pediatr Endocrinol Metab. 2022;35:373-385. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 8. | Yu T, Fu H, Yang A, Liang Y. Clinical and Functional Characterization of Novel AGL Variants in Two Families with Glycogen Storage Disease Type III. Int J Endocrinol. 2023;2023:6679871. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 9. | Liu B, Wu B, Lu Y, Zhang P, Xiao F, Li G, Wang H, Dong X, Liu R, Li Y, Xie X, Zhou W, Wang J, Lu Y. A Novel, Recurrent, 3.6-kb Deletion in the PYGL Gene Contributes to Glycogen Storage Disease Type VI. J Mol Diagn. 2020;22:1373-1382. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 2] [Cited by in RCA: 2] [Article Influence: 0.4] [Reference Citation Analysis (0)] |
| 10. | Beyzaei Z, Ezgu F, Geramizadeh B, Imanieh MH, Haghighat M, Dehghani SM, Honar N, Zahmatkeshan M, Jassbi A, Mahboubifar M, Alborzi A. Clinical and genetic spectrum of glycogen storage disease in Iranian population using targeted gene sequencing. Sci Rep. 2021;11:7040. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 17] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 11. | Kishnani PS, Austin SL, Abdenur JE, Arn P, Bali DS, Boney A, Chung WK, Dagli AI, Dale D, Koeberl D, Somers MJ, Wechsler SB, Weinstein DA, Wolfsdorf JI, Watson MS; American College of Medical Genetics and Genomics. Diagnosis and management of glycogen storage disease type I: a practice guideline of the American College of Medical Genetics and Genomics. Genet Med. 2014;16:e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 216] [Cited by in RCA: 298] [Article Influence: 29.8] [Reference Citation Analysis (0)] |
| 12. | Kishnani PS, Austin SL, Arn P, Bali DS, Boney A, Case LE, Chung WK, Desai DM, El-Gharbawy A, Haller R, Smit GP, Smith AD, Hobson-Webb LD, Wechsler SB, Weinstein DA, Watson MS; ACMG. Glycogen storage disease type III diagnosis and management guidelines. Genet Med. 2010;12:446-463. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 167] [Cited by in RCA: 177] [Article Influence: 11.8] [Reference Citation Analysis (0)] |
| 13. | Koch RL, Soler-Alfonso C, Kiely BT, Asai A, Smith AL, Bali DS, Kang PB, Landstrom AP, Akman HO, Burrow TA, Orthmann-Murphy JL, Goldman DS, Pendyal S, El-Gharbawy AH, Austin SL, Case LE, Schiffmann R, Hirano M, Kishnani PS. Diagnosis and management of glycogen storage disease type IV, including adult polyglucosan body disease: A clinical practice resource. Mol Genet Metab. 2023;138:107525. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 24] [Reference Citation Analysis (0)] |
| 14. | Kishnani PS, Goldstein J, Austin SL, Arn P, Bachrach B, Bali DS, Chung WK, El-Gharbawy A, Brown LM, Kahler S, Pendyal S, Ross KM, Tsilianidis L, Weinstein DA, Watson MS; ACMG Work Group on Diagnosis and Management of Glycogen Storage Diseases Type VI and IX. Diagnosis and management of glycogen storage diseases type VI and IX: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG). Genet Med. 2019;21:772-789. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 57] [Cited by in RCA: 81] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 15. | Sintusek P, Phewplung T, Sanpavat A, Poovorawan Y. Liver tumors in children with chronic liver diseases. World J Gastrointest Oncol. 2021;13:1680-1695. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 6] [Cited by in RCA: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 16. | Jones MA, Bhide S, Chin E, Ng BG, Rhodenizer D, Zhang VW, Sun JJ, Tanner A, Freeze HH, Hegde MR. Targeted polymerase chain reaction-based enrichment and next generation sequencing for diagnostic testing of congenital disorders of glycosylation. Genet Med. 2011;13:921-932. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 64] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 17. | DaRe JT, Vasta V, Penn J, Tran NT, Hahn SH. Targeted exome sequencing for mitochondrial disorders reveals high genetic heterogeneity. BMC Med Genet. 2013;14:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 51] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 18. | Fernández-Marmiesse A, Morey M, Pineda M, Eiris J, Couce ML, Castro-Gago M, Fraga JM, Lacerda L, Gouveia S, Pérez-Poyato MS, Armstrong J, Castiñeiras D, Cocho JA. Assessment of a targeted resequencing assay as a support tool in the diagnosis of lysosomal storage disorders. Orphanet J Rare Dis. 2014;9:59. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 36] [Cited by in RCA: 39] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 19. | Vega AI, Medrano C, Navarrete R, Desviat LR, Merinero B, Rodríguez-Pombo P, Vitoria I, Ugarte M, Pérez-Cerdá C, Pérez B. Molecular diagnosis of glycogen storage disease and disorders with overlapping clinical symptoms by massive parallel sequencing. Genet Med. 2016;18:1037-1043. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 25] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 20. | Tagliaferri F, Massese M, Russo L, Commone A, Gasperini S, Pretese R, Dionisi-Vici C, Maiorana A. Hepatic glycogen storage diseases type 0, VI and IX: description of an italian cohort. Orphanet J Rare Dis. 2022;17:285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 6] [Reference Citation Analysis (0)] |
| 21. | Zheng BX, Lin Q, Li M, Jin Y. Three novel mutations of the G6PC gene identified in Chinese patients with glycogen storage disease type Ia. Eur J Pediatr. 2015;174:59-63. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 9] [Article Influence: 0.9] [Reference Citation Analysis (0)] |
| 22. | Perveen S, Gupta N, Kumar M, Kaur P, Chowdhury MR, Kabra M. Spectrum of amyloglucosidase mutations in Asian Indian patients with Glycogen storage disease type III. Am J Med Genet A. 2020;182:1190-1200. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 23. | Giannakis M, Mu XJ, Shukla SA, Qian ZR, Cohen O, Nishihara R, Bahl S, Cao Y, Amin-Mansour A, Yamauchi M, Sukawa Y, Stewart C, Rosenberg M, Mima K, Inamura K, Nosho K, Nowak JA, Lawrence MS, Giovannucci EL, Chan AT, Ng K, Meyerhardt JA, Van Allen EM, Getz G, Gabriel SB, Lander ES, Wu CJ, Fuchs CS, Ogino S, Garraway LA. Genomic Correlates of Immune-Cell Infiltrates in Colorectal Carcinoma. Cell Rep. 2016;17:1206. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 148] [Cited by in RCA: 199] [Article Influence: 22.1] [Reference Citation Analysis (0)] |
| 24. | Luo X, Duan Y, Fang D, Sun Y, Xiao B, Zhang H, Han L, Liang L, Gong Z, Gu X, Yu Y, Qiu W. Diagnosis and follow-up of glycogen storage disease (GSD) type VI from the largest GSD center in China. Hum Mutat. 2022;43:557-567. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 25. | Beauchamp NJ, Dalton A, Ramaswami U, Niinikoski H, Mention K, Kenny P, Kolho KL, Raiman J, Walter J, Treacy E, Tanner S, Sharrard M. Glycogen storage disease type IX: High variability in clinical phenotype. Mol Genet Metab. 2007;92:88-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 93] [Cited by in RCA: 87] [Article Influence: 4.8] [Reference Citation Analysis (0)] |