Published online Jun 9, 2024. doi: 10.5409/wjcp.v13.i2.92737
Revised: April 23, 2024
Accepted: May 6, 2024
Published online: June 9, 2024
Processing time: 124 Days and 3.1 Hours
Autism spectrum disorder (ASD) is a neurodevelopmental condition characterized by deficits in social communication and repetitive behaviors. Metabolomic profiling has emerged as a valuable tool for understanding the underlying metabolic dysregulations associated with ASD.
To comprehensively explore metabolomic changes in children with ASD, integrating findings from various research articles, reviews, systematic reviews, meta-analyses, case reports, editorials, and a book chapter.
A systematic search was conducted in electronic databases, including PubMed, PubMed Central, Cochrane Library, Embase, Web of Science, CINAHL, Scopus, LISA, and NLM catalog up until January 2024. Inclusion criteria encompassed research articles (83), review articles (145), meta-analyses (6), systematic reviews (6), case reports (2), editorials (2), and a book chapter (1) related to metabolomic changes in children with ASD. Exclusion criteria were applied to ensure the relevance and quality of included studies.
The systematic review identified specific metabolites and metabolic pathways showing consistent differences in children with ASD compared to typically developing individuals. These metabolic biomarkers may serve as objective measures to support clinical assessments, improve diagnostic accuracy, and inform personalized treatment approaches. Metabolomic profiling also offers insights into the metabolic alterations associated with comorbid conditions commonly observed in individuals with ASD.
Integration of metabolomic changes in children with ASD holds promise for enhancing diagnostic accuracy, guiding personalized treatment approaches, monitoring treatment response, and improving outcomes. Further research is needed to validate findings, establish standardized protocols, and overcome technical challenges in metabolomic analysis. By advancing our understanding of metabolic dysregulations in ASD, clinicians can improve the lives of affected individuals and their families.
Core Tip: This systematic review examines metabolic changes in individuals with autism spectrum disorder (ASD) by integrating various sources of evidence. Through an extensive search, the review explores factors influencing metabolomic changes in children with ASD, such as age, genetics, diet, gut microbiota, and medical interventions. The systematic review identifies common metabolic dysregulations, including abnormalities in energy metabolism, oxidative stress, mitochondrial dysfunction, and neurotransmitter metabolism. Despite limitations, integrating metabolomic changes in ASD holds promise for improving diagnosis and treatment approaches.
- Citation: Al-Beltagi M, Saeed NK, Bediwy AS, Elbeltagi R. Metabolomic changes in children with autism. World J Clin Pediatr 2024; 13(2): 92737
- URL: https://www.wjgnet.com/2219-2808/full/v13/i2/92737.htm
- DOI: https://dx.doi.org/10.5409/wjcp.v13.i2.92737
Metabolites are crucial molecules that are essential to the body's metabolism. These molecules are produced during the breakdown of substances and help create energy and other vital compounds necessary for various cellular functions. Metabolites come in different sizes and involve a variety of biochemical reactions that take place within the cells. Metabolites are also crucial in drug biotransformation and can result from microbial activity, which can have both beneficial and harmful effects on the human body[1]. The metabolome refers to the complete set of small molecules, or metabolites, present within a biological sample (such as a cell, tissue, organ, or organism) at a given time. These include sugars, amino acids, lipids, and other compounds that are fundamental to the body's functioning and can act as signaling molecules, energy sources, building blocks for larger molecules, or waste products that are subsequently eliminated[2]. Metabolomes can change dramatically based on external and internal factors like nutrient availability and medications. Metabolomics is a field of study that involves analyzing the various metabolites present in a biological sample using specialized techniques like mass spectrometry. For example, metabolomics can tell us about antioxidants, their types and quantities, and the factors influencing their trends to maintain good health. Advancements in metabolomics research hold great promise for improving medicine and enhancing human health[3].
Metabolomic changes refer to variations in the levels or patterns of small molecules or metabolites within a biological system, which can occur due to various factors[4]. Different diseases or health conditions can result in distinctive changes in metabolite levels, which can help diagnose or monitor diseases. External factors like diet, exercise, exposure to toxins, or medications can alter metabolite concentrations or profiles[5]. Genetic differences can also influence the body's metabolism, leading to individual metabolomic profile differences. In addition, metabolomic profiles can change during different stages of life, such as during growth, aging, or in response to hormonal changes[6]. Analyzing these changes through metabolomics can provide valuable insights into the biochemical pathways affected by various conditions, potentially aiding in disease diagnosis, understanding biological mechanisms, and developing targeted treatments or interventions[7].
Autism spectrum disorder (ASD) is a complex neurodevelopmental disorder that is characterized by difficulties in social interaction, communication, and repetitive behaviors[8]. Although the exact causes of autism are not fully understood, there is a growing interest in exploring its metabolic aspects to gain insights into its underlying mechanisms and enhance its management. Several studies have reported abnormalities in amino acid metabolism in children with autism. These abnormalities include imbalances in amino acid levels, particularly elevated levels of specific amino acids such as tryptophan and phenylalanine[9]. These changes may relate to disruptions in neurotransmitter synthesis and signaling pathways. Moreover, there are also differences in energy metabolism in children with autism, which may involve altered mitochondrial function. Mitochondria are critical for cell energy production; hence, any changes in their function can have significant consequences[10]. Some metabolomic studies have suggested a connection between the gut microbiome and autism[11].
Changes in metabolites produced by gut bacteria, such as short-chain fatty acids (SCFAs) and certain neurotransmitter-related compounds, have been observed in children with autism[12]. In addition, aberrations in purine and pyrimidine metabolism have been identified in metabolomic analyses of children with autism. These metabolic pathways involve DNA and RNA synthesis and various cellular processes[13].
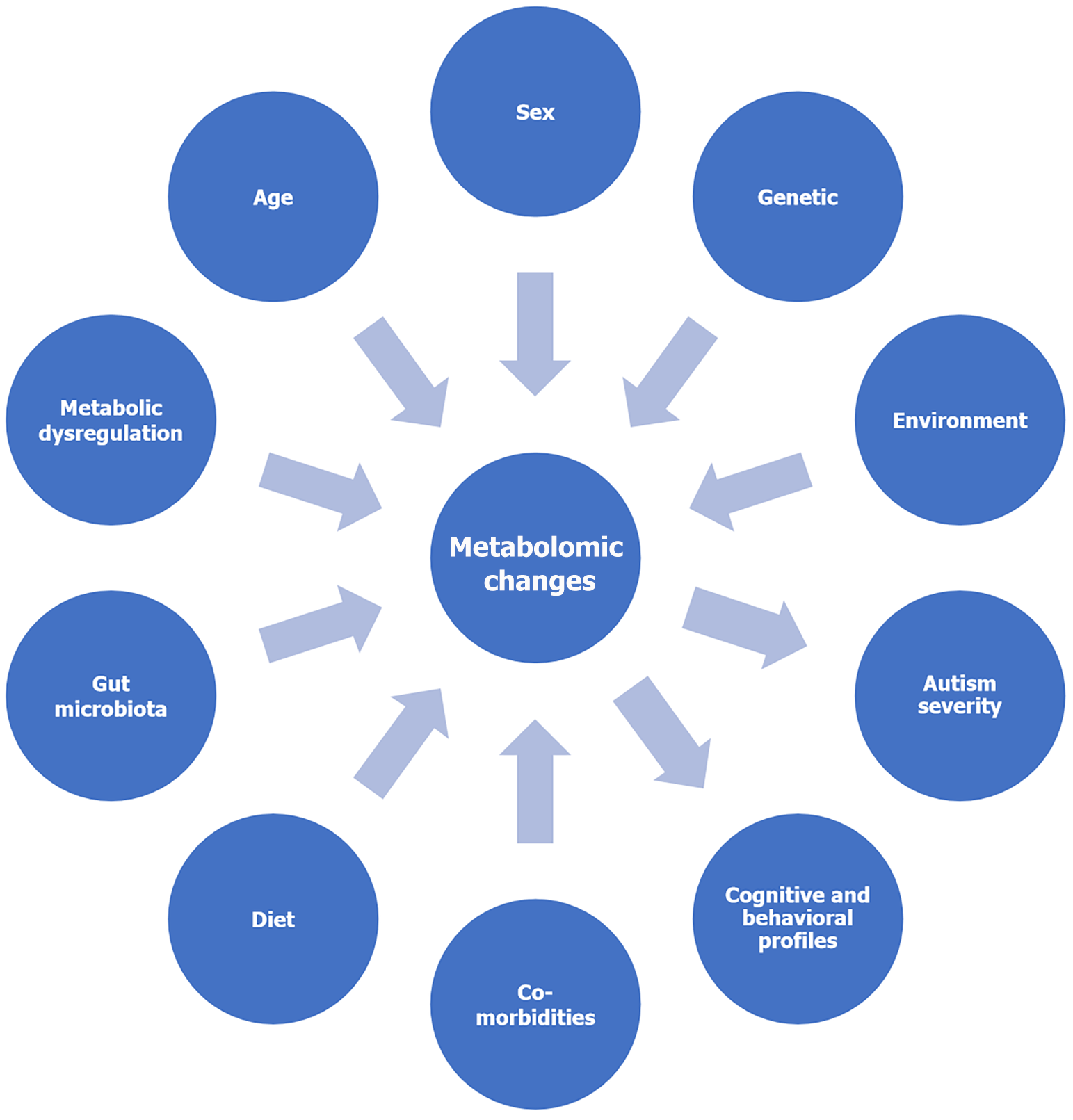
Alterations in lipid metabolism, including changes in phospholipids and fatty acid profiles, have been reported. Lipids are essential components of cell membranes and play crucial roles in brain development and function[14]. Some studies have indicated increased oxidative stress and differences in antioxidant metabolism in children with autism. Oxidative stress refers to an imbalance between the production of reactive oxygen species (ROS) and the body's ability to neutralize them, potentially leading to cellular damage[15]. However, metabolomic findings in children with autism can vary between individuals and may be influenced by factors such as age, sex, genetics, and environmental factors. Additionally, interpreting these findings is complex, and more research is needed to fully understand the significance of these metabolic differences in the context of autism. Metabolomics is just one piece of the puzzle in understanding the intricate mechanisms underlying autism. Integrating metabolomic data with other omics data (genomics, transcriptomics, proteomics) and clinical information can provide a more comprehensive view of the disorder and potentially lead to identifying biomarkers for early diagnosis, subtyping, and personalized interventions[16]. Metabolic changes that occur early in life could help in the early detection of autism in a clinical-stage called pre-autism[17]. This systematic review aims to discuss the various metabolomic changes that can affect the degree of autism and help in diagnosis and management.
We conducted a comprehensive systematic literature review until January 2024 to investigate the changes in metabolites found in children with ASD. Our search included research articles, review articles, meta-analyses, systematic reviews, case reports, editorials, and a book chapter published up until January 2024, written in English and focusing on metabolomic alterations in children with ASD. We searched across electronic databases such as PubMed, PubMed Central, Cochrane Library, Embase, Web of Science, CINAHL, Scopus, LISA, and NLM catalog using relevant keywords related to ASD, metabolomics, metabolic dysregulation, amino acids, lipids, oxidative stress, gut microbiota, and children. After screening based on titles, abstracts, and full-text reviews, we extracted data from selected articles, including study design, sample characteristics, metabolomic techniques, key findings, and implications. Exclusion criteria were applied to ensure the relevance and quality of included studies. We excluded articles not written in English, studies not focusing on metabolomic changes in children with ASD, studies not accessible through electronic databases, or not available in full-text format.
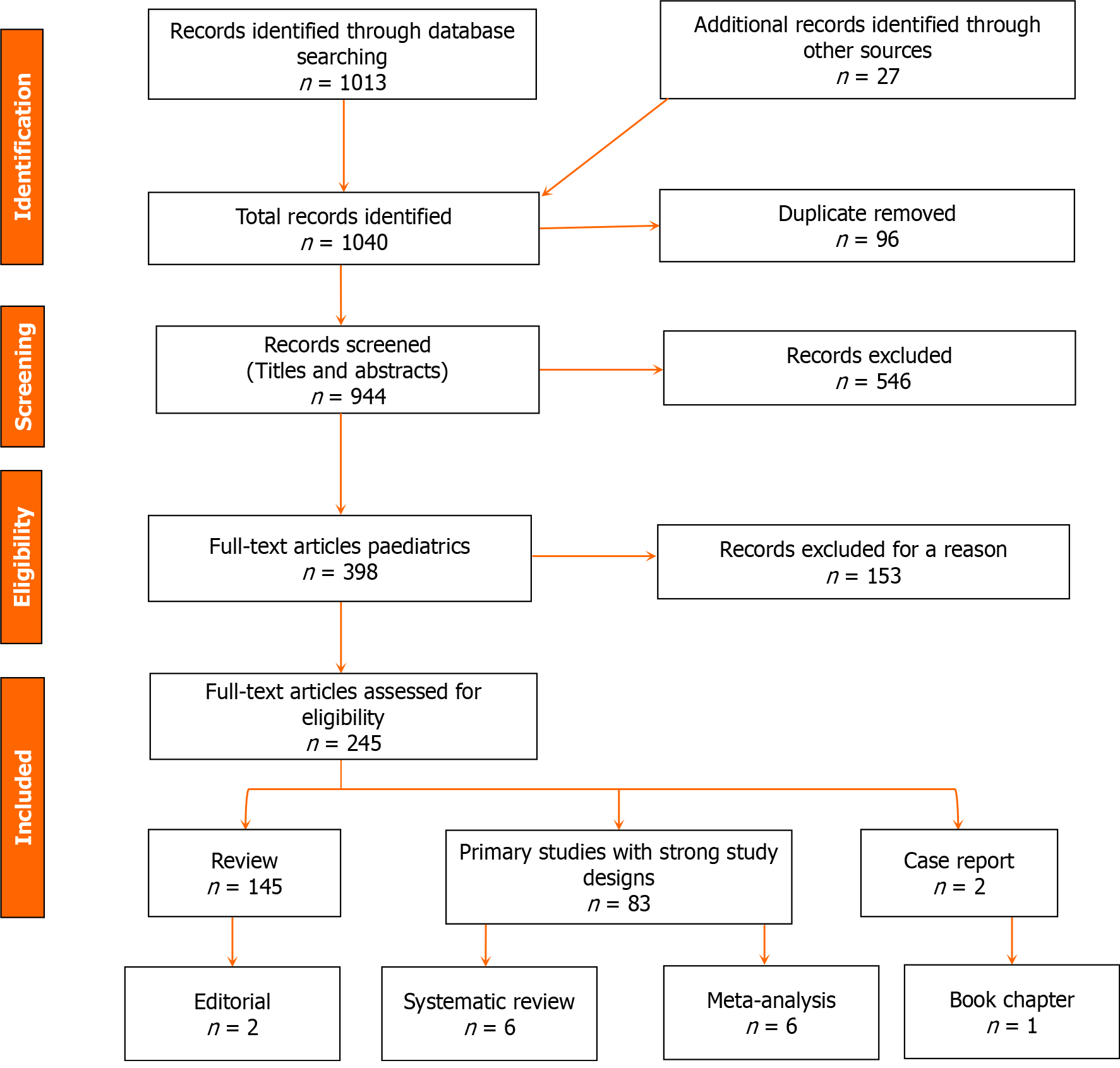
The retrieved data was synthesized and analyzed to identify potential biomarkers, underlying metabolic dysregulations, clinical implications, and patterns. We conducted a quality assessment of the included studies using appropriate tools and adhered to ethical guidelines and standards for systematic reviews. Findings were reported following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines, while limitations, such as publication bias and study quality variations, were acknowledged. Please refer to Figure 1 for the flow chart of the study.
The comprehensive literature review identified 83 research articles, 145 review articles, 6 meta-analyses, 6 systematic reviews, 2 case reports, 2 editorials, and 1 book chapter that meet the inclusion criteria (Figure 1). The search of electronic databases found relevant publications until January 2024, focusing on the metabolomic changes in children diagnosed with ASD. The data synthesis revealed several key themes, including age-related differences in metabolomic profiles, sex-specific metabolic variations, genetic influences on metabolism, environmental factors that impact metabolomic alterations, dietary patterns affecting metabolic pathways, associations between the severity of ASD symptoms and metabolic dysregulations, and the role of gut microbiota in modulating metabolomic profiles. Moreover, metabolic dysregulations related to energy metabolism, oxidative stress, mitochondrial dysfunction, neurotransmitter metabolism, and comorbid medical conditions were identified. These findings provide insights into the complex metabolic landscape of ASD and highlight the potential for personalized diagnostic and therapeutic approaches targeting specific metabolic pathways. The limitations of the reviewed studies, such as variations in methodology, sample size, and study design, were noted, underscoring the need for further research to validate findings and establish standardized protocols in the field of ASD metabolomics.
ASD is a complex condition that affects neurodevelopment. It is characterized by social communication and interaction challenges and restricted and repetitive behaviors[18]. While the exact causes of autism are not yet clear, research suggests that it is a multifactorial condition that involves genetic, environmental, and neurobiological factors. The pathophysiology of autism is complex and involves interactions between genetic predisposition, environmental influences, brain development, and neural functioning[19]. Genetics plays a significant role in autism, and studies have shown that certain genetic mutations or variations can increase the risk of developing ASD. Some cases of autism are associated with spontaneous mutations that occur in the sperm or egg cells or early in fetal development rather than being inherited from parents[20]. Certain genetic syndromes, such as Fragile X syndrome or Rett syndrome, have a higher prevalence of ASD. However, a straightforward genetic pattern is not always present, and multiple genes interacting with environmental factors might be involved. In addition, changes in gene expression without altering the DNA sequence, known as epigenetic modifications, are also being investigated concerning ASD. Environmental factors might influence these modifications and could contribute to ASD risk[21].
Environmental factors may have a significant effect on autism manifestations. Exposures during pregnancy, such as maternal infections, certain medications, or prenatal complications, have been suggested as potential environmental factors contributing to autism[22]. Postnatal environmental influences might also contribute, including exposure to toxins, pollutants, or certain medications during early development. Altered brain development is a key aspect of the pathophysiology of autism[23]. Studies using neuroimaging techniques have shown differences in brain structure and connectivity in individuals with ASD, particularly in areas involved in social cognition and communication. Imbalances in neurotransmitters [such as serotonin, dopamine, and gamma-aminobutyric acid (GABA)] have been implicated in ASD, affecting neural signaling and communication between brain regions[24].
Several studies suggest that individuals with ASD may have immune system dysregulation and inflammation, which could play a role in the development of the condition[25]. Chronic inflammation in the brain has also been linked to ASD, which genetic factors, environmental triggers, or a combination of both may cause. Differences in the structure and function of the brain have been observed in individuals with ASD, including atypical neuronal pruning, abnormal connectivity between brain regions, and changes in the size and shape of certain brain structures[26]. Irregularities in synaptic function and signaling pathways have also been suggested as contributors to ASD. Developmental alteration of synapse formation, function, and pruning could affect neural circuitry. Furthermore, researchers are investigating disruptions in the balance between excitation and inhibition in neuronal networks and differences in long-range and local connectivity in the brain as potential factors in autism[27].
Many individuals with ASD experience atypical sensory processing, such as heightened or reduced sensitivity to sensory stimuli. They might be hypersensitive or hyposensitive to sensory stimuli like light, sound, touch, taste, or smell, which can influence their behavior and reactions to the environment[28]. Variability in cognitive abilities, including strengths in specific areas (such as visual thinking or pattern recognition) and challenges in others (such as social cognition), is a hallmark of ASD. Many individuals with ASD also experience gastrointestinal problems, such as constipation, diarrhea, and bloating. It is not yet clear whether these problems are a direct cause of ASD or a symptom of the underlying condition[29].
Research into the metabolic profiles of children with ASD has revealed significant differences compared to those of neurotypical children. These differences can alter various metabolic pathways, including amino acid, energy, and neurotransmitter metabolism[30]. By understanding the underlying metabolic mechanisms, researchers may be able to pinpoint reliable biomarkers for early diagnosis of ASD. In addition, understanding these metabolic disturbances could open avenues for targeted interventions, highlighting the complex interplay between genetic predispositions and environmental factors in shaping metabolic profiles associated with ASD[31]. Solving the metabolomic derangement puzzles in patients with ASD could lead to the development of personalized and targeted interventions and treatments to correct metabolic imbalances and improve their outcomes. However, identifying specific metabolomic signatures as potential biomarkers for ASD remains a challenge due to heterogeneity in ASD presentations and variations among individuals[16].
Research into the metabolic profiles of children with ASD has found significant differences when compared to the metabolic profiles of neurotypical children. These differences can affect various metabolic pathways, including amino acid, energy, and neurotransmitter metabolism. By understanding the underlying metabolic mechanisms, researchers may be able to identify reliable biomarkers for early diagnosis of ASD, especially for at-risk populations[17]. Furthermore, understanding these metabolic discrepancies can open up avenues for highlighting how genetic predispositions and environmental factors interact to shape the metabolic profiles associated with ASD[32]. Solving the metabo
Research has shown that there are differences in the amino acid profiles of children with ASD compared to those without ASD. Some studies have identified specific changes in amino acid metabolism in individuals with ASD, which could have implications for their neurological function and behavior. These changes may include increases, decreases, or disruptions to the metabolic pathways of certain amino acids. For instance, children with ASD may have elevated levels of phenylalanine and glutamate but decreased levels of glutathione and creatine. These alterations can cause neurotransmitter imbalances, which are important for brain communication. They can also contribute to oxidative stress, damage brain cells, and disrupt energy production, which can affect the proper functioning of the brain[34].
Tryptophan, an essential aromatic amino acid, is a precursor for quinolinic and kynurenic acid serotonin, a neurotransmitter implicated in mood regulation and social behavior. We obtain tryptophan through protein-rich foods like dairy, oats, bananas, peanuts, dried prunes, bread, eggs, tuna, fish, cheese, poultry, and chocolate[35]. Once ingested, tryptophan is absorbed in the gut, transported to the liver, and then to various organs, including the brain. In the brain, tryptophan gets metabolized through two main pathways: The kynurenine pathway and the serotonin (5-hydroxytryptamine) pathway[36]. The Kynurenine pathway generates various metabolites, including kynurenic acid and quinolinic acid, which influence brain function and immunity. In contrast, the serotonin pathway produces serotonin, a vital neurotransmitter involved in mood, sleep, and social behavior[37].
Some research indicates disruptions in the tryptophan pathway in individuals with ASD. These disruptions might reduce blood and CSF tryptophan levels, inducing altered serotonin levels. Serotonin is crucial in social interaction and emotional regulation[38]. Therefore, the reduction of serotonin could potentially influence mood, social interactions, and other behaviors associated with ASD. Some studies have reported lower levels of tryptophan, the precursor for serotonin, in individuals with ASD compared to neurotypical individuals[39]. Boccuto et al[40] found that patients with ASD have lower levels of tryptophan, which is linked to the behavioral traits associated with autism, regardless of their genetic background. Ormstad et al[41] showed elevated brain-derived neurotrophic factor levels and lower tryptophan and kynurenic acid levels in children with ASD and intellectual disability disorder. At the same time, they found elevated tryptophan and lower serotonin synthesis in patients with Asperger syndrome. This reduction might affect serotonin synthesis, potentially impacting mood regulation, social behavior, and emotional processing, all of which are areas often affected in ASD. However, a meta-analysis by Almulla et al[42] found no abnormalities in peripheral blood tryptophan metabolism, indoleamine 2,3-dioxygenase enzyme activity, or tryptophan catabolite production in ASD.
Tryptophan can also be metabolized through the kynurenine pathway, producing various metabolites, including kynurenic acid and quinolinic acid, which influence brain function and immunity. Kynurenic acid might have excitatory effects on brain cells, potentially contributing to repetitive behaviors[43]. Imbalances in the kynurenine pathway have been observed in individuals with ASD, with some studies reporting abnormalities in kynurenine pathway metabolites, such as increased levels of certain kynurenine pathway metabolites or imbalances in the ratios between these metabolites[44].
Serotonin is crucial in various developmental processes, including cell proliferation, migration, and differentiation. It affects various brain regions, including those involved in mood, social behavior, sensory processing, and repetitive behaviors, as it plays an important neurotrophic role during brain development. Disruptions in serotonin pathways during critical developmental periods may affect brain wiring, potentially contributing to the development of ASD traits[45]. Early serotonin system disturbance affects cortical development and thalamocortical innervation maturation and development, a potential mechanism common to autism and pediatric epilepsies associated with cortical dysplasia. Studies have shown variations in serotonin levels in the blood, brain, and gastrointestinal tract of individuals with ASD compared to those without ASD. However, these differences aren't consistent across all individuals with ASD, suggesting that serotonin dysregulation might be only one of many factors contributing to ASD[46,47]. In addition, serotonin interacts with various other neurotransmitter systems and affects multiple physiological processes, which might contribute to the diverse array of symptoms and traits seen in ASD[48].
A high level of serotonin in the blood, known as hyperserotonemia, is the first biomarker identified in ASD. This condition is found in more than 25% of children with ASD, and there is an inverse relationship between serotonin levels and patient intelligence[39]. Genetic linkage and association studies have shown that hyperserotonemia and ASD risk are linked to the chromosomal region containing the serotonin transporter gene in males but not females[49]. Studies in the Hutterite population have also revealed associations between genes like integrin β3 subunit (ITGB3) and the Vitamin D receptor gene with serotonin levels. Further investigations in males have shown that ITGB3 and serotonin transporter (SLC6A4) genes are linked to 5-HT levels in the blood. These genes were found to interact, influencing platelet serotonin uptake and potentially relating to autism susceptibility genes[50,51]. However, despite its strong and specific association with ASD, no prospective study has yet assessed whether hyperserotonemia may predict ASD risk in infants, including baby siblings of children with ASD[52].
While direct serotonin measurements in the brain are challenging, studies examining peripheral markers (like blood or platelet levels) of serotonin or its metabolites have shown mixed results. Some individuals with ASD have displayed alterations in these peripheral markers, suggesting potential dysregulation in serotonin signaling[53]. Additionally, medications that target serotonin pathways (like selective serotonin reuptake inhibitors - SSRIs) have been explored in managing certain symptoms associated with ASD, although their effectiveness varies among individuals[54].
Many factors can influence tryptophan levels in individuals with ASD. Deficiencies in certain vitamins and minerals, like vitamin B6 and zinc, can hinder tryptophan metabolism. Therefore, consuming adequate protein sources naturally containing tryptophan can support healthy levels. Dietary supplementation with vitamins B and magnesium influences tryptophan levels due to their impact on its metabolic homeostasis[38]. The gut microbiome plays a role in tryptophan metabolism, and imbalances in gut bacteria might impact its absorption and utilization. Gut dysbiosis is common in patients with autism. Gut dysbiosis, in turn, causes impaired tryptophan metabolism and consequently impairs cognitive functions in patients with ASD[55]. Certain genetic variations might influence genes related to tryptophan metabolism, contributing to potential differences in individuals with ASD. Higazi et al[56] found a significant decrease in the expression levels of monoamine oxygenase A, 3-hydroxy anthranilate oxygenase, and aminoadipate aminotransferase genes in Egyptian children with ASD compared to individuals without ASD. They were negatively correlated to ASD scoring. Solute carrier transporter 7a5 (SLC7A5), a large neutral amino acid transporter localized at the blood-brain barrier (BBB), is essential in maintaining normal levels of brain-branched-chain amino acids. Tărlungeanu et al[57] found that the deletion of Slc7a5 from the endothelial cells of the BBB leads to atypical brain amino acid profile, abnormal mRNA translation, and severe neurological abnormalities. Many patients with autistic traits and motor delay carry deleterious homozygous mutations in the SLC7A5 gene. Most plasma tryptophane is bound to albumin and hence is unavailable for transport into the brain. Therefore, the plasma levels of free tryptophan depend on the albumin level and can be affected by the different causes of hypoalbuminemia. It should also be noted that a lack of tryptophan leads to a lack of albumin synthesis[58]. Chronic stress and anxiety can deplete serotonin levels, potentially impacting tryptophan utilization[59]. Disrupted sleep patterns, common in ASD, can affect the synthesis and breakdown of serotonin, influencing tryptophan availability[60]. Certain medications, like some antidepressants, might interact with tryptophan metabolism, requiring careful monitoring[61].
The metabolism of phenylalanine and tyrosine is a complex process that plays a vital role in human health. Phenylalanine is an essential amino acid obtained from our diet, which the body processes through several enzymatic steps. One crucial process is when phenylalanine hydroxylase (PAH), an enzyme, converts phenylalanine into tyrosine. This conversion is necessary to maintain appropriate levels of tyrosine in the body[62]. Tyrosine is another amino acid that is a precursor for many essential compounds. It is a critical building block for producing neurotransmitters such as dopamine, nor
The connection between phenylalanine and tyrosine metabolism and ASD has been an area of research interest, though it's complex and not fully understood. Some studies have explored metabolic pathways involving phenylalanine and tyrosine in individuals with ASD[32]. Alterations in these metabolic pathways, including abnormalities in the levels or ratios of these amino acids and their metabolites, have been observed in some individuals with ASD. Research has suggested that disruptions in the phenylalanine-to-tyrosine conversion pathway might affect neurotransmitter synthesis, particularly dopamine and serotonin. Dopamine and serotonin play essential roles in regulating mood, behavior, and social interactions, aspects often affected in individuals with ASD[65]. Naushad et al[66] showed a significant decrease in levels of essential amino acids tryptophan, phenylalanine, and methionine and reduced amino acids precursors of neurotransmitters such as tyrosine and tryptophan in children with autism than in neurotypical children. Moreover, some individuals with ASD reportedly have abnormalities in enzymes involved in phenylalanine and tyrosine metabolism, leading to variations in amino acid levels[67]. These variations may contribute to differences in neurotransmitter levels or function, potentially impacting behavior and cognitive functioning associated with ASD. Other studies claimed that gut microbiota's disturbed metabolic action on phenylalanine and tyrosine produced different metabolites that can be used as biomarkers for ASD[68].
However, the phenylalanine to tyrosine ratio is more predictive of cerebral glucose metabolism than the phenylalanine level alone. Some researchers have proposed that imbalances or abnormalities in the phenylalanine-to-tyrosine ratio might affect neurotransmitter levels, potentially contributing to certain aspects of ASD symptomatology. The high ratio observed in children with autism is due to the failure of phenylalanine to be converted to tyrosine and, consequently, to dopamine in the brain[69]. The high levels of phenylalanine and tyrosine were not observed only in children with ASD but also observed in their parents and siblings[70]. Increased phenylalanine levels in children with ASD could be related to increased intakes. Arum et al[71] observed high phenylalanine and tryptophan intake in children with autism and hyperactivity, and the level of intake is significantly associated with the severity of hyperactivity.
The relationship between phenylalanine, tyrosine metabolism, and ASD is not yet fully established and remains complex. Not all individuals with ASD show abnormalities in these metabolic pathways[72]. Further investigation is required to determine if these metabolic differences play a causative role in ASD development. It's important to note that ASD is a multifactorial condition influenced by a combination of genetic, environmental, and neurological factors[73]. Phenylalanine and tyrosine metabolism are just a part of the broader spectrum of factors that researchers are studying to understand the complexities of ASD[65]. More studies are necessary to determine the specific contributions, if any, of phenylalanine and tyrosine metabolism to the development or characteristics of ASD. Additionally, it is vital to deter
The methionine cycle is an essential biochemical pathway that plays a vital role in various biological processes in the body. This pathway involves sulfur metabolism and synthesizes certain amino acids, proteins, and other molecules critical for cellular function[74]. Methionine, an essential amino acid needed by the body, is produced in cells through a series of reactions that start with the amino acid homocysteine and use molecules like adenosine triphosphate (ATP) and methyl donors like S-adenosylmethionine (SAM)[75]. The transsulfuration pathway connects the methionine cycle to the synthesis of cysteine, another important sulfur-containing amino acid and precursor of glutathione. Homocysteine, derived from the methionine cycle, can convert to cysteine through several enzymatic reactions[76]. It can also be re-methylated into methionine by methionine-synthase using vitamin B12 as a cofactor. SAM is a critical molecule formed during the methionine cycle. It acts as a methyl donor in various biological methylation reactions, contributing to modifying DNA, RNA, proteins, and other molecules[77]. Enzymes involved in the methionine cycle and sulfur metabolism are regulated to maintain a balance of sulfur-containing molecules and prevent the accumulation of potentially harmful intermediates[78]. Disruptions or deficiencies in the methionine cycle and sulfur metabolism can lead to various health issues. For instance, deficiencies in enzymes involved in these pathways can cause homocystinuria, a genetic disorder characterized by the buildup of homocysteine in the body[79].
Disorders in the metabolic pathways of the methionine cycle might play a role in the development of ASD in a subset of individuals. The methionine cycle is involved in methylation reactions, which regulate gene expression through epigenetic modifications. Altered methylation patterns have been observed in some individuals with ASD[80]. Changes in DNA methylation could potentially influence brain development and function, including neurogenesis, neuronal differentiation, synaptogenesis, learning, and memory[81].
Disruptions or abnormalities in the methionine cycle and related metabolic pathways may cause imbalances in essential molecules like SAM (S-adenosylmethionine), affecting various cellular processes[82]. Studies have reported differences in methionine metabolism in individuals with ASD compared to neurotypical individuals. For instance, Indika et al[83] found reduced methionine and SAM levels in children with ASD, which may reflect the impaired remethylation pathway. Similarly, Geier et al[84] observed reduced levels of serum glutathione, cysteine, methionine, cystathionine, and homocysteine in pre-pubertal children with ASD compared to typically developed children. They also noted elevated levels of serum dehydroepiandrosterone and total testosterone relative to the sex and age-specific normal ranges, which could indicate a possible cyclical relationship between the androgen pathways and methionine cycle-transsulfuration in some children with ASD. Additionally, the intracellular concentration of methionine and its metabolites is also reduced in children with ASD. Suh et al[85] found a significant reduction of intracellular cysteine, glutathione, and S-adenosylmethionine and elevated intracellular homocysteine levels in leukocytes of children with ASD than in children with normal neurodevelopment. These intracellular changes in leukocytes of children with ASD could explain the immune disorders that are common in children with ASD. A meta-analysis by Guo et al[86] found impaired methylation capacity with significantly decreased levels of Methionine, S-adenosylmethionine, and S-adenosylmethionine/S-adenosylhomocysteine ratio and significantly increased levels of S-adenosylhomocysteine in children with ASD.
Both genetic and environmental factors contribute to the development of ASD. Genetic variations in genes involved in the methionine cycle and related pathways concerning ASD have been studied. genetic mutations and epimutation of DNA methylation can be seen in children with ASD on multiple levels from fetal life throughout postnatal life, affecting both embryonic brain development and early childhood synaptogenesis[87,88]. Haghiri et al[89] found a significant increase in the incidence of genetic mutations of methionine synthase in children with ASD than in controls. Mutations in the methionine synthase reductase gene were also more common among Iranian children with ASD than in typically developed controls. The methylenetetrahydrofolate reductase (MTHFR) gene produces an enzyme involved in the methionine cycle[90]. Variations in the MTHFR gene, particularly a common polymorphism known as the MTHFR C677T variant, have been studied concerning ASD[91]. Some studies have suggested an association between the MTHFR C677T variant and an increased risk of ASD[92,93], while others have found no significant association[94]. The catechol-O-methyltransferase (COMT) gene regulates the production of the COMT enzyme that breaks neurotransmitters like dopamine. Variations in the COMT gene have been associated with altered dopamine levels and cognitive impairments. Some studies have found an association between COMT gene variants and ASD, particularly with social and communication difficulties[95,96]. Furthermore, genes involved in the folate pathway, such as the folate receptor alpha (FOLR1) gene, have also been extensively studied in patients with ASD. Folate is essential for DNA methylation and other methylation processes, and disruptions in folate metabolism have been linked to neurodevelopmental disorders, including autism. Variations in the FOLR1 gene have been implicated in ASD susceptibility in some studies[97].
Environmental factors, such as nutritional influences or exposure to certain substances, might also impact these metabolic pathways. Sulfate deficiency in both the mother and the child due to excess exposure to environmental toxins and inadequate skin exposure to sunlight leads to widespread hypomethylation in the fetal brain with devastating consequences, including increased incidence of ASD[98]. In addition, the methionine cycle is directly influenced by the availability of methionine in the diet. Methionine-rich foods like certain meats and dairy products contribute to the amino acid pool[99]. The methionine cycle requires essential cofactors such as folate and B vitamins (especially B6, B12, and folate) to function properly. Inadequate intake of these vitamins can disrupt the normal functioning of the pathway[100]. The composition of gut microbiota can also affect methionine metabolism. Interactions between the host and gut bacteria can impact the availability and utilization of methionine[101].
Physical activity can likewise affect methionine metabolism. Exercise-induced changes in metabolism may influence the demand for sulfur-containing amino acids like methionine[102]. Chronic stress or psychological factors, including the methionine cycle, can impact overall metabolism. Stress hormones may affect the regulation of enzymes involved in these pathways[103]. Certain medications can interfere with methionine metabolism. For example, drugs that affect folate or vitamin B12 absorption can indirectly impact the methionine cycle. Exposure to heavy metals such as lead, cadmium, or mercury can also interfere with enzymes involved in the methionine cycle and disrupt sulfur metabolism[104]. Certain environmental toxins or pollutants may also adversely affect the methionine metabolic pathways. This can include exposure to industrial chemicals or pollutants in the air or water[105]. Table 1 summarizes some studies concerned with the alteration of the amino acid metabolic pathway in individuals with ASD.
| Ref. | Study methods | Amino acid pathway | Changes in ASD |
| Boccuto et al[40] | Blood or CSF sample analysis | phenylalanine and glutamate | Elevated phenylalanine and glutamate levels |
| Ormstad et al[41] | Metabolomic profiling | phenylalanine and glutamate | Decreased glutathione and creatine levels |
| Almulla et al[42] | Peripheral blood analysis | Tryptophan pathway | Disrupted tryptophan metabolism |
| Higazi et al[56] | Genetic analysis of MAOA, HAAO and AADAT genes using real-time RT-qPCR. | tryptophan pathway | significant decrease in the expression of the selected genes within ASD children relative to children with learning disabilities and healthy controls |
| Naushad et al[66] | Genetic analysis | Tryptophan pathway | Lower serotonin synthesis |
| Naushad et al[66] | Enzyme activity measurement | Phenylalanine and tyrosine metabolism | Reduced tyrosine synthesis |
| Arum et al[71] | Dietary intake assessment | Phenylalanine and tyrosine metabolism | Abnormal phenylalanine-to-tyrosine ratio |
| Geier et al[84] | Peripheral blood analysis | Methionine and sulfur metabolism | reduced levels of serum glutathione, cysteine, methionine, cystathionine, and homocysteine in pre-pubertal children |
| Suh et al[85] | Genetic analysis | Methionine and sulfur metabolism | Reduced SAM levels |
| Guo et al[86] | Meta-analysis of metabolomic profiling | Methionine and sulfur metabolism | Impaired methylation capacity |
| Haghiri et al[89] | Genetic analysis | Methionine metabolism | mutations of methionine synthase in children with ASD |
Purine and pyrimidine nucleotides serve as crucial building blocks for nucleic acid synthesis, yet their roles extend beyond this primary function. Purines function as metabolic signals, provide energy, regulate cell growth, participate in coenzymes, contribute to sugar transport, and donate phosphate groups in phosphorylation reactions[106]. On the other hand, pyrimidines play roles in biosynthesizing polysaccharides and phospholipids, participating in detoxification processes, and contributing to protein and lipid glycosylation[107]. A substantial amount of ATP in the nervous tissue is produced, primarily supporting energy needs for membrane-active pumps like Na+/K+ ATPase. This energy is vital for sustaining synaptic transmission and facilitating collaboration between neurons and glial cells[108]. Perturbations in purine and pyrimidine metabolism can potentially impact brain function and contribute to the development or manifestation of ASD symptoms (Table 2).
| Aspect | Purine metabolism | Pyrimidine metabolism |
| Functions | Building blocks for nucleic acid synthesis; Metabolic signals; Provide energy; Regulate cell growth; Participate in coenzymes; Contribute to sugar transport; Donate phosphate groups in phosphorylation reactions | Synthesizing DNA and RNA; Energy metabolism; Neurotransmitter signaling |
| Specific roles | Provide energy for membrane-active pumps like Na+/K+ ATPase; Vital for sustaining synaptic transmission; Facilitate collaboration between neurons and glial cells | Biosynthesis of polysaccharides and phospholipids; Participate in detoxification processes; Contribute to protein and lipid glycosylation |
| Impact on brain function | Altered purine metabolism may impact brain function and contribute to ASD symptoms; Adenosine acts as a neuromodulator, inhibiting neurotransmitter release and regulating sleep-wake cycles | Abnormalities in pyrimidine metabolism may be linked to ASD and neurodevelopmental issues; Disturbances in uracil metabolism could contribute to mitochondrial dysfunction in ASD |
| Potential biomarkers | Elevated adenosine levels and altered ADA activity observed in ASD; Abnormal levels of purine metabolites such as uric acid reported in ASD | Altered uracil levels and abnormal ratios of uracil to other pyrimidine bases reported in ASD; Abnormal levels of pyrimidine nucleotides observed in ASD |
| Genetic | Mutations in genes encoding enzymes involved in purine metabolism found in individuals with ASD | Genetic mutations in the gene encoding DPD identified in individuals with ASD |
| Therapeutic implications | Modulating adenosine signaling and targeting enzymes involved in purine metabolism could potentially improve neurochemical functioning in ASD | Supplementation with pyrimidine precursors such as uridine was explored as a possible intervention to improve mitochondrial function and neurodevelopmental outcomes in ASD |
Adenosine is a purine nucleoside that acts as a neuromodulator in the brain. It acts as an inhibitory neurotransmitter, inhibiting the release of neurotransmitters like dopamine, norepinephrine, serotonin, acetylcholine, and glutamate. It also regulates sleep-wake cycles by promoting sleepiness and initiating the sleep phase[109]. It helps to mitigate the harmful effects of excessive neuronal activity or excitotoxicity and acts as an anticonvulsant and a vasodilator, influencing blood flow in the brain. Adenosine has anti-inflammatory effects and is involved in the modulation of immune responses in the central nervous system. It can help regulate the inflammatory environment in the brain[110]. Adenosine interacts with neurotransmitters, including glutamate, dopamine, and serotonin, to regulate the development of oligodendroglia and myelination[111].
Altered adenosine signaling has been observed in individuals with ASD. Adenosine receptors have been implicated in regulating neurotransmitter release, synaptic plasticity, and neuronal excitability, all of which are relevant to ASD pathophysiology. Tanimura et al[112] showed that a combination of adenosine 2A receptor agonist and adenosine 1 receptor agonist reduces repetitive behaviors selectively and could aid with this core symptom in ASD. In contrast, either drug alone induces no or non-selective effect on behaviors, respectively. Some studies have reported elevated adenosine levels in the brains of individuals with ASD. Higher adenosine concentrations could influence neuronal activity and neurotransmitter release, potentially affecting cognitive and behavioral processes. Adenosine Deaminase (ADA) is an enzyme involved in purine metabolism that converts adenosine to inosine[113]. Altered ADA activity has been reported in some children with ASD. Dysregulation of ADA could impact adenosine levels and contribute to the neurochemical imbalances observed in ASD[114].
Abnormal levels of purine metabolites, such as uric acid, have been reported in individuals with ASD. Uric acid is the end product of purine degradation, and its dysregulation may reflect disturbances in the purine metabolic pathway[13]. Various studies analyzing metabolites have found an important reduction in uric acid levels in the urine of individuals diagnosed with ASD, especially those with food selectivity[115]. However, a study conducted by Page et al[116] found that some patients with certain types of autism showed high levels of uric acid, which was attributed to increased production of purines in these individuals. Certain genetic variations related to purine metabolism have been associated with an increased risk of ASD[117]. For example, mutations in genes encoding enzymes involved in purine metabolism, such as adenylosuccinate lyase, adenosine deaminase, and 5-aminoimidazole-4-carboxamide ribonucleotide formyltransferase/IMP cyclohydrolase (ATIC), have been identified in individuals with ASD[13]. Abnormalities in purine metabolism pathways may offer potential targets for therapeutic interventions in ASD. Modulating adenosine signaling or targeting enzymes involved in purine metabolism could potentially restore balance and improve neurochemical functioning.
Pyrimidines are molecules that play a vital role in synthesizing DNA and RNA, energy metabolism, and neurotransmitter signaling. While research on purine metabolism in children with ASD is more extensive, studies on pyrimidine metabolism are limited. Nevertheless, some evidence suggests that abnormalities in pyrimidine metabolism may be linked to ASD and neurodevelopmental and behavioral issues[118]. Uracil is a pyrimidine base, and disturbances in its metabolism have been observed in individuals with ASD. Uracil is involved in nucleotide synthesis, which is the building blocks of DNA and RNA. Disturbances in pyrimidine metabolism, including uracil, could potentially affect various cellular processes, including brain development and function[106]. Uracil is normally converted to thymine through the action of the enzyme dihydropyrimidine dehydrogenase (DPD). Altered DPD activity and imbalances in the uracil-to-thymine ratio have been reported in some children with ASD[119]. Genetic mutations in the gene encoding the enzyme DPD, which is involved in pyrimidine catabolism, have been found in some individuals with ASD[120].
Mitochondrial dysfunction has been implicated in a subset of individuals with autism. Since uracil and pyrimidine metabolism are closely linked to mitochondrial function, disturbances in uracil metabolism might contribute to mitochondrial dysfunction observed in some cases of ASD[10,121]. Additionally, abnormalities in folate metabolism, which is interconnected with pyrimidine synthesis, have been implicated in ASD[122]. Some studies have explored the possibility of using uracil disturbances as potential biomarkers for autism. Altered uracil levels or abnormal ratios of uracil to other pyrimidine bases have been reported in certain individuals with ASD[123]. However, further research is needed to validate these findings and determine their clinical significance. In addition, studies have reported alterations in the levels of pyrimidine nucleotides, such as uridine, cytidine, and deoxycytidine, in individuals with ASD[124]. These nucleotides are crucial for DNA and RNA synthesis and have important roles in neuronal development and function. Modulating pyrimidine metabolism pathways could potentially have therapeutic implications for ASD[125]. For instance, supplementation with certain pyrimidine precursors, such as uridine, has been explored as a possible intervention to improve mitochondrial function and neurodevelopmental outcomes in ASD[126].
Mitochondria is a tiny structure located within nearly all body cells. It is the cellular powerhouse responsible for generating energy in the form of ATP. Due to their high energy needs, the muscle and the brain cells have a particularly high mitochondrial density to support their energy needs[127]. There is growing evidence of an increased prevalence of impaired functioning of the mitochondria in patients with ASD compared to the general population. This dysfunction can result from genetic mutations, environmental factors, or a combination of both[128]. A groundbreaking study by Giulivi et al[129] at the University of California in 2010 showed that 80% of the children with ASD enrolled in their study had blood tests indicating mitochondrial dysfunction, mtDNA overreplication, and mtDNA deletions. However, the estimates of the co-occurrence of mitochondrial disorders in individuals with ASD range from 5% to 80%[130]. Many factors increase the prevalence of mitochondrial dysfunction in children with autism, such as genetic mutations, dietary deficiencies of vitamins and minerals in the diet, certain chemicals and heavy metals exposure, some drugs, certain bacterial and viral infections, and stressful conditions[131]. Mitochondrial dysfunction can arise from genetic mutations in mitochondrial DNA (mtDNA) or nuclear DNA, affecting the function of mitochondrial proteins and enzymes[132,133]. Some studies have identified specific genetic variations associated with mitochondrial dysfunction in individuals with ASD, suggesting a potential genetic overlap between these conditions[134].
It's important to note that while mitochondrial dysfunction may be more common in individuals with ASD, it is not a defining feature of ASD. A significant number of people with ASD do not have mitochondrial dysfunction. However, mitochondrial dysfunction can lead to inadequate production of ATP, which is the energy currency of cells, including neurons in the brain. Insufficient energy can affect various organs and cellular processes, including synaptic plasticity, neuronal development, signaling, and maintenance, which may contribute to atypical brain functioning observed in individuals with ASD[135,136]. Mitochondrial dysfunction can also cause an imbalance between the production of ROS and the cellular antioxidant defense mechanisms, leading to oxidative stress[137]. Increased oxidative stress can damage cellular components, such as lipids, proteins, and DNA, impacting neuronal function and development[138]. Additionally, mitochondrial dysfunction can disrupt metabolic processes, such as abnormal amino acid metabolism, impaired fatty acid oxidation, and dysregulated carbohydrate metabolism. These metabolic alterations may further contribute to the development of ASD and impact overall cellular energy balance[139].
Diagnosing mitochondrial dysfunction in individuals with ASD can be complex. Mitochondrial dysfunction is highly heterogeneous and can lead to a wide range of symptoms that may contribute to the development or severity of ASD symptoms. Although the manifestation of mitochondrial dysfunction in children with autism can vary widely, there is evidence to suggest that some individuals with ASD may experience abnormalities in mitochondrial function[140]. Children with mitochondrial dysfunction and autism may experience delays in reaching developmental milestones, such as walking, talking, or social interactions[141]. They may also have impaired language development and communication skills, including delayed speech, limited vocabulary, difficulties with expressive and receptive language, and challenges in social communication. Some children with mitochondrial dysfunction and autism may experience motor difficulties, including poor muscle tone (hypotonia), coordination problems, and gross or fine motor skill deficits. Mitochondrial dysfunction can also impact cognitive abilities, leading to intellectual disabilities, learning difficulties, or problems with attention and executive functioning[142]. Behavioral abnormalities, such as hyperactivity, repetitive behaviors, anxiety, aggression, or self-injurious behaviors, are also common in children with both mitochondrial dysfunction and autism[143]. Epileptic seizures may occur in some children with both conditions[144]. Gastrointestinal problems, such as chronic constipation, diarrhea, or gastrointestinal inflammation, have been reported in individuals with mitochondrial dysfunction and autism[145]. Children may also demonstrate heightened sensitivities or aversions to sensory stimuli, such as loud noises, bright lights, certain textures, or specific tastes or smells. It's important to note that these symptoms can also be present in individuals with autism without mitochondrial dysfunction[28].
Comprehensive evaluations involving clinical assessments, biochemical analyses, and genetic testing are often required. Specific diagnostic criteria, such as the "Mitochondrial Disease Criteria" or the "Mitochondrial Autism Criteria," have been proposed to aid in identifying individuals with both mitochondrial dysfunction and ASD[146]. As such, a comprehensive evaluation by healthcare professionals with expertise in both autism and mitochondrial disorders is necessary for accurate diagnosis and appropriate management[147]. Each child's experience with mitochondrial dysfunction and autism can be unique, and the severity and specific symptoms can vary. Early diagnosis and intervention, along with a multidisciplinary approach involving healthcare professionals from various specialties, can help address the specific needs of children with co-occurring mitochondrial dysfunction and autism[143].
Several laboratory tests can provide valuable insights into mitochondrial function and potential dysfunction. These may include blood tests to assess lactate, pyruvate, amino acids, creatine kinase, ammonia, total and free carnitine, and an acylcarnitine profile. Additionally, urine testing for organic acids can be performed[148]. Mitochondrial DNA analysis and nuclear DNA sequencing may be employed to identify specific genetic mutations associated with mitochondrial disorders[149]. Brain imaging techniques like magnetic resonance spectroscopy (MRS) can be used to assess brain chemistry and detect elevated lactate levels, which can indicate mitochondrial dysfunction[150]. In some cases, a muscle biopsy may be recommended to evaluate mitochondrial function directly. This involves obtaining a small sample of muscle tissue under local anesthesia for laboratory analysis[151]. Diagnosis of mitochondrial dysfunction in children with autism can be challenging due to the overlap of symptoms and the lack of specific diagnostic criteria. Therefore, a multidisciplinary approach involving pediatricians, neurologists, geneticists, and metabolic specialists is often necessary to arrive at an accurate diagnosis. Treatment strategies may include dietary interventions (such as specific nutritional supplements or ketogenic diets), vitamin and cofactor supplementation, antioxidants, and medications targeted at specific symptoms[152]. However, the effectiveness of these interventions in improving ASD symptoms associated with mitochondrial dysfunction is still an area of ongoing research[153,154]. Table 3 provides a concise summary of the prevalence, causes, impact, symptoms, diagnosis, management, challenges, and ongoing research related to mito
| Aspect | Mitochondrial metabolic disorders in ASD |
| Overview | Mitochondria are cellular structures responsible for generating energy (ATP). High mitochondrial density in muscle and brain cells |
| Prevalence and causes | Increased prevalence of mitochondrial dysfunction in ASD compared to the general population. Can result from genetic mutations, environmental factors, or both |
| Evidence | About 80% of children with ASD show blood test indications of mitochondrial dysfunction and DNA abnormalities. Estimates of co-occurrence range from 5% to 80% |
| Contributing factors | Genetic mutations; Dietary deficiencies; Chemical and heavy metal exposure; Certain drugs; Bacterial and viral infections; Stressful conditions |
| Impact on ASD | Insufficient ATP production can affect synaptic plasticity, neuronal development, signaling, and maintenance. Oxidative stress and damage to cellular components may occur. Disruption of metabolic processes can further impact ASD development |
| Symptoms and diagnosis | Symptoms include delays in developmental milestones, impaired language and communication, motor difficulties, cognitive impairments, behavioral abnormalities, seizures, and gastrointestinal issues. Diagnosis involves comprehensive clinical assessments, biochemical analyses, genetic testing, and specific diagnostic criteria. Laboratory tests may include blood tests, urine tests, DNA analysis, brain imaging, and muscle biopsy |
| Management and treatment | Treatment strategies may include dietary interventions, nutritional supplements, antioxidants, and medications targeted at specific symptoms. A multidisciplinary approach involving healthcare professionals from various specialties is necessary for accurate diagnosis and management |
| Challenges and ongoing research | Diagnosis can be challenging due to overlapping symptoms and lack of specific criteria. The effectiveness of interventions in improving ASD symptoms associated with mitochondrial dysfunction is still under research |
Antioxidant defense mechanisms counteract oxidative stress and minimize its harmful effects. These mechanisms include enzymatic and non-enzymatic antioxidants. Superoxide dismutase, catalase, glutathione peroxidase, and others are enzymes that neutralize ROS and protect against oxidative damage. Vitamins C and E, glutathione, coenzyme Q10, and various phytochemicals act as antioxidants, scavenging free radicals and preventing oxidative damage. Antioxidant enzymes work together in a coordinated manner to maintain redox balance and protect against oxidative stress[155,156]. Research has shown that oxidative stress and antioxidant metabolism are important factors in understanding ASD. Oxidative stress occurs when there is an imbalance between the production of ROS and the body's antioxidant defense systems. ROS are byproducts of cellular metabolism that include free radicals and other molecules. When ROS production exceeds the body's antioxidant capacity, oxidative stress occurs[157]. Several studies have found increased oxidative stress markers in children with ASD, indicating their role in the development of the disorder[158-160].
There are various reasons why children with autism may experience oxidative stress. The interaction between oxidative stress, mitochondrial dysfunction, inflammation, and immune dysregulation can contribute to the development of ASD[161]. One of the primary reasons is impaired mitochondrial function, which is frequently observed in individuals with autism. Mitochondria are crucial in generating cellular energy and are also a significant source of ROS. When mitochondrial function is compromised, it can lead to excessive ROS production, causing oxidative stress[162]. Additionally, chronic inflammation and immune system dysregulation, which are common in individuals with autism, can also contribute to oxidative stress[163]. Altered antioxidant metabolism, such as low levels of antioxidants like glutathione or reduced antioxidant enzyme activity, can weaken the body's ability to combat oxidative stress[164]. Bjørklund et al[165] showed that individuals with ASD have reduced glutathione levels, indicating a potential imbalance in their antioxidant metabolism. Additionally, some studies have found lower levels of vitamins C and E in children with ASD, which may affect their antioxidant capacity[166]. Furthermore, exposure to environmental toxins like heavy metals, pesticides, and pollutants can also increase oxidative stress levels in individuals with autism[167].
Oxidative stress is a harmful process affecting different cellular aspects and vital processes. ROS can cause damage to lipids, leading to harmful byproducts that disrupt cell membranes and hinder cellular function[168]. Oxidative stress can also alter proteins, affecting their structure and function and impacting enzymatic activity, signaling pathways, and cellular vital processes[169]. Furthermore, ROS can cause DNA damage, including DNA strand breaks and modifications, leading to genomic instability and impaired cellular function[170]. The brain is especially vulnerable to oxidative stress due to its high energy demand, high lipid content, and limited antioxidant defenses. This type of stress can disrupt neurodevelopmental processes, synaptic plasticity, neurotransmitter balance, and overall brain function, which may potentially contribute to the core symptoms of ASD[171].
Given the association between oxidative stress and ASD, there has been interest in exploring antioxidant interventions as a potential therapeutic approach[172]. However, clinical trials examining the efficacy of antioxidant supplementation in ASD have yielded mixed results. While some studies have shown improvements in certain symptoms, others have reported no significant effects[173]. The effectiveness of antioxidant therapies may depend on factors such as the specific antioxidants used, the individual's antioxidant status, underlying genetic factors, and the presence of other co-occurring conditions[173]. Further research is needed to understand better the role of oxidative stress and antioxidant metabolism in ASD. This includes investigating the underlying mechanisms and identifying reliable biomarkers of oxidative stress in individuals with autism[174]. Additionally, studies exploring personalized antioxidant therapies and interventions targeting mitochondrial dysfunction and inflammation in ASD are warranted.
Lipid metabolism encompasses the body's synthesis, breakdown, and transportation of fats. Lipids are important components of cell membranes, energy storage, and signaling molecules. Abnormalities in lipid metabolism have been observed in children with ASD, suggesting a potential link between lipid dysregulation and the pathophysiology of the disorder[175]. Dyslipidemia, characterized by abnormal lipid levels in the blood, has been reported in several studies involving children with ASD[176]. These studies have revealed changes in various lipid parameters. For instance, some studies have found elevated levels of total cholesterol in children with ASD compared to typically developing individuals[177]. However, a Tunisian study found that decreased levels of total cholesterol and erythrocyte magnesium are risk factors for ASD[178]. Similarly, increased levels of low-density lipoprotein cholesterol (LDL-C), often referred to as "bad" cholesterol, have been observed in some individuals with ASD[176]. Conversely, reduced maternal postpartum plasma LDL levels were associated with an increased risk of ASD among children born to overweight or obese mothers, as found by Park et al[179]. Additionally, children with autism may exhibit decreased levels of high-density lipoprotein cholesterol (HDL-C), often called "good" cholesterol[180]. Elevated levels of triglycerides have also been observed in some individuals with ASD[181]. Furthermore, an increased ratio of LDL-C to HDL-C, considered a marker of increased cardiovascular risk, has been identified in children with autism[177]. Oxidative stress, a state of imbalance between the production of ROS and the body's antioxidant defenses, is frequently observed in individuals with ASD. This oxidative stress can lead to lipid peroxidation and the generation of oxidized lipids, which are increased in children with autism, suggesting heightened oxidative stress[182].
Several factors may contribute to lipid metabolism abnormalities in children with autism. Genetic variations in lipid metabolism-related genes (e.g., EFR3A4) have been associated with an increased risk of non-syndromic idiopathic ASD. These genetic variants may influence lipid metabolism pathways and contribute to lipid dysregulation. Genetic syndromes of lipid metabolism, such as Smith–Melli–Opitz syndrome, are frequently associated with neurodevelopmental delay[183]. Oxidative stress, characterized by an imbalance between the production of ROS and the body's antioxidant defenses, is often observed in individuals with ASD. Oxidative stress can lead to lipid peroxidation and the generation of oxidized lipids[184]. Chronic inflammation is commonly reported in individuals with ASD and has been associated with altered lipid metabolism. Inflammatory processes can disrupt lipid homeostasis and contribute to dyslipidemia[185]. Emerging evidence suggests a link between gut microbiota composition and lipid metabolism. Alterations in the gut microbiota, which have been observed in some individuals with ASD, may influence lipid metabolism patterns[186]. Dietary factors, such as nutrient composition and quality, may influence lipid metabolism and contribute to lipid abnormalities in children with autism[176].
Since lipids play essential roles in brain development and function, abnormalities in lipid metabolism can have implications for neuronal membrane composition, myelination, synaptogenesis, and neurotransmitter signaling, potentially contributing to the neurodevelopmental abnormalities observed in ASD[187]. Lipid disorders, including dyslipidemia and increased oxidative stress, have been associated with chronic inflammation, a common feature in individuals with ASD[15]. Disruptions in lipid metabolism can also affect cellular energy production and utilization, potentially impacting brain function and development in individuals with ASD[126]. Oxidative stress resulting from lipid disorders can lead to lipid peroxidation and the generation of harmful byproducts, which may contribute to neuroinflammation and neuronal damage. In light of the potential role of lipid metabolism abnormalities in ASD, there is interest in exploring lipid-based interventions as potential therapeutic strategies[188]. However, further research is needed to determine the effectiveness of such interventions and their impact on lipid profiles and ASD symptoms. It is important to note that while there is evidence suggesting associations between lipid disorders and ASD, not all individuals with ASD will have lipid abnormalities, and not all individuals with lipid disorders will have ASD[189]. The relationship between these two conditions is complex and likely influenced by various genetic, environmental, and metabolic factors. Continued research is necessary to fully understand the mechanisms underlying lipid disorders in ASD and their specific effects on the disorder. Table 4 summarizes the prevalence, contributing factors, impact, therapeutic implications, and research needs related to lipid metabolism abnormalities in individuals with ASD.
| Aspect | Lipid metabolism in ASD |
| Overview | Lipid metabolism involves the synthesis, breakdown, and transportation of fats, which are crucial for cell membranes and energy |
| Dyslipidemia in ASD | Abnormal lipid levels observed in children with ASD. Variations include elevated total cholesterol and LDL-C, reduced HDL-C, and increased triglycerides. Increased LDL-C to HDL-C ratio, a marker of cardiovascular risk |
| Factors contributing to abnormalities | Genetic variations in lipid metabolism-related genes. Syndromes like Smith–Melli–Opitz syndrome linked to lipid metabolism and neurodevelopmental delay. Oxidative stress and chronic inflammation are common in ASD, affecting lipid metabolism. Gut microbiota alterations and dietary factors are also implicated |
| Impact on ASD | Abnormal lipid metabolism can affect brain development, myelination, synaptogenesis, and neurotransmitter signaling in ASD. Disruptions may lead to oxidative stress, neuroinflammation, and neuronal damage. Potential implications for cellular energy production and utilization in the brain |
| Therapeutic implications | Interest in lipid-based interventions for ASD, but effectiveness needs further research. Potential therapeutic targets to address lipid disorders and associated symptoms in ASD. Complex relationship between lipid disorders and ASD, influenced by genetic, environmental, and metabolic factors |
| Research needs | Further research is needed to understand the mechanisms underlying lipid disorders in ASD. Investigation required into the effectiveness of lipid-based interventions on ASD symptoms and lipid profiles. Recognition that not all individuals with ASD have lipid abnormalities, and vice versa |
Extensive research has highlighted the potential implications of Omega-3 fatty acids for ASD. These essential fatty acids, which are mostly found in fish oil, have gained a lot of attention due to their supposed effects on neurodevelopment and cognitive function[190]. Omega-3s are a type of polyunsaturated fatty acids that play crucial roles in various biological processes. The two principal types of Omega-3s, eicosapentaenoic acid and docosahexaenoic acid (DHA), are essential components that help maintain neuronal membrane integrity, modulate neurotransmission, regulate inflammation, and promote overall brain health[191]. Omega-3 fatty acids play a crucial role in the early development of the brain as they help in the formation and functioning of neural cells. DHA, which is present in large amounts in the brain, is responsible for creating synapses and forming myelin sheaths around neurons[192]. These are essential processes that enable proper brain connectivity and communication. Additionally, omega-3s impact neurotransmitter signaling, which affects the production and release of key neurotransmitters like serotonin, dopamine, and GABA[193]. These neurotransmitters regulate mood, behavior, and cognitive function, which are often affected in individuals with ASD.
Inflammation and immune dysregulation have been implicated in ASD pathophysiology. Omega-3 fatty acids possess anti-inflammatory properties and can modulate immune responses, potentially mitigating neuroinflammatory processes associated with ASD[194]. While research on omega-3 supplementation's effects on ASD behavioral symptoms yields mixed findings, some studies report enhancements in social interaction, communication, and reduction in repetitive behaviors, emphasizing the variability in individual responses and the necessity for further investigation to delineate consistent effects[195]. Research suggests that inflammation and immune dysregulation may contribute to the development of ASD. Omega-3 fatty acids have anti-inflammatory properties and can regulate immune responses, potentially mitigating neuroinflammatory processes associated with ASD. However, studies on omega-3 supplementation's effects on ASD behavioral symptoms have yielded mixed results[196]. While some studies report impro
ASD frequently co-occurs with conditions like attention deficit hyperactivity disorder (ADHD) and anxiety. Omega-3 fatty acids have been explored for their potential therapeutic benefits in managing these comorbidities, showing promise in reducing ADHD symptoms and enhancing attention, as well as ameliorating anxiety symptoms in individuals with ASD[198]. Nonetheless, further investigation is warranted to validate these findings. Omega-3 fatty acid supplementation is considered safe and well-tolerated[199]. However, caution should be exercised regarding high doses, which may lead to mild side effects such as gastrointestinal disturbances or increased bleeding risk, especially in individuals taking anticoagulant medications[200]. Consequently, consulting with healthcare professionals before initiating any supplementation regimen is advisable to ensure safety and efficacy.
Teeming with diverse bacteria, the gut microbiome significantly influences a child's physical and mental development. This impact is particularly pronounced when the brain undergoes rapid growth during the perinatal period. These gut microbes influence the production of crucial neurotransmitters like GABA and serotonin through the gut-brain axis, playing a vital role in brain wiring and synaptogenesis. Serotonin, primarily produced by gut bacteria, heavily influences brain development and mood regulation. While it can't directly reach the brain, it still sends powerful signals through the peripheral nervous system[201]. Individuals with ASD exhibit a distinct gut microbiome composition in comparison to individuals without the disorder. Research has found that changes in the gut microbiome, including a reduction in microbial diversity, can impact gut health and potentially influence ASD development and symptoms[145]. Some bacterial groups, such as Clostridium, non-spore-forming anaerobes, Faecalibacterium, and microaerophilic bacteria such as Desulfovibrio, consistently increase in number, while beneficial species, such as Bifidobacterium, decrease. These changes can affect the production of metabolites like SCFAs and hydrogen sulfide, which can have both positive and negative effects on the host. Furthermore, it has been observed that the proportion of Desulfovibrio is correlated with the severity of autism symptoms[201,202].
The gut microbiome has a direct communication channel with the brain, known as the gut-brain axis. This two-way pathway involves neural, immune, and endocrine signals. The gut produces various metabolites like SCFAs, neurotransmitters, and neuroactive compounds, which can influence neuronal signaling, inflammation, and immune responses[203]. These effects can ultimately have an impact on behavior and cognition. Moreover, the gut-brain axis plays a significant role in regulating the immune system. Therefore, any disruptions in the gut microbiome can have serious consequences[204]. One such disruption is known as "leaky gut," which occurs when there is increased intestinal permeability. This condition can lead to reduced microbial diversity, pathogen overgrowth, and weakening of the mucus layer. As a result, tight junctions between intestinal cells get disrupted, allowing microbial products and inflammatory molecules to seep into the bloodstream, causing systemic inflammation. This inflammation can potentially affect brain function[205].
Several metabolic changes occur in the gut microbiome of children with ASD. These changes involve the gut bacteria's production and processing of various metabolites. SCFAs, which are produced by the fermentation of dietary fiber by gut bacteria, play important roles in energy metabolism and gut health. Studies have shown that individuals with ASD may have altered levels of SCFAs, including lower levels of butyrate, propionate, and acetate[206]. These changes in SCFA production may have implications for energy metabolism and gut-brain communication. Gut bacteria are involved in the metabolism of amino acids, which are crucial for various physiological processes[207].
Altered amino acid metabolism has been observed in individuals with ASD, where there may be disruptions in the breakdown of certain amino acids, such as tryptophan and phenylalanine, leading to imbalances in their levels. These imbalances can affect neurotransmitter synthesis and potentially contribute to ASD symptoms[208]. Gut bacteria can produce and modulate neurotransmitters like serotonin, dopamine, and GABA. Altered neurotransmitter metabolism in the gut microbiome has been associated with ASD. For instance, imbalances in the production of serotonin, a neurotransmitter involved in mood regulation, have been observed in individuals with ASD. These imbalances may influence brain function and behavior[208].
Gut bacteria can also metabolize bile acids, which play a role in fat digestion and absorption. Studies have found differences in bile acid metabolism in individuals with ASD, including changes in the composition and abundance of specific bile acid species[209]. These alterations may affect fat metabolism and potentially contribute to gastrointestinal symptoms in ASD. Individuals with ASD may have increased oxidative stress, which is an imbalance between the production of reactive oxygen species and the body's ability to neutralize them with antioxidants[210]. The gut microbiome can influence oxidative stress through its metabolic activities. Altered antioxidant metabolism and imbalances in oxidative stress markers have been reported in individuals with ASD, suggesting a potential role of the gut microbiome in these processes[211].
Understanding these gut microbiome changes holds significant clinical weight. Gastrointestinal symptoms like pain, constipation, and diarrhea are common in ASD, and research suggests gut microbiome alterations may contribute[212]. Modulating the gut microbiome through dietary interventions or targeted therapies could potentially alleviate these symptoms and improve overall well-being. Furthermore, evidence suggests a link between gut microbiome alterations and behavioral symptoms in ASD, highlighting the potential of gut-based interventions to improve cognitive and behavioral outcomes[213].
Several dietary modifications targeting the gut microbiome have been studied in individuals with ASD. These interventions aim to alter the composition and function of the gut microbiome and include approaches such as probiotics, prebiotics, and dietary restrictions[211]. Probiotics are live bacteria or yeasts that are believed to have beneficial effects on the gut microbiome. Prebiotics are substances that promote the growth of beneficial bacteria in the gut. Some studies have shown positive effects of probiotic supplementation on gastrointestinal symptoms and behavioral outcomes in individuals with ASD[214]. However, the findings have been inconsistent, and further research is needed to determine the specific strains, dosages, and duration of treatment that may be most effective.
There have been studies conducted on individuals with ASD who have dietary restrictions, such as gluten-free and casein-free diets. Gluten is a protein commonly found in wheat, barley, and rye, while casein is found in milk and dairy products[215]. These diets aim to remove any possible dietary triggers that could worsen gastrointestinal symptoms or affect the behavior of individuals with ASD. Some studies have reported that a subset of individuals with ASD who follow these dietary restrictions have experienced improvements in their behavior and gastrointestinal symptoms[216]. However, the evidence is limited, and further research is necessary to understand the mechanisms better and identify which individuals may benefit from these dietary interventions.
Ongoing research is exploring the role of gut microbiome in ASD. Long-term studies can provide valuable insights into ASD development and progression by examining dynamic changes in the gut microbiome throughout life. Personalized interventions based on individual gut microbiome profiles show promise. However, well-designed clinical trials are crucial to assess the efficacy and safety of microbiome-based therapies and establish evidence-based guidelines for their use in ASD. By integrating gut microbiome research with genetics, epigenetics, and other omics approaches, we can better understand ASD and develop new therapeutic strategies.
A complex interplay of various factors, including age, sex, genetic background, environmental interactions, dietary factors, gut microbiota, metabolic dysregulation, comorbid medical conditions, and pharmacological interventions, influences the rate of metabolomic changes in children with ASD (Figure 2).
The metabolic processes in children change dynamically during childhood, which can impact the rate of metabolomic changes in children with ASD. Studies have shown age-related differences in metabolomic profiles, indicating that metabolic pathways evolve as children grow and develop. For example, Sharma et al[217] showed that children with autism below five years had a greater brain metabolism than older children with autism with a linear decrease in brain metabolism with aging. These differences may be attributed to developmental changes in various metabolic processes, such as energy metabolism, neurotransmitter synthesis, and amino acid metabolism. Understanding the specific metabolic pathways that undergo age-related changes can offer insights into the metabolic dysregulations associated with ASD at different developmental stages[125,218].
Metabolomic profiles in children with ASD can be influenced by both sex and genetic background. Studies in metabolomics have revealed sex-specific differences in the metabolomes of individuals with ASD, suggesting the possibility of sex-specific metabolic dysregulation. For example, Xiong et al[219] found significantly increased adenine, creatinine, 2-methylguanosine, and 7-alpha-hydroxytestololactone levels and reduced creatine in females with ASD. These differences might be attributable to factors such as sex-specific hormonal influences, genetic variations in sex chromosomes, or sex-specific interactions between genes and the environment. Genetic factors also play a significant role in shaping metabolism and metabolomic profiles. Genome-wide association studies have identified genetic variants associated with ASD that are involved in metabolic pathways and neurotransmitter metabolism. Mutations in genes involved in metabolic pathways or regulatory genes can impact the rate and nature of metabolomic changes associated with ASD[71,220].
Environmental factors can interact with genetic factors and affect metabolic changes in children with ASD. Prenatal and postnatal environmental exposures have been found to alter metabolomic profiles. Prenatal factors, like maternal diet, infections, or exposure to toxins, can influence fetal development and program metabolic pathways in the child. Postnatal factors, including dietary patterns, medication exposure, toxins, pollutants, and gut microbiota, can further modify metabolomic profiles. These environmental factors can disrupt metabolic homeostasis, leading to alterations in metabolomic profiles associated with ASD[221,222].
The diet plays a significant role in shaping the metabolome, and the dietary patterns directly impact metabolic profiles in children with ASD. Studies on metabolomics have identified differences in nutrient intake and metabolism between individuals with ASD and typically developing individuals[223,224]. In individuals with ASD, amino acid metabolism, lipid metabolism, and energy metabolism alterations have been observed[225]. The imbalance in essential nutrients or amino acids can lead to metabolic dysregulations and contribute to differences in metabolomic profiles. Additionally, food sensitivities and intolerances that are commonly observed in individuals with ASD can impact metabolomic profiles by influencing nutrient absorption and gut function[226].
Research has shown that the severity of autism symptoms can affect the metabolic profiles of individuals. Specifically, there are associations between the severity of core symptoms of ASD, such as deficits in social communication, restricted and repetitive behaviors, and certain metabolic alterations[30]. Studies have observed differences in metabolite levels and metabolic pathways between individuals with severe ASD and those with milder forms of the condition[126]. It is believed that the severity of ASD symptoms may reflect underlying metabolic dysregulation, which contributes to variations in metabolomic profiles[227]. Differences in cognitive and behavioral patterns found in individuals with ASD can lead to changes in their metabolomic profiles. Specific cognitive abilities, such as language skills or intellectual functioning, have been linked to distinct metabolomic alterations[226]. Moreover, particular behavioral features, such as self-injurious behaviors or hyperactivity, may be associated with specific metabolic dysregulations and metabolomic profiles[228]. In addition, certain behaviors can modify metabolic activity in children with autism. Ranieri et al[229] showed that exercise interventions and physical activity help to improve communication, social interaction and behaviors, and gut in children with autism.
The gut microbiota has emerged as a key metabolism modulator and can influence metabolomic changes in individuals with ASD. Metabolomics studies have identified differences in microbial-derived metabolites between individuals with ASD and neurotypical individuals[11]. Alterations in the gut microbiota composition, known as dysbiosis, are commonly observed in individuals with ASD. Dysbiosis can affect metabolic pathways by producing metabolites, such as short-chain fatty acids, neurotransmitters, and bile acids, which can have systemic effects on the host metabolome[230]. These microbial-derived metabolites can influence immune function, gut barrier integrity, and neurotransmitter synthesis, thereby impacting metabolomic profiles in children with ASD[12].
Metabolomics studies have consistently identified metabolic dysregulations in individuals with ASD. Abnormalities in energy metabolism, oxidative stress, mitochondrial dysfunction, and neurotransmitter metabolism have been reported[144]. These metabolic perturbations can lead to alterations in metabolomic profiles. For example, disruptions in energy metabolism can result in changes in metabolites involved in glucose metabolism, lipid metabolism, and the tricarboxylic acid cycle[231]. Mitochondrial dysfunction can lead to perturbations in energy production and the generation of reactive oxygen species, which can further impact metabolomic profiles through oxidative stress-related pathways[232]. These metabolic dysregulations are thought to contribute to the pathophysiology of ASD and may be linked to the core symptoms and associated comorbidities.
Children with ASD often present with comorbid medical conditions, including gastrointestinal disorders, immune dysregulation, and metabolic disorders. These comorbidities can influence metabolomic profiles and contribute to the heterogeneity of metabolomic changes observed in children with ASD[8]. For example, gastrointestinal disorders, such as inflammatory bowel disease or gastrointestinal inflammation, can alter nutrient absorption and metabolism, leading to changes in metabolomic profiles. Immune dysregulation and chronic inflammation can also impact metabolic pathways and metabolomic profiles[233]. Metabolic disorders, such as abnormalities in lipid metabolism or energy metabolism, can directly affect metabolomic profiles in children with ASD[234].
Pharmacological interventions and treatments commonly used in children with ASD can influence metabolic pathways and contribute to changes in metabolomic profiles[235]. Medications, including psychotropic drugs, may directly impact metabolic processes by modulating neurotransmitter metabolism or affecting energy metabolism[236]. These medications can alter metabolomic profiles by influencing the levels of specific metabolites or by modulating metabolic pathways. Dietary supplements and specialized diets, such as the ketogenic diet or gluten-free/casein-free diet, can also affect metabolomic profiles by altering nutrient availability and metabolic pathways[237]. Behavioral interventions to improve dietary habits or lifestyle factors can indirectly influence metabolism and metabolomic profiles[238].
Understanding these factors and their specific effects on metabolomic profiles can provide valuable insights into the underlying metabolic dysregulations associated with ASD. It may help inform personalized treatment approaches for affected individuals. Further research is needed to elucidate the causal relationships and temporal dynamics between these factors and metabolomic changes in ASD and identify potential biomarkers for early detection and therapeutic monitoring.
Metabolomic profiling is an active area of research in children with ASD, with significant potential for improving diagnostic and therapeutic approaches. By comparing the metabolomic profiles of individuals with ASD to those of typically developing individuals, researchers have identified specific metabolites and metabolic pathways that show consistent differences[239]. These metabolic biomarkers may serve as objective measures to support clinical assessments and improve diagnostic accuracy, aiding in early detection, diagnosis, and subtyping of ASD[240].
Metabolomic analyses provide insights into the underlying metabolic dysregulations associated with ASD. This knowledge can help unravel the complex etiology of the disorder and inform the development of targeted interventions. Metabolomic profiling has the potential to enable personalized medicine approaches in the management of ASD[241]. By characterizing the unique metabolic profiles of individuals with ASD, clinicians may be able to tailor interventions and treatments to address specific metabolic dysregulations, potentially enhancing treatment efficacy and minimizing adverse effects[242].
Metabolomic profiling can also be used to monitor treatment response and assess the effectiveness of interventions in children with ASD[239]. By tracking changes in metabolomic profiles over time, clinicians can evaluate the impact of therapeutic interventions on metabolic dysregulations, guiding treatment adjustments and optimizing therapeutic strategies for individual patients[243]. The identified metabolic pathways may be potential therapeutic targets for developing novel interventions. Modulating the identified metabolic pathways through dietary modifications, supplementation, or pharmacological interventions may help restore metabolic balance and improve ASD symptoms[244].
Metabolomic profiling may also provide insights into the metabolic alterations associated with comorbid conditions commonly observed in individuals with ASD, such as gastrointestinal issues, epilepsy, or sleep disturbances[226]. Understanding these metabolic connections can guide the management of co-occurring conditions and inform strategies for comprehensive care[8]. Additionally, examining the metabolomic profiles of children with ASD at an early age may identify markers associated with different developmental trajectories, guiding early intervention strategies and providing prognostic indicators for personalized treatment planning[245]. While metabolomic profiling shows great potential, further research is needed to validate findings, establish standardized protocols, and overcome technical and analytical challenges. Nonetheless, the integration of metabolomic changes in children with ASD has the potential to enhance diagnostic accuracy, inform personalized treatment approaches, and improve outcomes for individuals with ASD[239].
Despite the comprehensive nature of this systematic review, several limitations should be acknowledged. Firstly, the inclusion criteria focused specifically on metabolomic changes in children with ASD, potentially excluding relevant studies that examined metabolomic alterations in other age groups or neurodevelopmental disorders. Additionally, although efforts were made to ensure a thorough search across various electronic databases up until January 2024, it is possible that some relevant studies may have been missed. The reliance on electronic databases may have also introduced publication bias, as studies not indexed in these databases were not included. Furthermore, the heterogeneity among the included studies, such as differences in methodology, sample size, and study design, may have influenced the interpretation and synthesis of results. Moreover, while the review aimed to provide a comprehensive overview of metabolomic changes in ASD, the complexity of metabolomic data and the diversity of factors influencing metabolism necessitate cautious interpretation. Finally, the absence of a quality assessment of included studies and the lack of meta-analytical techniques limit the robustness of the synthesized findings. Despite these limitations, this review provides valuable insights into the current understanding of metabolomic alterations in children with ASD and highlights avenues for future research.
Although we tried our best to conduct a comprehensive systematic review, it has several limitations that need to be acknowledged. Firstly, the inclusion criteria only focused on metabolomic changes in children with ASD, which means that relevant studies examining metabolomic alterations in other age groups or neurodevelopmental disorders may have been excluded. Additionally, even though efforts were made to ensure a thorough search across various electronic databases up until January 2024, it is likely that some relevant studies may have been missed. This reliance on electronic databases may have also introduced publication bias, as studies not indexed in these databases were not included. Furthermore, heterogeneity among the included studies, including differences in methodology, sample size, and study design, may have influenced the interpretation and synthesis of results. While the review aimed to provide a comprehensive systematic overview of metabolomic changes in ASD, the complexity of metabolomic data and the diversity of factors influencing metabolism necessitate cautious interpretation. Finally, the absence of a quality assessment of included studies and the lack of meta-analytical techniques limit the robustness of the synthesized findings. Nevertheless, this systematic review provides valuable insights into the current understanding of metabolomic alterations in children with ASD and highlights areas for future research.
This systematic review provides an in-depth analysis of the changes in metabolism that occur in children with ASD. It combines the findings of various research studies, including reviews, meta-analyses, systematic reviews, case reports, editorials, and a book chapter, to offer a comprehensive overview of the topic. The review highlights the complex factors that contribute to metabolic changes in ASD and identifies potential biomarkers that could be used for diagnosis and personalized treatment. However, the review also acknowledges that there are limitations to the available literature, such as variations in study design and the possibility of publication bias. Despite these limitations, the review emphasizes the importance of further research to validate the findings, establish standardized protocols, and overcome technical challenges. By incorporating metabolomic changes into clinical practice, healthcare professionals can improve diagnosing, treating, and monitoring children with ASD. The review concludes that continued research into metabolomic alterations in ASD holds promise for advancing our understanding of the disorder and improving the lives of affected individuals and their families.
We thank the anonymous referees and editors for their valuable suggestions.
| 1. | Chen H, Rosen CE, González-Hernández JA, Song D, Potempa J, Ring AM, Palm NW. Highly multiplexed bioactivity screening reveals human and microbiota metabolome-GPCRome interactions. Cell. 2023;186:3095-3110.e19. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 38] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 2. | Eghbalnia HR, Romero PR, Westler WM, Baskaran K, Ulrich EL, Markley JL. Increasing rigor in NMR-based metabolomics through validated and open source tools. Curr Opin Biotechnol. 2017;43:56-61. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 16] [Cited by in RCA: 17] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 3. | Robinson O, Lau CE. How do metabolic processes age: Evidence from human metabolomic studies. Curr Opin Chem Biol. 2023;76:102360. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 4. | Qiu S, Cai Y, Yao H, Lin C, Xie Y, Tang S, Zhang A. Small molecule metabolites: discovery of biomarkers and therapeutic targets. Signal Transduct Target Ther. 2023;8:132. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 168] [Cited by in RCA: 329] [Article Influence: 164.5] [Reference Citation Analysis (0)] |
| 5. | Gonzalez-Covarrubias V, Martínez-Martínez E, Del Bosque-Plata L. The Potential of Metabolomics in Biomedical Applications. Metabolites. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 8] [Cited by in RCA: 102] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 6. | Balashova EE, Maslov DL, Trifonova OP, Lokhov PG, Archakov AI. Metabolome Profiling in Aging Studies. Biology (Basel). 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 7. | Tounta V, Liu Y, Cheyne A, Larrouy-Maumus G. Metabolomics in infectious diseases and drug discovery. Mol Omics. 2021;17:376-393. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 72] [Cited by in RCA: 71] [Article Influence: 17.8] [Reference Citation Analysis (0)] |
| 8. | Al-Beltagi M. Autism medical comorbidities. World J Clin Pediatr. 2021;10:15-28. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 151] [Cited by in RCA: 134] [Article Influence: 33.5] [Reference Citation Analysis (12)] |
| 9. | Liu A, Zhou W, Qu L, He F, Wang H, Wang Y, Cai C, Li X, Wang M. Altered Urinary Amino Acids in Children With Autism Spectrum Disorders. Front Cell Neurosci. 2019;13:7. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 28] [Cited by in RCA: 43] [Article Influence: 7.2] [Reference Citation Analysis (0)] |
| 10. | Siddiqui MF, Elwell C, Johnson MH. Mitochondrial Dysfunction in Autism Spectrum Disorders. Autism Open Access. 2016;6. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 52] [Cited by in RCA: 71] [Article Influence: 7.9] [Reference Citation Analysis (0)] |
| 11. | Mehra A, Arora G, Sahni G, Kaur M, Singh H, Singh B, Kaur S. Gut microbiota and Autism Spectrum Disorder: From pathogenesis to potential therapeutic perspectives. J Tradit Complement Med. 2023;13:135-149. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 45] [Reference Citation Analysis (0)] |
| 12. | Peralta-Marzal LN, Prince N, Bajic D, Roussin L, Naudon L, Rabot S, Garssen J, Kraneveld AD, Perez-Pardo P. The Impact of Gut Microbiota-Derived Metabolites in Autism Spectrum Disorders. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 23] [Cited by in RCA: 33] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 13. | Dai S, Lin J, Hou Y, Luo X, Shen Y, Ou J. Purine signaling pathway dysfunction in autism spectrum disorders: Evidence from multiple omics data. Front Mol Neurosci. 2023;16:1089871. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 13] [Reference Citation Analysis (0)] |
| 14. | Brigandi SA, Shao H, Qian SY, Shen Y, Wu BL, Kang JX. Autistic children exhibit decreased levels of essential Fatty acids in red blood cells. Int J Mol Sci. 2015;16:10061-10076. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 15. | Liu X, Lin J, Zhang H, Khan NU, Zhang J, Tang X, Cao X, Shen L. Oxidative Stress in Autism Spectrum Disorder-Current Progress of Mechanisms and Biomarkers. Front Psychiatry. 2022;13:813304. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 16. | Smith AM, Donley ELR, Ney DM, Amaral DG, Burrier RE, Natowicz MR. Metabolomic biomarkers in autism: identification of complex dysregulations of cellular bioenergetics. Front Psychiatry. 2023;14:1249578. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 17. | Al-Beltagi M. Pre-autism: What a paediatrician should know about early diagnosis of autism. World J Clin Pediatr. 2023;12:273-294. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 1] [Reference Citation Analysis (0)] |
| 18. | Elbeltagi R, Al-Beltagi M, Saeed NK, Alhawamdeh R. Play therapy in children with autism: Its role, implications, and limitations. World J Clin Pediatr. 2023;12:1-22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 33] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (14)] |
| 19. | Chaste P, Leboyer M. Autism risk factors: genes, environment, and gene-environment interactions. Dialogues Clin Neurosci. 2012;14:281-292. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 355] [Cited by in RCA: 440] [Article Influence: 36.7] [Reference Citation Analysis (0)] |
| 20. | Escher J, Yan W, Rissman EF, Wang HV, Hernandez A, Corces VG. Beyond Genes: Germline Disruption in the Etiology of Autism Spectrum Disorders. J Autism Dev Disord. 2022;52:4608-4624. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 21. | Eshraghi AA, Liu G, Kay SS, Eshraghi RS, Mittal J, Moshiree B, Mittal R. Epigenetics and Autism Spectrum Disorder: Is There a Correlation? Front Cell Neurosci. 2018;12:78. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 58] [Article Influence: 8.3] [Reference Citation Analysis (0)] |
| 22. | Lyall K, Schmidt RJ, Hertz-Picciotto I. Maternal lifestyle and environmental risk factors for autism spectrum disorders. Int J Epidemiol. 2014;43:443-464. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 245] [Cited by in RCA: 281] [Article Influence: 25.5] [Reference Citation Analysis (0)] |
| 23. | Karimi P, Kamali E, Mousavi SM, Karahmadi M. Environmental factors influencing the risk of autism. J Res Med Sci. 2017;22:27. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 137] [Cited by in RCA: 127] [Article Influence: 15.9] [Reference Citation Analysis (0)] |
| 24. | Quaak I, Brouns MR, Van de Bor M. The dynamics of autism spectrum disorders: how neurotoxic compounds and neurotransmitters interact. Int J Environ Res Public Health. 2013;10:3384-3408. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 54] [Cited by in RCA: 64] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 25. | Robinson-Agramonte MLA, Noris García E, Fraga Guerra J, Vega Hurtado Y, Antonucci N, Semprún-Hernández N, Schultz S, Siniscalco D. Immune Dysregulation in Autism Spectrum Disorder: What Do We Know about It? Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 26. | Maximo JO, Cadena EJ, Kana RK. The implications of brain connectivity in the neuropsychology of autism. Neuropsychol Rev. 2014;24:16-31. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 198] [Cited by in RCA: 174] [Article Influence: 15.8] [Reference Citation Analysis (0)] |
| 27. | Zikopoulos B, Barbas H. Altered neural connectivity in excitatory and inhibitory cortical circuits in autism. Front Hum Neurosci. 2013;7:609. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 185] [Cited by in RCA: 216] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 28. | Marco EJ, Hinkley LB, Hill SS, Nagarajan SS. Sensory processing in autism: a review of neurophysiologic findings. Pediatr Res. 2011;69:48R-54R. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 779] [Cited by in RCA: 619] [Article Influence: 44.2] [Reference Citation Analysis (0)] |
| 29. | Al-Beltagi M, Saeed NK, Bediwy AS, Elbeltagi R, Alhawamdeh R. Role of gastrointestinal health in managing children with autism spectrum disorder. World J Clin Pediatr. 2023;12:171-196. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 23] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (4)] |
| 30. | Adams JB, Audhya T, McDonough-Means S, Rubin RA, Quig D, Geis E, Gehn E, Loresto M, Mitchell J, Atwood S, Barnhouse S, Lee W. Nutritional and metabolic status of children with autism vs. neurotypical children, and the association with autism severity. Nutr Metab (Lond). 2011;8:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 281] [Cited by in RCA: 270] [Article Influence: 19.3] [Reference Citation Analysis (0)] |
| 31. | Jensen AR, Lane AL, Werner BA, McLees SE, Fletcher TS, Frye RE. Modern Biomarkers for Autism Spectrum Disorder: Future Directions. Mol Diagn Ther. 2022;26:483-495. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 38] [Reference Citation Analysis (0)] |
| 32. | Senarathne UD, Indika NR, Jezela-Stanek A, Ciara E, Frye RE, Chen C, Stepien KM. Biochemical, Genetic and Clinical Diagnostic Approaches to Autism-Associated Inherited Metabolic Disorders. Genes (Basel). 2023;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 33. | Smith AM, Natowicz MR, Braas D, Ludwig MA, Ney DM, Donley ELR, Burrier RE, Amaral DG. A Metabolomics Approach to Screening for Autism Risk in the Children's Autism Metabolome Project. Autism Res. 2020;13:1270-1285. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 17] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 34. | Nisar S, Bhat AA, Masoodi T, Hashem S, Akhtar S, Ali TA, Amjad S, Chawla S, Bagga P, Frenneaux MP, Reddy R, Fakhro K, Haris M. Genetics of glutamate and its receptors in autism spectrum disorder. Mol Psychiatry. 2022;27:2380-2392. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 83] [Article Influence: 27.7] [Reference Citation Analysis (0)] |
| 35. | Richard DM, Dawes MA, Mathias CW, Acheson A, Hill-Kapturczak N, Dougherty DM. L-Tryptophan: Basic Metabolic Functions, Behavioral Research and Therapeutic Indications. Int J Tryptophan Res. 2009;2:45-60. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 469] [Cited by in RCA: 409] [Article Influence: 25.6] [Reference Citation Analysis (0)] |
| 36. | Gao K, Mu CL, Farzi A, Zhu WY. Tryptophan Metabolism: A Link Between the Gut Microbiota and Brain. Adv Nutr. 2020;11:709-723. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 170] [Cited by in RCA: 502] [Article Influence: 100.4] [Reference Citation Analysis (0)] |
| 37. | Campbell BM, Charych E, Lee AW, Möller T. Kynurenines in CNS disease: regulation by inflammatory cytokines. Front Neurosci. 2014;8:12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 218] [Cited by in RCA: 285] [Article Influence: 25.9] [Reference Citation Analysis (0)] |
| 38. | Kałużna-Czaplińska J, Jóźwik-Pruska J, Chirumbolo S, Bjørklund G. Tryptophan status in autism spectrum disorder and the influence of supplementation on its level. Metab Brain Dis. 2017;32:1585-1593. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 43] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 39. | Muller CL, Anacker AMJ, Veenstra-VanderWeele J. The serotonin system in autism spectrum disorder: From biomarker to animal models. Neuroscience. 2016;321:24-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 350] [Cited by in RCA: 366] [Article Influence: 40.7] [Reference Citation Analysis (0)] |
| 40. | Boccuto L, Chen CF, Pittman AR, Skinner CD, McCartney HJ, Jones K, Bochner BR, Stevenson RE, Schwartz CE. Decreased tryptophan metabolism in patients with autism spectrum disorders. Mol Autism. 2013;4:16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 98] [Cited by in RCA: 118] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 41. | Ormstad H, Bryn V, Verkerk R, Skjeldal OH, Halvorsen B, Saugstad OD, Isaksen J, Maes M. Serum Tryptophan, Tryptophan Catabolites and Brain-derived Neurotrophic Factor in Subgroups of Youngsters with Autism Spectrum Disorders. CNS Neurol Disord Drug Targets. 2018;17:626-639. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 14] [Cited by in RCA: 26] [Article Influence: 4.3] [Reference Citation Analysis (0)] |
| 42. | Almulla AF, Thipakorn Y, Tunvirachaisakul C, Maes M. The tryptophan catabolite or kynurenine pathway in autism spectrum disorder; a systematic review and meta-analysis. Autism Res. 2023;16:2302-2315. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 9] [Reference Citation Analysis (0)] |
| 43. | Tsuji A, Ikeda Y, Yoshikawa S, Taniguchi K, Sawamura H, Morikawa S, Nakashima M, Asai T, Matsuda S. The Tryptophan and Kynurenine Pathway Involved in the Development of Immune-Related Diseases. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 60] [Reference Citation Analysis (0)] |
| 44. | Myint AM, Halaris A. Imbalances in Kynurenines as Potential Biomarkers in the Diagnosis and Treatment of Psychiatric Disorders. Front Psychiatry. 2022;13:913303. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 45. | Garbarino VR, Gilman TL, Daws LC, Gould GG. Extreme enhancement or depletion of serotonin transporter function and serotonin availability in autism spectrum disorder. Pharmacol Res. 2019;140:85-99. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 64] [Cited by in RCA: 59] [Article Influence: 9.8] [Reference Citation Analysis (0)] |
| 46. | Chugani DC. Serotonin in autism and pediatric epilepsies. Ment Retard Dev Disabil Res Rev. 2004;10:112-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 62] [Cited by in RCA: 60] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 47. | Zhao F, Zhang H, Wang P, Cui W, Xu K, Chen D, Hu M, Li Z, Geng X, Wei S. Oxytocin and serotonin in the modulation of neural function: Neurobiological underpinnings of autism-related behavior. Front Neurosci. 2022;16:919890. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 48. | Abdulamir HA, Abdul-Rasheed OF, Abdulghani EA. Serotonin and serotonin transporter levels in autistic children. Saudi Med J. 2018;39:487-494. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 29] [Cited by in RCA: 43] [Article Influence: 6.1] [Reference Citation Analysis (0)] |
| 49. | Weiss LA, Abney M, Cook EH Jr, Ober C. Sex-specific genetic architecture of whole blood serotonin levels. Am J Hum Genet. 2005;76:33-41. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 106] [Cited by in RCA: 108] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 50. | Patrick RP, Ames BN. Vitamin D hormone regulates serotonin synthesis. Part 1: relevance for autism. FASEB J. 2014;28:2398-2413. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 212] [Cited by in RCA: 271] [Article Influence: 24.6] [Reference Citation Analysis (0)] |
| 51. | Carneiro AM, Cook EH, Murphy DL, Blakely RD. Interactions between integrin alphaIIbbeta3 and the serotonin transporter regulate serotonin transport and platelet aggregation in mice and humans. J Clin Invest. 2008;118:1544-1552. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 139] [Cited by in RCA: 142] [Article Influence: 8.4] [Reference Citation Analysis (0)] |
| 52. | Alon R. Predicting typically-developing siblings’ acceptance of their sibling with ASD during emerging adulthood. Res Autism Spectr Disord. 2022;99:102065. [DOI] [Full Text] |
| 53. | Gabriele S, Sacco R, Persico AM. Blood serotonin levels in autism spectrum disorder: a systematic review and meta-analysis. Eur Neuropsychopharmacol. 2014;24:919-929. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 280] [Cited by in RCA: 232] [Article Influence: 21.1] [Reference Citation Analysis (0)] |
| 54. | Lee A, Choo H, Jeon B. Serotonin Receptors as Therapeutic Targets for Autism Spectrum Disorder Treatment. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 55. | Agus A, Planchais J, Sokol H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe. 2018;23:716-724. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 834] [Cited by in RCA: 1802] [Article Influence: 300.3] [Reference Citation Analysis (1)] |
| 56. | Higazi AM, Kamel HM, Abdel-Naeem EA, Abdullah NM, Mahrous DM, Osman AM. Expression analysis of selected genes involved in tryptophan metabolic pathways in Egyptian children with Autism Spectrum Disorder and learning disabilities. Sci Rep. 2021;11:6931. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 12] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 57. | Tărlungeanu DC, Deliu E, Dotter CP, Kara M, Janiesch PC, Scalise M, Galluccio M, Tesulov M, Morelli E, Sonmez FM, Bilguvar K, Ohgaki R, Kanai Y, Johansen A, Esharif S, Ben-Omran T, Topcu M, Schlessinger A, Indiveri C, Duncan KE, Caglayan AO, Gunel M, Gleeson JG, Novarino G. Impaired Amino Acid Transport at the Blood Brain Barrier Is a Cause of Autism Spectrum Disorder. Cell. 2016;167:1481-1494.e18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 178] [Cited by in RCA: 258] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 58. | Yap SH, Hafkenscheid JC, van Tongeren JH. Important role of tryptophan on albumin synthesis in patients suffering from anorexia nervosa and hypoalbuminemia. Am J Clin Nutr. 1975;28:1356-1363. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 10] [Article Influence: 0.2] [Reference Citation Analysis (0)] |
| 59. | Schopman SME, Bosman RC, Muntingh ADT, van Balkom AJLM, Batelaan NM. Effects of tryptophan depletion on anxiety, a systematic review. Transl Psychiatry. 2021;11:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 4] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 60. | Elmenhorst D, Kroll T, Matusch A, Bauer A. Sleep deprivation increases cerebral serotonin 2A receptor binding in humans. Sleep. 2012;35:1615-1623. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 48] [Cited by in RCA: 58] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 61. | Eskelund A, Li Y, Budac DP, Müller HK, Gulinello M, Sanchez C, Wegener G. Drugs with antidepressant properties affect tryptophan metabolites differently in rodent models with depression-like behavior. J Neurochem. 2017;142:118-131. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 24] [Cited by in RCA: 36] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 62. | Aliu E, Kanungo S, Arnold GL. Amino acid disorders. Ann Transl Med. 2018;6:471. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 47] [Cited by in RCA: 81] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 63. | Bloemendaal M, Froböse MI, Wegman J, Zandbelt BB, van de Rest O, Cools R, Aarts E. Neuro-Cognitive Effects of Acute Tyrosine Administration on Reactive and Proactive Response Inhibition in Healthy Older Adults. eNeuro. 2018;5. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 64. | Williams RA, Mamotte CD, Burnett JR. Phenylketonuria: an inborn error of phenylalanine metabolism. Clin Biochem Rev. 2008;29:31-41. [PubMed] |
| 65. | Randazzo M, Prato A, Messina M, Meli C, Casabona A, Rizzo R, Barone R. Neuroactive Amino Acid Profile in Autism Spectrum Disorder: Results from a Clinical Sample. Children (Basel). 2023;10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 66. | Naushad SM, Jain JM, Prasad CK, Naik U, Akella RR. Autistic children exhibit distinct plasma amino acid profile. Indian J Biochem Biophys. 2013;50:474-478. [PubMed] |
| 67. | Mandic-Maravic V, Grujicic R, Milutinovic L, Munjiza-Jovanovic A, Pejovic-Milovancevic M. Dopamine in Autism Spectrum Disorders-Focus on D2/D3 Partial Agonists and Their Possible Use in Treatment. Front Psychiatry. 2021;12:787097. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 35] [Article Influence: 8.8] [Reference Citation Analysis (0)] |
| 68. | Clayton TA. Metabolic differences underlying two distinct rat urinary phenotypes, a suggested role for gut microbial metabolism of phenylalanine and a possible connection to autism. FEBS Lett. 2012;586:956-961. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 68] [Article Influence: 5.2] [Reference Citation Analysis (0)] |
| 69. | McGinnity CJ, Riaño Barros DA, Guedj E, Girard N, Symeon C, Walker H, Barrington SF, Summers M, Pitkanen M, Rahman Y. A Retrospective Case Series Analysis of the Relationship Between Phenylalanine: Tyrosine Ratio and Cerebral Glucose Metabolism in Classical Phenylketonuria and Hyperphenylalaninemia. Front Neurosci. 2021;15:664525. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 70. | Aldred S, Moore KM, Fitzgerald M, Waring RH. Plasma amino acid levels in children with autism and their families. J Autism Dev Disord. 2003;33:93-97. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 126] [Cited by in RCA: 131] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 71. | Arum P, Amareta DI, Zannah F. Phenylalanine and Tryptophan Intake of Hyperactive Children with Autism. J Biomed Transl Res. 2017;3:34. [RCA] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.1] [Reference Citation Analysis (0)] |
| 72. | Žigman T, Petković Ramadža D, Šimić G, Barić I. Inborn Errors of Metabolism Associated With Autism Spectrum Disorders: Approaches to Intervention. Front Neurosci. 2021;15:673600. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 73. | Almandil NB, Alkuroud DN, AbdulAzeez S, AlSulaiman A, Elaissari A, Borgio JF. Environmental and Genetic Factors in Autism Spectrum Disorders: Special Emphasis on Data from Arabian Studies. Int J Environ Res Public Health. 2019;16. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 33] [Cited by in RCA: 58] [Article Influence: 9.7] [Reference Citation Analysis (0)] |
| 74. | Parkhitko AA, Jouandin P, Mohr SE, Perrimon N. Methionine metabolism and methyltransferases in the regulation of aging and lifespan extension across species. Aging Cell. 2019;18:e13034. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 155] [Cited by in RCA: 174] [Article Influence: 29.0] [Reference Citation Analysis (0)] |
| 75. | Škovierová H, Vidomanová E, Mahmood S, Sopková J, Drgová A, Červeňová T, Halašová E, Lehotský J. The Molecular and Cellular Effect of Homocysteine Metabolism Imbalance on Human Health. Int J Mol Sci. 2016;17. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 206] [Cited by in RCA: 283] [Article Influence: 31.4] [Reference Citation Analysis (0)] |
| 76. | Corona-Trejo A, Gonsebatt ME, Trejo-Solis C, Campos-Peña V, Quintas-Granados LI, Villegas-Vázquez EY, Daniel Reyes-Hernández O, Hernández-Abad VJ, Figueroa-González G, Silva-Adaya D. Transsulfuration pathway: a targeting neuromodulator in Parkinson's disease. Rev Neurosci. 2023;34:915-932. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 77. | Ouyang Y, Wu Q, Li J, Sun S. S-adenosylmethionine: A metabolite critical to the regulation of autophagy. Cell Prolif. 2020;53:e12891. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 67] [Cited by in RCA: 90] [Article Influence: 18.0] [Reference Citation Analysis (0)] |
| 78. | Stipanuk MH, Ueki I. Dealing with methionine/homocysteine sulfur: cysteine metabolism to taurine and inorganic sulfur. J Inherit Metab Dis. 2011;34:17-32. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 402] [Cited by in RCA: 338] [Article Influence: 24.1] [Reference Citation Analysis (0)] |
| 79. | Ganguly P, Alam SF. Role of homocysteine in the development of cardiovascular disease. Nutr J. 2015;14:6. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 490] [Cited by in RCA: 674] [Article Influence: 67.4] [Reference Citation Analysis (0)] |
| 80. | Melnyk S, Fuchs GJ, Schulz E, Lopez M, Kahler SG, Fussell JJ, Bellando J, Pavliv O, Rose S, Seidel L, Gaylor DW, James SJ. Metabolic imbalance associated with methylation dysregulation and oxidative damage in children with autism. J Autism Dev Disord. 2012;42:367-377. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 199] [Cited by in RCA: 175] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 81. | Xie J, Xie L, Wei H, Li XJ, Lin L. Dynamic Regulation of DNA Methylation and Brain Functions. Biology (Basel). 2023;12. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 22] [Cited by in RCA: 22] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 82. | Fukumoto K, Ito K, Saer B, Taylor G, Ye S, Yamano M, Toriba Y, Hayes A, Okamura H, Fustin JM. Excess S-adenosylmethionine inhibits methylation via catabolism to adenine. Commun Biol. 2022;5:313. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 83. | Indika NR, Deutz NEP, Engelen MPKJ, Peiris H, Wijetunge S, Perera R. Sulfur amino acid metabolism and related metabotypes of autism spectrum disorder: A review of biochemical evidence for a hypothesis. Biochimie. 2021;184:143-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 5] [Cited by in RCA: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 84. | Geier DA, Geier MR. A clinical and laboratory evaluation of methionine cycle-transsulfuration and androgen pathway markers in children with autistic disorders. Horm Res. 2006;66:182-188. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 53] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 85. | Suh JH, Walsh WJ, Mcginnis WR, Lewis A, Ames BN. Altered Sulfur Amino Acid Metabolism In Immune Cells of Children Diagnosed With Autism. American J of Biochemistry and Biotechnology. 2008;4:105-113. [RCA] [DOI] [Full Text] [Cited by in Crossref: 26] [Cited by in RCA: 27] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 86. | Guo BQ, Ding SB, Li HB. Blood biomarker levels of methylation capacity in autism spectrum disorder: a systematic review and meta-analysis. Acta Psychiatr Scand. 2020;141:492-509. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 21] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 87. | Roufael M, Bitar T, Sacre Y, Andres C, Hleihel W. Folate-Methionine Cycle Disruptions in ASD Patients and Possible Interventions: A Systematic Review. Genes (Basel). 2023;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 88. | Tremblay MW, Jiang YH. DNA Methylation and Susceptibility to Autism Spectrum Disorder. Annu Rev Med. 2019;70:151-166. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 83] [Article Influence: 13.8] [Reference Citation Analysis (0)] |
| 89. | Haghiri R, Mashayekhi F, Bidabadi E, Salehi Z. Analysis of methionine synthase (rs1805087) gene polymorphism in autism patients in Northern Iran. Acta Neurobiol Exp (Wars). 2016;76:318-323. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 11] [Cited by in RCA: 21] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 90. | Ajabi S, Mashayekhi F, Bidabadi E. A study of MTRR 66A>G gene polymorphism in patients with autism from northern Iran. Neurol Asia. 2017;22:59-64. |
| 91. | Zhang J, Ma X, Su Y, Wang L, Shang S, Yue W. Association Study of MTHFR C677T Polymorphism and Birth Body Mass With Risk of Autism in Chinese Han Population. Front Psychiatry. 2021;12:560948. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 92. | Li CX, Liu YG, Che YP, Ou JL, Ruan WC, Yu YL, Li HF. Association Between MTHFR C677T Polymorphism and Susceptibility to Autism Spectrum Disorders: A Meta-Analysis in Chinese Han Population. Front Pediatr. 2021;9:598805. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 93. | Sener EF, Oztop DB, Ozkul Y. MTHFR Gene C677T Polymorphism in Autism Spectrum Disorders. Genet Res Int. 2014;2014:698574. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 23] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 94. | Radoeva PD, Coman IL, Salazar CA, Gentile KL, Higgins AM, Middleton FA, Antshel KM, Fremont W, Shprintzen RJ, Morrow BE, Kates WR. Association between autism spectrum disorder in individuals with velocardiofacial (22q11.2 deletion) syndrome and PRODH and COMT genotypes. Psychiatr Genet. 2014;24:269-272. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 23] [Cited by in RCA: 26] [Article Influence: 2.6] [Reference Citation Analysis (0)] |
| 95. | Gadow KD, Roohi J, DeVincent CJ, Kirsch S, Hatchwell E. Association of COMT (Val158Met) and BDNF (Val66Met) gene polymorphisms with anxiety, ADHD and tics in children with autism spectrum disorder. J Autism Dev Disord. 2009;39:1542-1551. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 52] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 96. | Uddin MG, Siddiqui SA, Uddin MS, Aziz MA, Hussain MS, Furhatun-noor, Millat MS, Sen N, Muhuri B, Islam MS. Genetic variants of ZNF385B and COMT are associated with autism spectrum disorder in the Bangladeshi children. Meta Gene. 2020;26:100820. [RCA] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 6] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 97. | Rossignol DA, Frye RE. Cerebral Folate Deficiency, Folate Receptor Alpha Autoantibodies and Leucovorin (Folinic Acid) Treatment in Autism Spectrum Disorders: A Systematic Review and Meta-Analysis. J Pers Med. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 43] [Article Influence: 10.8] [Reference Citation Analysis (0)] |
| 98. | Hartzell S, Seneff S. Impaired Sulfate Metabolism and Epigenetics: Is There a Link in Autism? Entropy. 2012;14:1953-1977. [RCA] [DOI] [Full Text] [Cited by in Crossref: 15] [Cited by in RCA: 15] [Article Influence: 1.2] [Reference Citation Analysis (0)] |
| 99. | Ungvari A, Gulej R, Csik B, Mukli P, Negri S, Tarantini S, Yabluchanskiy A, Benyo Z, Csiszar A, Ungvari Z. The Role of Methionine-Rich Diet in Unhealthy Cerebrovascular and Brain Aging: Mechanisms and Implications for Cognitive Impairment. Nutrients. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 10] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 100. | Lyon P, Strippoli V, Fang B, Cimmino L. B Vitamins and One-Carbon Metabolism: Implications in Human Health and Disease. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 120] [Cited by in RCA: 197] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 101. | Wu X, Han Z, Liu B, Yu D, Sun J, Ge L, Tang W, Liu S. Gut microbiota contributes to the methionine metabolism in host. Front Microbiol. 2022;13:1065668. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 17] [Reference Citation Analysis (0)] |
| 102. | Lee S, Olsen T, Vinknes KJ, Refsum H, Gulseth HL, Birkeland KI, Drevon CA. Plasma Sulphur-Containing Amino Acids, Physical Exercise and Insulin Sensitivity in Overweight Dysglycemic and Normal Weight Normoglycemic Men. Nutrients. 2018;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 33] [Article Influence: 4.7] [Reference Citation Analysis (0)] |
| 103. | Poplawski J, Radmilovic A, Montina TD, Metz GAS. Cardiorenal metabolic biomarkers link early life stress to risk of non-communicable diseases and adverse mental health outcomes. Sci Rep. 2020;10:13295. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 20] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 104. | Ledda C, Cannizzaro E, Lovreglio P, Vitale E, Stufano A, Montana A, Li Volti G, Rapisarda V. Exposure to Toxic Heavy Metals Can Influence Homocysteine Metabolism? Antioxidants (Basel). 2019;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 13] [Cited by in RCA: 15] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 105. | Le Magueresse-Battistoni B, Vidal H, Naville D. Environmental Pollutants and Metabolic Disorders: The Multi-Exposure Scenario of Life. Front Endocrinol (Lausanne). 2018;9:582. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 66] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 106. | Fumagalli M, Lecca D, Abbracchio MP, Ceruti S. Pathophysiological Role of Purines and Pyrimidines in Neurodevelopment: Unveiling New Pharmacological Approaches to Congenital Brain Diseases. Front Pharmacol. 2017;8:941. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 56] [Cited by in RCA: 80] [Article Influence: 10.0] [Reference Citation Analysis (0)] |
| 107. | Löffler M, Carrey EA, Zameitat E. Orotic Acid, More Than Just an Intermediate of Pyrimidine de novo Synthesis. J Genet Genomics. 2015;42:207-219. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 45] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 108. | Harris JJ, Jolivet R, Attwell D. Synaptic energy use and supply. Neuron. 2012;75:762-777. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 894] [Cited by in RCA: 1093] [Article Influence: 84.1] [Reference Citation Analysis (0)] |
| 109. | Pasquini S, Contri C, Merighi S, Gessi S, Borea PA, Varani K, Vincenzi F. Adenosine Receptors in Neuropsychiatric Disorders: Fine Regulators of Neurotransmission and Potential Therapeutic Targets. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 110. | Pasquini S, Contri C, Borea PA, Vincenzi F, Varani K. Adenosine and Inflammation: Here, There and Everywhere. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 111. | Shen HY, Huang N, Reemmer J, Xiao L. Adenosine Actions on Oligodendroglia and Myelination in Autism Spectrum Disorder. Front Cell Neurosci. 2018;12:482. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 11] [Cited by in RCA: 15] [Article Influence: 2.1] [Reference Citation Analysis (0)] |
| 112. | Tanimura Y, Vaziri S, Lewis MH. Indirect basal ganglia pathway mediation of repetitive behavior: attenuation by adenosine receptor agonists. Behav Brain Res. 2010;210:116-122. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 73] [Cited by in RCA: 68] [Article Influence: 4.5] [Reference Citation Analysis (0)] |
| 113. | Cristalli G, Costanzi S, Lambertucci C, Lupidi G, Vittori S, Volpini R, Camaioni E. Adenosine deaminase: functional implications and different classes of inhibitors. Med Res Rev. 2001;21:105-128. [PubMed] [DOI] [Full Text] |
| 114. | Bottini N, De Luca D, Saccucci P, Fiumara A, Elia M, Porfirio MC, Lucarelli P, Curatolo P. Autism: evidence of association with adenosine deaminase genetic polymorphism. Neurogenetics. 2001;3:111-113. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 45] [Article Influence: 1.9] [Reference Citation Analysis (0)] |
| 115. | Mussap M, Siracusano M, Noto A, Fattuoni C, Riccioni A, Rajula HSR, Fanos V, Curatolo P, Barberini L, Mazzone L. The Urine Metabolome of Young Autistic Children Correlates with Their Clinical Profile Severity. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 116. | Page T, Coleman M. Purine metabolism abnormalities in a hyperuricosuric subclass of autism. Biochim Biophys Acta. 2000;1500:291-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 42] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 117. | Cohen D, Pichard N, Tordjman S, Baumann C, Burglen L, Excoffier E, Lazar G, Mazet P, Pinquier C, Verloes A, Héron D. Specific genetic disorders and autism: clinical contribution towards their identification. J Autism Dev Disord. 2005;35:103-116. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 204] [Cited by in RCA: 181] [Article Influence: 9.1] [Reference Citation Analysis (0)] |
| 118. | Micheli V, Camici M, Tozzi MG, Ipata PL, Sestini S, Bertelli M, Pompucci G. Neurological disorders of purine and pyrimidine metabolism. Curr Top Med Chem. 2011;11:923-947. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 77] [Cited by in RCA: 89] [Article Influence: 6.4] [Reference Citation Analysis (0)] |
| 119. | Chen BC, Mohd Rawi R, Meinsma R, Meijer J, Hennekam RC, van Kuilenburg AB. Dihydropyrimidine dehydrogenase deficiency in two malaysian siblings with abnormal MRI findings. Mol Syndromol. 2014;5:299-303. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 7] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 120. | Carter MT, Nikkel SM, Fernandez BA, Marshall CR, Noor A, Lionel AC, Prasad A, Pinto D, Joseph-George AM, Noakes C, Fairbrother-Davies C, Roberts W, Vincent J, Weksberg R, Scherer SW. Hemizygous deletions on chromosome 1p21.3 involving the DPYD gene in individuals with autism spectrum disorder. Clin Genet. 2011;80:435-443. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 43] [Cited by in RCA: 45] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 121. | Sprenger HG, MacVicar T, Bahat A, Fiedler KU, Hermans S, Ehrentraut D, Ried K, Milenkovic D, Bonekamp N, Larsson NG, Nolte H, Giavalisco P, Langer T. Cellular pyrimidine imbalance triggers mitochondrial DNA-dependent innate immunity. Nat Metab. 2021;3:636-650. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 39] [Cited by in RCA: 93] [Article Influence: 23.3] [Reference Citation Analysis (0)] |
| 122. | Frye RE, Slattery JC, Quadros EV. Folate metabolism abnormalities in autism: potential biomarkers. Biomark Med. 2017;11:687-699. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 42] [Cited by in RCA: 58] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 123. | Campistol J, Díez-Juan M, Callejón L, Fernandez-De Miguel A, Casado M, Garcia Cazorla A, Lozano R, Artuch R. Inborn error metabolic screening in individuals with nonsyndromic autism spectrum disorders. Dev Med Child Neurol. 2016;58:842-847. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 27] [Cited by in RCA: 24] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 124. | Kurochkin I, Khrameeva E, Tkachev A, Stepanova V, Vanyushkina A, Stekolshchikova E, Li Q, Zubkov D, Shichkova P, Halene T, Willmitzer L, Giavalisco P, Akbarian S, Khaitovich P. Metabolome signature of autism in the human prefrontal cortex. Commun Biol. 2019;2:234. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 41] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 125. | Geryk J, Krsička D, Vlčková M, Havlovicová M, Macek M Jr, Kremlíková Pourová R. The Key Role of Purine Metabolism in the Folate-Dependent Phenotype of Autism Spectrum Disorders: An In Silico Analysis. Metabolites. 2020;10. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 126. | Cheng N, Rho JM, Masino SA. Metabolic Dysfunction Underlying Autism Spectrum Disorder and Potential Treatment Approaches. Front Mol Neurosci. 2017;10:34. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 82] [Cited by in RCA: 93] [Article Influence: 11.6] [Reference Citation Analysis (0)] |
| 127. | Rossmann MP, Dubois SM, Agarwal S, Zon LI. Mitochondrial function in development and disease. Dis Model Mech. 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 61] [Cited by in RCA: 99] [Article Influence: 24.8] [Reference Citation Analysis (0)] |
| 128. | Zanon A, Pramstaller PP, Hicks AA, Pichler I. Environmental and Genetic Variables Influencing Mitochondrial Health and Parkinson's Disease Penetrance. Parkinsons Dis. 2018;2018:8684906. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 11] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 129. | Giulivi C, Zhang YF, Omanska-Klusek A, Ross-Inta C, Wong S, Hertz-Picciotto I, Tassone F, Pessah IN. Mitochondrial dysfunction in autism. JAMA. 2010;304:2389-2396. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 284] [Cited by in RCA: 318] [Article Influence: 21.2] [Reference Citation Analysis (0)] |
| 130. | Frye RE, Rossignol DA. Mitochondrial dysfunction can connect the diverse medical symptoms associated with autism spectrum disorders. Pediatr Res. 2011;69:41R-47R. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 147] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 131. | Nabi SU, Rehman MU, Arafah A, Taifa S, Khan IS, Khan A, Rashid S, Jan F, Wani HA, Ahmad SF. Treatment of Autism Spectrum Disorders by Mitochondrial-targeted Drug: Future of Neurological Diseases Therapeutics. Curr Neuropharmacol. 2023;21:1042-1064. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 11] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 132. | Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet. 2005;6:389-402. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1215] [Cited by in RCA: 1329] [Article Influence: 66.5] [Reference Citation Analysis (0)] |
| 133. | Stoccoro A, Coppedè F. Mitochondrial DNA Methylation and Human Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 110] [Article Influence: 27.5] [Reference Citation Analysis (0)] |
| 134. | Varga NÁ, Pentelényi K, Balicza P, Gézsi A, Reményi V, Hársfalvi V, Bencsik R, Illés A, Prekop C, Molnár MJ. Mitochondrial dysfunction and autism: comprehensive genetic analyses of children with autism and mtDNA deletion. Behav Brain Funct. 2018;14:4. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 35] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 135. | Golpich M, Amini E, Mohamed Z, Azman Ali R, Mohamed Ibrahim N, Ahmadiani A. Mitochondrial Dysfunction and Biogenesis in Neurodegenerative diseases: Pathogenesis and Treatment. CNS Neurosci Ther. 2017;23:5-22. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 324] [Cited by in RCA: 397] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 136. | Johri A, Beal MF. Mitochondrial dysfunction in neurodegenerative diseases. J Pharmacol Exp Ther. 2012;342:619-630. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 573] [Cited by in RCA: 533] [Article Influence: 41.0] [Reference Citation Analysis (0)] |
| 137. | Kowalczyk P, Sulejczak D, Kleczkowska P, Bukowska-Ośko I, Kucia M, Popiel M, Wietrak E, Kramkowski K, Wrzosek K, Kaczyńska K. Mitochondrial Oxidative Stress-A Causative Factor and Therapeutic Target in Many Diseases. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 5] [Cited by in RCA: 229] [Article Influence: 57.3] [Reference Citation Analysis (0)] |
| 138. | Omotosho IO, Akinade AO, Lagunju IA, Yakubu MA. Oxidative stress indices in ASD children in Sub-Sahara Africa. J Neurodev Disord. 2021;13:50. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 139. | Moos WH, Faller DV, Glavas IP, Harpp DN, Kamperi N, Kanara I, Kodukula K, Mavrakis AN, Pernokas J, Pernokas M, Pinkert CA, Powers WR, Steliou K, Tamvakopoulos C, Vavvas DG, Zamboni RJ, Sampani K. Pathogenic mitochondrial dysfunction and metabolic abnormalities. Biochem Pharmacol. 2021;193:114809. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 7] [Cited by in RCA: 25] [Article Influence: 6.3] [Reference Citation Analysis (0)] |
| 140. | Rossignol DA, Frye RE. Mitochondrial dysfunction in autism spectrum disorders: a systematic review and meta-analysis. Mol Psychiatry. 2012;17:290-314. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 675] [Cited by in RCA: 606] [Article Influence: 46.6] [Reference Citation Analysis (0)] |
| 141. | Gliga T, Jones EJ, Bedford R, Charman T, Johnson MH. From early markers to neuro-developmental mechanisms of autism. Dev Rev. 2014;34:189-207. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 87] [Cited by in RCA: 90] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 142. | Klein IL, van de Loo KFE, Smeitink JAM, Janssen MCH, Kessels RPC, van Karnebeek CD, van der Veer E, Custers JAE, Verhaak CM. Cognitive functioning and mental health in mitochondrial disease: A systematic scoping review. Neurosci Biobehav Rev. 2021;125:57-77. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 11] [Article Influence: 2.8] [Reference Citation Analysis (0)] |
| 143. | Genovese A, Butler MG. Clinical Assessment, Genetics, and Treatment Approaches in Autism Spectrum Disorder (ASD). Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 116] [Cited by in RCA: 126] [Article Influence: 25.2] [Reference Citation Analysis (0)] |
| 144. | Frye RE. Metabolic and mitochondrial disorders associated with epilepsy in children with autism spectrum disorder. Epilepsy Behav. 2015;47:147-157. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 67] [Cited by in RCA: 73] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 145. | Wasilewska J, Klukowski M. Gastrointestinal symptoms and autism spectrum disorder: links and risks - a possible new overlap syndrome. Pediatric Health Med Ther. 2015;6:153-166. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 108] [Cited by in RCA: 99] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 146. | Morava E, van den Heuvel L, Hol F, de Vries MC, Hogeveen M, Rodenburg RJ, Smeitink JA. Mitochondrial disease criteria: diagnostic applications in children. Neurology. 2006;67:1823-1826. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 176] [Cited by in RCA: 214] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 147. | Gevezova M, Sbirkov Y, Sarafian V, Plaimas K, Suratanee A, Maes M. Autistic spectrum disorder (ASD) - Gene, molecular and pathway signatures linking systemic inflammation, mitochondrial dysfunction, transsynaptic signalling, and neurodevelopment. Brain Behav Immun Health. 2023;30:100646. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 21] [Reference Citation Analysis (0)] |
| 148. | Muraresku CC, McCormick EM, Falk MJ. Mitochondrial Disease: Advances in clinical diagnosis, management, therapeutic development, and preventative strategies. Curr Genet Med Rep. 2018;6:62-72. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 27] [Article Influence: 3.9] [Reference Citation Analysis (0)] |
| 149. | Stenton SL, Prokisch H. Genetics of mitochondrial diseases: Identifying mutations to help diagnosis. EBioMedicine. 2020;56:102784. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 74] [Cited by in RCA: 155] [Article Influence: 31.0] [Reference Citation Analysis (0)] |
| 150. | Lunsing RJ, Strating K, de Koning TJ, Sijens PE. Diagnostic value of MRS-quantified brain tissue lactate level in identifying children with mitochondrial disorders. Eur Radiol. 2017;27:976-984. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 25] [Cited by in RCA: 27] [Article Influence: 3.4] [Reference Citation Analysis (0)] |
| 151. | Lai RE, Holman ME, Chen Q, Rivers J, Lesnefsky EJ, Gorgey AS. Assessment of mitochondrial respiratory capacity using minimally invasive and noninvasive techniques in persons with spinal cord injury. PLoS One. 2022;17:e0265141. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 4] [Reference Citation Analysis (0)] |
| 152. | Frye RE. A Personalized Multidisciplinary Approach to Evaluating and Treating Autism Spectrum Disorder. J Pers Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 153. | Camp KM, Krotoski D, Parisi MA, Gwinn KA, Cohen BH, Cox CS, Enns GM, Falk MJ, Goldstein AC, Gopal-Srivastava R, Gorman GS, Hersh SP, Hirano M, Hoffman FA, Karaa A, MacLeod EL, McFarland R, Mohan C, Mulberg AE, Odenkirchen JC, Parikh S, Rutherford PJ, Suggs-Anderson SK, Tang WH, Vockley J, Wolfe LA, Yannicelli S, Yeske PE, Coates PM. Nutritional interventions in primary mitochondrial disorders: Developing an evidence base. Mol Genet Metab. 2016;119:187-206. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 41] [Cited by in RCA: 38] [Article Influence: 4.2] [Reference Citation Analysis (0)] |
| 154. | Pfeffer G, Majamaa K, Turnbull DM, Thorburn D, Chinnery PF. Treatment for mitochondrial disorders. Cochrane Database Syst Rev. 2012;2012:CD004426. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 111] [Cited by in RCA: 170] [Article Influence: 13.1] [Reference Citation Analysis (0)] |
| 155. | Birben E, Sahiner UM, Sackesen C, Erzurum S, Kalayci O. Oxidative stress and antioxidant defense. World Allergy Organ J. 2012;5:9-19. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2413] [Cited by in RCA: 2936] [Article Influence: 225.8] [Reference Citation Analysis (0)] |
| 156. | Pizzino G, Irrera N, Cucinotta M, Pallio G, Mannino F, Arcoraci V, Squadrito F, Altavilla D, Bitto A. Oxidative Stress: Harms and Benefits for Human Health. Oxid Med Cell Longev. 2017;2017:8416763. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1959] [Cited by in RCA: 2332] [Article Influence: 291.5] [Reference Citation Analysis (0)] |
| 157. | Teleanu DM, Niculescu AG, Lungu II, Radu CI, Vladâcenco O, Roza E, Costăchescu B, Grumezescu AM, Teleanu RI. An Overview of Oxidative Stress, Neuroinflammation, and Neurodegenerative Diseases. Int J Mol Sci. 2022;23. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 9] [Cited by in RCA: 449] [Article Influence: 149.7] [Reference Citation Analysis (0)] |
| 158. | Lin X, Zhou Y, Li S, Zhou H, Ma B, Zhang Z, Liang J. Markers related to oxidative stress in peripheral blood in children with autism spectrum disorder. Res Autism Spectr Disord. 2022;99:102067. [DOI] [Full Text] |
| 159. | Chen L, Shi XJ, Liu H, Mao X, Gui LN, Wang H, Cheng Y. Oxidative stress marker aberrations in children with autism spectrum disorder: a systematic review and meta-analysis of 87 studies (N = 9109). Transl Psychiatry. 2021;11:15. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 95] [Article Influence: 23.8] [Reference Citation Analysis (0)] |
| 160. | El-Ansary A, Bjørklund G, Chirumbolo S, Alnakhli OM. Predictive value of selected biomarkers related to metabolism and oxidative stress in children with autism spectrum disorder. Metab Brain Dis. 2017;32:1209-1221. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 44] [Cited by in RCA: 56] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 161. | Pangrazzi L, Balasco L, Bozzi Y. Oxidative Stress and Immune System Dysfunction in Autism Spectrum Disorders. Int J Mol Sci. 2020;21. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 52] [Cited by in RCA: 122] [Article Influence: 24.4] [Reference Citation Analysis (0)] |
| 162. | Hassan H, Zakaria F, Makpol S, Karim NA. A Link between Mitochondrial Dysregulation and Idiopathic Autism Spectrum Disorder (ASD): Alterations in Mitochondrial Respiratory Capacity and Membrane Potential. Curr Issues Mol Biol. 2021;43:2238-2252. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 8] [Reference Citation Analysis (0)] |
| 163. | Erbescu A, Papuc SM, Budisteanu M, Arghir A, Neagu M. Re-emerging concepts of immune dysregulation in autism spectrum disorders. Front Psychiatry. 2022;13:1006612. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 164. | Sharifi-Rad M, Anil Kumar NV, Zucca P, Varoni EM, Dini L, Panzarini E, Rajkovic J, Tsouh Fokou PV, Azzini E, Peluso I, Prakash Mishra A, Nigam M, El Rayess Y, Beyrouthy ME, Polito L, Iriti M, Martins N, Martorell M, Docea AO, Setzer WN, Calina D, Cho WC, Sharifi-Rad J. Lifestyle, Oxidative Stress, and Antioxidants: Back and Forth in the Pathophysiology of Chronic Diseases. Front Physiol. 2020;11:694. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1271] [Cited by in RCA: 913] [Article Influence: 182.6] [Reference Citation Analysis (0)] |
| 165. | Bjørklund G, Tinkov AA, Hosnedlová B, Kizek R, Ajsuvakova OP, Chirumbolo S, Skalnaya MG, Peana M, Dadar M, El-Ansary A, Qasem H, Adams JB, Aaseth J, Skalny AV. The role of glutathione redox imbalance in autism spectrum disorder: A review. Free Radic Biol Med. 2020;160:149-162. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 54] [Cited by in RCA: 95] [Article Influence: 19.0] [Reference Citation Analysis (0)] |
| 166. | Krajcovicova-Kudlackova M, Valachovicova M, Mislanova C, Hudecova Z, Sustrova M, Ostatnikova D. Plasma concentrations of selected antioxidants in autistic children and adolescents. Bratisl Lek Listy. 2009;110:247-250. [PubMed] |
| 167. | Rossignol DA, Genuis SJ, Frye RE. Environmental toxicants and autism spectrum disorders: a systematic review. Transl Psychiatry. 2014;4:e360. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 371] [Cited by in RCA: 317] [Article Influence: 28.8] [Reference Citation Analysis (0)] |
| 168. | Su LJ, Zhang JH, Gomez H, Murugan R, Hong X, Xu D, Jiang F, Peng ZY. Reactive Oxygen Species-Induced Lipid Peroxidation in Apoptosis, Autophagy, and Ferroptosis. Oxid Med Cell Longev. 2019;2019:5080843. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1314] [Cited by in RCA: 1197] [Article Influence: 199.5] [Reference Citation Analysis (0)] |
| 169. | Cecarini V, Gee J, Fioretti E, Amici M, Angeletti M, Eleuteri AM, Keller JN. Protein oxidation and cellular homeostasis: Emphasis on metabolism. Biochim Biophys Acta. 2007;1773:93-104. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 159] [Cited by in RCA: 179] [Article Influence: 9.4] [Reference Citation Analysis (0)] |
| 170. | Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis. 2009;30:2-10. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 394] [Cited by in RCA: 473] [Article Influence: 27.8] [Reference Citation Analysis (0)] |
| 171. | Biswas K, Alexander K, Francis MM. Reactive Oxygen Species: Angels and Demons in the Life of a Neuron. Neuro Sci. 2022;3:130-145. [RCA] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 37] [Article Influence: 12.3] [Reference Citation Analysis (0)] |
| 172. | Castejon AM, Spaw JA. Autism and Oxidative Stress Interventions: Impact on Autistic Behavior. Austin J Pharmacol Ther. 2014;2:1015. |
| 173. | Pangrazzi L, Balasco L, Bozzi Y. Natural Antioxidants: A Novel Therapeutic Approach to Autism Spectrum Disorders? Antioxidants (Basel). 2020;9. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 174. | Membrino V, Di Paolo A, Alia S, Papiri G, Vignini A. The Role of Oxidative Stress in Autism Spectrum Disorder: A Narrative Literature Review. Oxygen. 2023;3:34-44. [RCA] [DOI] [Full Text] [Cited by in Crossref: 9] [Cited by in RCA: 6] [Article Influence: 3.0] [Reference Citation Analysis (0)] |
| 175. | Usui N, Iwata K, Miyachi T, Takagai S, Wakusawa K, Nara T, Tsuchiya KJ, Matsumoto K, Kurita D, Kameno Y, Wakuda T, Takebayashi K, Iwata Y, Fujioka T, Hirai T, Toyoshima M, Ohnishi T, Toyota T, Maekawa M, Yoshikawa T, Nakamura K, Tsujii M, Sugiyama T, Mori N, Matsuzaki H. VLDL-specific increases of fatty acids in autism spectrum disorder correlate with social interaction. EBioMedicine. 2020;58:102917. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 14] [Cited by in RCA: 33] [Article Influence: 6.6] [Reference Citation Analysis (0)] |
| 176. | Esposito CM, Buoli M, Ciappolino V, Agostoni C, Brambilla P. The Role of Cholesterol and Fatty Acids in the Etiology and Diagnosis of Autism Spectrum Disorders. Int J Mol Sci. 2021;22. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 6] [Cited by in RCA: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 177. | Błażewicz A, Szymańska I, Astel A, Stenzel-Bembenek A, Dolliver WR, Makarewicz A. Assessment of Changes over Time of Lipid Profile, C-Reactive Protein Level and Body Mass Index in Teenagers and Young Adults on Different Diets Belonging to Autism Spectrum Disorder. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 3] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 178. | Slama S, Bahia W, Soltani I, Gaddour N, Ferchichi S. Risk factors in autism spectrum disorder: A Tunisian case-control study. Saudi J Biol Sci. 2022;29:2749-2755. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 179. | Park BY, Yao R, Tierney E, Brucato M, Hong X, Wang G, Ji Y, Pearson C, Fallin MD, Wang X, Volk H. The association between maternal lipid profile after birth and offspring risk of autism spectrum disorder. Ann Epidemiol. 2021;53:50-55.e1. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 9] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 180. | Kim EK, Neggers YH, Shin CS, Kim E, Kim EM. Alterations in lipid profile of autistic boys: a case control study. Nutr Res. 2010;30:255-260. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 30] [Cited by in RCA: 35] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 181. | Luçardo JDC, Monk GF, Dias MDS, Martins-Silva T, Fernandes MP, Maia JC, Valle SC, Vaz JDS. Interest in food and triglyceride concentrations in children and adolescents with autistic spectrum disorder. J Pediatr (Rio J). 2021;97:103-108. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 8] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 182. | Chauhan A, Chauhan V, Brown WT, Cohen I. Oxidative stress in autism: increased lipid peroxidation and reduced serum levels of ceruloplasmin and transferrin--the antioxidant proteins. Life Sci. 2004;75:2539-2549. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 261] [Cited by in RCA: 278] [Article Influence: 13.2] [Reference Citation Analysis (0)] |
| 183. | Yap CX, Henders AK, Alvares GA, Giles C, Huynh K, Nguyen A, Wallace L, McLaren T, Yang Y, Hernandez LM, Gandal MJ, Hansell NK, Cleary D, Grove R, Hafekost C, Harun A, Holdsworth H, Jellett R, Khan F, Lawson LP, Leslie J, Levis Frenk M, Masi A, Mathew NE, Muniandy M, Nothard M, Miller JL, Nunn L, Strike LT, Cadby G, Moses EK; Busselton Health Study Investigators, de Zubicaray GI, Thompson PM, McMahon KL, Wright MJ, Visscher PM, Dawson PA, Dissanayake C, Eapen V, Heussler HS, Whitehouse AJO, Meikle PJ, Wray NR, Gratten J. Interactions between the lipidome and genetic and environmental factors in autism. Nat Med. 2023;29:936-949. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Cited by in RCA: 21] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 184. | Forman HJ, Zhang H. Targeting oxidative stress in disease: promise and limitations of antioxidant therapy. Nat Rev Drug Discov. 2021;20:689-709. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1299] [Cited by in RCA: 1452] [Article Influence: 363.0] [Reference Citation Analysis (0)] |
| 185. | Pandey MK. Uncovering the Lipid Web: Discovering the Multifaceted Roles of Lipids in Human Diseases and Therapeutic Opportunities. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Cited by in RCA: 3] [Reference Citation Analysis (0)] |
| 186. | Fattorusso A, Di Genova L, Dell'Isola GB, Mencaroni E, Esposito S. Autism Spectrum Disorders and the Gut Microbiota. Nutrients. 2019;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 331] [Cited by in RCA: 273] [Article Influence: 45.5] [Reference Citation Analysis (1)] |
| 187. | Hussain G, Wang J, Rasul A, Anwar H, Imran A, Qasim M, Zafar S, Kamran SKS, Razzaq A, Aziz N, Ahmad W, Shabbir A, Iqbal J, Baig SM, Sun T. Role of cholesterol and sphingolipids in brain development and neurological diseases. Lipids Health Dis. 2019;18:26. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 184] [Cited by in RCA: 245] [Article Influence: 40.8] [Reference Citation Analysis (0)] |
| 188. | Shichiri M. The role of lipid peroxidation in neurological disorders. J Clin Biochem Nutr. 2014;54:151-160. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 154] [Cited by in RCA: 191] [Article Influence: 17.4] [Reference Citation Analysis (0)] |
| 189. | Wong C, Crawford DA. Lipid Signalling in the Pathology of Autism Spectrum Disorders. Comprehensive Guide to Autism. 2014;. [DOI] [Full Text] |
| 190. | DiNicolantonio JJ, O'Keefe JH. The Importance of Marine Omega-3s for Brain Development and the Prevention and Treatment of Behavior, Mood, and Other Brain Disorders. Nutrients. 2020;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 21] [Cited by in RCA: 76] [Article Influence: 15.2] [Reference Citation Analysis (0)] |
| 191. | Su KP, Matsuoka Y, Pae CU. Omega-3 Polyunsaturated Fatty Acids in Prevention of Mood and Anxiety Disorders. Clin Psychopharmacol Neurosci. 2015;13:129-137. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 70] [Cited by in RCA: 77] [Article Influence: 7.7] [Reference Citation Analysis (0)] |
| 192. | Zhang W, Li P, Hu X, Zhang F, Chen J, Gao Y. Omega-3 polyunsaturated fatty acids in the brain: metabolism and neuroprotection. Front Biosci (Landmark Ed). 2011;16:2653-2670. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 60] [Cited by in RCA: 61] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 193. | Zhou L, Xiong JY, Chai YQ, Huang L, Tang ZY, Zhang XF, Liu B, Zhang JT. Possible antidepressant mechanisms of omega-3 polyunsaturated fatty acids acting on the central nervous system. Front Psychiatry. 2022;13:933704. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 38] [Article Influence: 12.7] [Reference Citation Analysis (0)] |
| 194. | Calder PC. Omega-3 fatty acids and inflammatory processes. Nutrients. 2010;2:355-374. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 507] [Cited by in RCA: 679] [Article Influence: 45.3] [Reference Citation Analysis (0)] |
| 195. | Doaei S, Bourbour F, Teymoori Z, Jafari F, Kalantari N, Abbas Torki S, Ashoori N, Nemat Gorgani S, Gholamalizadeh M. The effect of omega-3 fatty acids supplementation on social and behavioral disorders of children with autism: a randomized clinical trial. Pediatr Endocrinol Diabetes Metab. 2021;27:12-18. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 196. | Veselinović A, Petrović S, Žikić V, Subotić M, Jakovljević V, Jeremić N, Vučić V. Neuroinflammation in Autism and Supplementation Based on Omega-3 Polyunsaturated Fatty Acids: A Narrative Review. Medicina (Kaunas). 2021;57. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 16] [Article Influence: 4.0] [Reference Citation Analysis (0)] |
| 197. | Sherzai D, Moness R, Sherzai S, Sherzai A. A Systematic Review of Omega-3 Fatty Acid Consumption and Cognitive Outcomes in Neurodevelopment. Am J Lifestyle Med. 2023;17:649-685. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Cited by in RCA: 1] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 198. | Königs A, Kiliaan AJ. Critical appraisal of omega-3 fatty acids in attention-deficit/hyperactivity disorder treatment. Neuropsychiatr Dis Treat. 2016;12:1869-1882. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 27] [Cited by in RCA: 29] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 199. | Bradberry JC, Hilleman DE. Overview of omega-3 Fatty Acid therapies. P T. 2013;38:681-691. [PubMed] |
| 200. | Villani AM, Crotty M, Cleland LG, James MJ, Fraser RJ, Cobiac L, Miller MD. Fish oil administration in older adults: is there potential for adverse events? A systematic review of the literature. BMC Geriatr. 2013;13:41. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 30] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 201. | Saeed NK, Al-Beltagi M, Bediwy AS, El-Sawaf Y, Toema O. Gut microbiota in various childhood disorders: Implication and indications. World J Gastroenterol. 2022;28:1875-1901. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in CrossRef: 72] [Cited by in RCA: 62] [Article Influence: 20.7] [Reference Citation Analysis (5)] |
| 202. | Roussin L, Prince N, Perez-Pardo P, Kraneveld AD, Rabot S, Naudon L. Role of the Gut Microbiota in the Pathophysiology of Autism Spectrum Disorder: Clinical and Preclinical Evidence. Microorganisms. 2020;8. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 24] [Cited by in RCA: 34] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 203. | Chen Y, Xu J, Chen Y. Regulation of Neurotransmitters by the Gut Microbiota and Effects on Cognition in Neurological Disorders. Nutrients. 2021;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 45] [Cited by in RCA: 415] [Article Influence: 103.8] [Reference Citation Analysis (1)] |
| 204. | Appleton J. The Gut-Brain Axis: Influence of Microbiota on Mood and Mental Health. Integr Med (Encinitas). 2018;17:28-32. [PubMed] |
| 205. | Christovich A, Luo XM. Gut Microbiota, Leaky Gut, and Autoimmune Diseases. Front Immunol. 2022;13:946248. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 42] [Cited by in RCA: 90] [Article Influence: 30.0] [Reference Citation Analysis (0)] |
| 206. | Xiong RG, Zhou DD, Wu SX, Huang SY, Saimaiti A, Yang ZJ, Shang A, Zhao CN, Gan RY, Li HB. Health Benefits and Side Effects of Short-Chain Fatty Acids. Foods. 2022;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 191] [Cited by in RCA: 176] [Article Influence: 58.7] [Reference Citation Analysis (0)] |
| 207. | Silva YP, Bernardi A, Frozza RL. The Role of Short-Chain Fatty Acids From Gut Microbiota in Gut-Brain Communication. Front Endocrinol (Lausanne). 2020;11:25. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 780] [Cited by in RCA: 1598] [Article Influence: 319.6] [Reference Citation Analysis (0)] |
| 208. | Jennings L, Basiri R. Amino Acids, B Vitamins, and Choline May Independently and Collaboratively Influence the Incidence and Core Symptoms of Autism Spectrum Disorder. Nutrients. 2022;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 15] [Article Influence: 5.0] [Reference Citation Analysis (0)] |
| 209. | Staley C, Weingarden AR, Khoruts A, Sadowsky MJ. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl Microbiol Biotechnol. 2017;101:47-64. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 367] [Cited by in RCA: 397] [Article Influence: 49.6] [Reference Citation Analysis (0)] |
| 210. | Jurnak F. The Pivotal Role of Aldehyde Toxicity in Autism Spectrum Disorder: The Therapeutic Potential of Micronutrient Supplementation. Nutr Metab Insights. 2015;8:57-77. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 5] [Article Influence: 0.6] [Reference Citation Analysis (0)] |
| 211. | Hu T, Dong Y, He C, Zhao M, He Q. The Gut Microbiota and Oxidative Stress in Autism Spectrum Disorders (ASD). Oxid Med Cell Longev. 2020;2020:8396708. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 10] [Cited by in RCA: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 212. | Vuong HE, Hsiao EY. Emerging Roles for the Gut Microbiome in Autism Spectrum Disorder. Biol Psychiatry. 2017;81:411-423. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 333] [Cited by in RCA: 368] [Article Influence: 46.0] [Reference Citation Analysis (0)] |
| 213. | Larroya A, Pantoja J, Codoñer-Franch P, Cenit MC. Towards Tailored Gut Microbiome-Based and Dietary Interventions for Promoting the Development and Maintenance of a Healthy Brain. Front Pediatr. 2021;9:705859. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 19] [Cited by in RCA: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 214. | Plaza-Diaz J, Ruiz-Ojeda FJ, Gil-Campos M, Gil A. Mechanisms of Action of Probiotics. Adv Nutr. 2019;10:S49-S66. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 901] [Cited by in RCA: 689] [Article Influence: 114.8] [Reference Citation Analysis (0)] |
| 215. | Baspinar B, Yardimci H. Gluten-Free Casein-Free Diet for Autism Spectrum Disorders: Can It Be Effective in Solving Behavioural and Gastrointestinal Problems? Eurasian J Med. 2020;52:292-297. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 12] [Cited by in RCA: 27] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 216. | Önal S, Sachadyn-Król M, Kostecka M. A Review of the Nutritional Approach and the Role of Dietary Components in Children with Autism Spectrum Disorders in Light of the Latest Scientific Research. Nutrients. 2023;15. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 17] [Cited by in RCA: 15] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 217. | Sharma A, Gokulchandran N, Sane H, Nivins S, Paranjape A, Badhe P. The Baseline Pattern and Age-related Developmental Metabolic Changes in the Brain of Children with Autism as Measured on Positron Emission Tomography/Computed Tomography Scan. World J Nucl Med. 2018;17:94-101. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 2] [Cited by in RCA: 5] [Reference Citation Analysis (0)] |
| 218. | Ming X, Stein TP, Barnes V, Rhodes N, Guo L. Metabolic perturbance in autism spectrum disorders: a metabolomics study. J Proteome Res. 2012;11:5856-5862. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 163] [Cited by in RCA: 176] [Article Influence: 13.5] [Reference Citation Analysis (0)] |
| 219. | Xiong X, Liu D, He W, Sheng X, Zhou W, Xie D, Liang H, Zeng T, Li T, Wang Y. Identification of gender-related metabolic disturbances in autism spectrum disorders using urinary metabolomics. Int J Biochem Cell Biol. 2019;115:105594. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 8] [Cited by in RCA: 13] [Article Influence: 2.2] [Reference Citation Analysis (0)] |
| 220. | James SJ, Melnyk S, Jernigan S, Cleves MA, Halsted CH, Wong DH, Cutler P, Bock K, Boris M, Bradstreet JJ, Baker SM, Gaylor DW. Metabolic endophenotype and related genotypes are associated with oxidative stress in children with autism. Am J Med Genet B Neuropsychiatr Genet. 2006;141B:947-956. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 440] [Cited by in RCA: 397] [Article Influence: 20.9] [Reference Citation Analysis (0)] |
| 221. | Qutranji L, Alkayyali T, Alkhateeb W, Sapmaz A, Aleter A, Almoustafa A, Aci Z, Hammad A, Nakhala A, Altunc U. Evaluation of the Relationship Between Environmental Factors, Nutrition, and Metabolic Changes in Children Diagnosed With Autism in North Cyprus: A Case-Control Study. Cureus. 2021;13:e17016. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 222. | Grabrucker AM. Environmental factors in autism. Front Psychiatry. 2012;3:118. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 80] [Cited by in RCA: 117] [Article Influence: 9.0] [Reference Citation Analysis (0)] |
| 223. | Qureshi F, Adams JB, Audhya T, Hahn J. Multivariate Analysis of Metabolomic and Nutritional Profiles among Children with Autism Spectrum Disorder. J Pers Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Reference Citation Analysis (0)] |
| 224. | Chalifour B, Holzhausen EA, Lim JJ, Yeo EN, Shen N, Jones DP, Peterson BS, Goran MI, Liang D, Alderete TL. The potential role of early life feeding patterns in shaping the infant fecal metabolome: implications for neurodevelopmental outcomes. NPJ Metab Health Dis. 2023;1. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 5] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 225. | Zhu J, Hua X, Yang T, Guo M, Li Q, Xiao L, Li L, Chen J, Li T. Alterations in Gut Vitamin and Amino Acid Metabolism are Associated with Symptoms and Neurodevelopment in Children with Autism Spectrum Disorder. J Autism Dev Disord. 2022;52:3116-3128. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 26] [Cited by in RCA: 22] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 226. | Siracusano M, Arturi L, Riccioni A, Noto A, Mussap M, Mazzone L. Metabolomics: Perspectives on Clinical Employment in Autism Spectrum Disorder. Int J Mol Sci. 2023;24. [RCA] [PubMed] [DOI] [Full Text] [Reference Citation Analysis (0)] |
| 227. | Laghi L, Mastromarino P, Prosperi M, Morales MA, Calderoni S, Santocchi E, Muratori F, Guiducci L. Are Fecal Metabolome and Microbiota Profiles Correlated with Autism Severity? A Cross-Sectional Study on ASD Preschoolers. Metabolites. 2021;11. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 1] [Cited by in RCA: 6] [Article Influence: 1.5] [Reference Citation Analysis (0)] |
| 228. | Soke GN, Rosenberg SA, Hamman RF, Fingerlin T, Rosenberg CR, Carpenter L, Lee LC, Giarelli E, Wiggins LD, Durkin MS, Reynolds A, DiGuiseppi C. Factors Associated with Self-Injurious Behaviors in Children with Autism Spectrum Disorder: Findings from Two Large National Samples. J Autism Dev Disord. 2017;47:285-296. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 34] [Cited by in RCA: 45] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 229. | Ranieri A, Mennitti C, Falcone N, La Monica I, Di Iorio MR, Tripodi L, Gentile A, Vitale M, Pero R, Pastore L, D'Argenio V, Scudiero O, Lombardo B. Positive effects of physical activity in autism spectrum disorder: how influences behavior, metabolic disorder and gut microbiota. Front Psychiatry. 2023;14:1238797. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 7] [Reference Citation Analysis (0)] |
| 230. | Kandpal M, Indari O, Baral B, Jakhmola S, Tiwari D, Bhandari V, Pandey RK, Bala K, Sonawane A, Jha HC. Dysbiosis of Gut Microbiota from the Perspective of the Gut-Brain Axis: Role in the Provocation of Neurological Disorders. Metabolites. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 41] [Cited by in RCA: 60] [Article Influence: 20.0] [Reference Citation Analysis (0)] |
| 231. | Brister D, Werner BA, Gideon G, McCarty PJ, Lane A, Burrows BT, McLees S, Adelson PD, Arango JI, Marsh W, Flores A, Pankratz MT, Ly NH, Flood M, Brown D, Carpentieri D, Jin Y, Gu H, Frye RE. Central Nervous System Metabolism in Autism, Epilepsy and Developmental Delays: A Cerebrospinal Fluid Analysis. Metabolites. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 12] [Cited by in RCA: 16] [Article Influence: 5.3] [Reference Citation Analysis (0)] |
| 232. | Mahony C, O'Ryan C. A molecular framework for autistic experiences: Mitochondrial allostatic load as a mediator between autism and psychopathology. Front Psychiatry. 2022;13:985713. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 4] [Reference Citation Analysis (0)] |
| 233. | Aldars-García L, Gisbert JP, Chaparro M. Metabolomics Insights into Inflammatory Bowel Disease: A Comprehensive Review. Pharmaceuticals (Basel). 2021;14. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 3] [Cited by in RCA: 45] [Article Influence: 11.3] [Reference Citation Analysis (0)] |
| 234. | Likhitweerawong N, Thonusin C, Boonchooduang N, Louthrenoo O, Nookaew I, Chattipakorn N, Chattipakorn SC. Profiles of urine and blood metabolomics in autism spectrum disorders. Metab Brain Dis. 2021;36:1641-1671. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 13] [Cited by in RCA: 29] [Article Influence: 7.3] [Reference Citation Analysis (0)] |
| 235. | Aishworiya R, Valica T, Hagerman R, Restrepo B. An Update on Psychopharmacological Treatment of Autism Spectrum Disorder. Neurotherapeutics. 2022;19:248-262. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 40] [Cited by in RCA: 89] [Article Influence: 29.7] [Reference Citation Analysis (0)] |
| 236. | Siafis S, Tzachanis D, Samara M, Papazisis G. Antipsychotic Drugs: From Receptor-binding Profiles to Metabolic Side Effects. Curr Neuropharmacol. 2018;16:1210-1223. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 93] [Cited by in RCA: 100] [Article Influence: 14.3] [Reference Citation Analysis (0)] |
| 237. | Matthews JS, Adams JB. Ratings of the Effectiveness of 13 Therapeutic Diets for Autism Spectrum Disorder: Results of a National Survey. J Pers Med. 2023;13. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in RCA: 15] [Reference Citation Analysis (0)] |
| 238. | Doreswamy S, Bashir A, Guarecuco JE, Lahori S, Baig A, Narra LR, Patel P, Heindl SE. Effects of Diet, Nutrition, and Exercise in Children With Autism and Autism Spectrum Disorder: A Literature Review. Cureus. 2020;12:e12222. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 7] [Cited by in RCA: 19] [Article Influence: 3.8] [Reference Citation Analysis (0)] |
| 239. | Brister D, Rose S, Delhey L, Tippett M, Jin Y, Gu H, Frye RE. Metabolomic Signatures of Autism Spectrum Disorder. J Pers Med. 2022;12. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 15] [Cited by in RCA: 21] [Article Influence: 7.0] [Reference Citation Analysis (0)] |
| 240. | Frye RE, Vassall S, Kaur G, Lewis C, Karim M, Rossignol D. Emerging biomarkers in autism spectrum disorder: a systematic review. Ann Transl Med. 2019;7:792. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 53] [Cited by in RCA: 100] [Article Influence: 16.7] [Reference Citation Analysis (0)] |
| 241. | Liu J, Tan Y, Zhang F, Wang Y, Chen S, Zhang N, Dai W, Zhou L, Li JC. Metabolomic analysis of plasma biomarkers in children with autism spectrum disorders. MedComm (2020). 2024;5:e488. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 3] [Reference Citation Analysis (0)] |
| 242. | Iyer SH, Yeh MY, Netzel L, Lindsey MG, Wallace M, Simeone KA, Simeone TA. Dietary and Metabolic Approaches for Treating Autism Spectrum Disorders, Affective Disorders and Cognitive Impairment Comorbid with Epilepsy: A Review of Clinical and Preclinical Evidence. Nutrients. 2024;16. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 1] [Reference Citation Analysis (0)] |
| 243. | Glinton KE, Elsea SH. Untargeted Metabolomics for Autism Spectrum Disorders: Current Status and Future Directions. Front Psychiatry. 2019;10:647. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 46] [Cited by in RCA: 52] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 244. | Hodges RE, Minich DM. Modulation of Metabolic Detoxification Pathways Using Foods and Food-Derived Components: A Scientific Review with Clinical Application. J Nutr Metab. 2015;2015:760689. [RCA] [PubMed] [DOI] [Full Text] [Full Text (PDF)] [Cited by in Crossref: 94] [Cited by in RCA: 117] [Article Influence: 11.7] [Reference Citation Analysis (0)] |
| 245. | Zwaigenbaum L, Bauman ML, Choueiri R, Fein D, Kasari C, Pierce K, Stone WL, Yirmiya N, Estes A, Hansen RL, McPartland JC, Natowicz MR, Buie T, Carter A, Davis PA, Granpeesheh D, Mailloux Z, Newschaffer C, Robins D, Smith Roley S, Wagner S, Wetherby A. Early Identification and Interventions for Autism Spectrum Disorder: Executive Summary. Pediatrics. 2015;136 Suppl 1:S1-S9. [RCA] [PubMed] [DOI] [Full Text] [Cited by in Crossref: 70] [Cited by in RCA: 68] [Article Influence: 6.8] [Reference Citation Analysis (0)] |